Epigenetic Effects and Potential Contributions of m6A Modification to Mammary Gland Development and Lactation of Dairy Goats Explored via MeRIP-seq
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Samples
2.2. RNA Extraction and Library Construction
2.3. Quality Control and Sequence Comparison of Sequencing Data
2.4. m6A Peak Identification of Mammary Gland Tissue at Different Lactation Periods
2.5. Functional Analysis of Differential Peaks Between Different Lactation Periods
2.6. Conjoint Analysis of RNA-seq and MeRIP-seq
2.7. Identification of Core Genes and Construction of PPI Regulatory Networks
3. Results
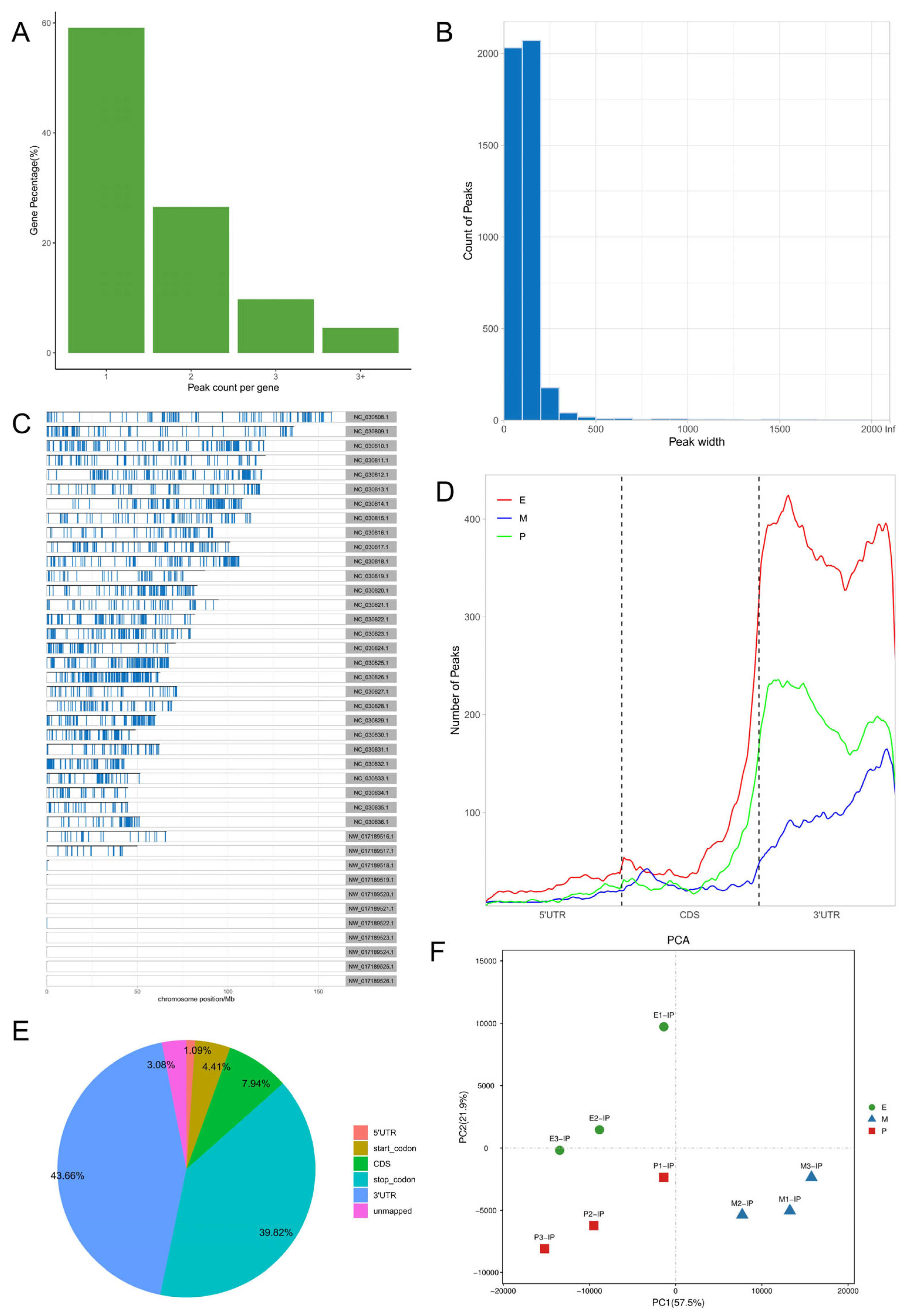
3.1. Sequencing Data Quality Assessment and Reference Genome Alignment
3.2. Identification and Analysis of m6A Methylase
3.3. m6A Peak Annotation and Motif Analysis
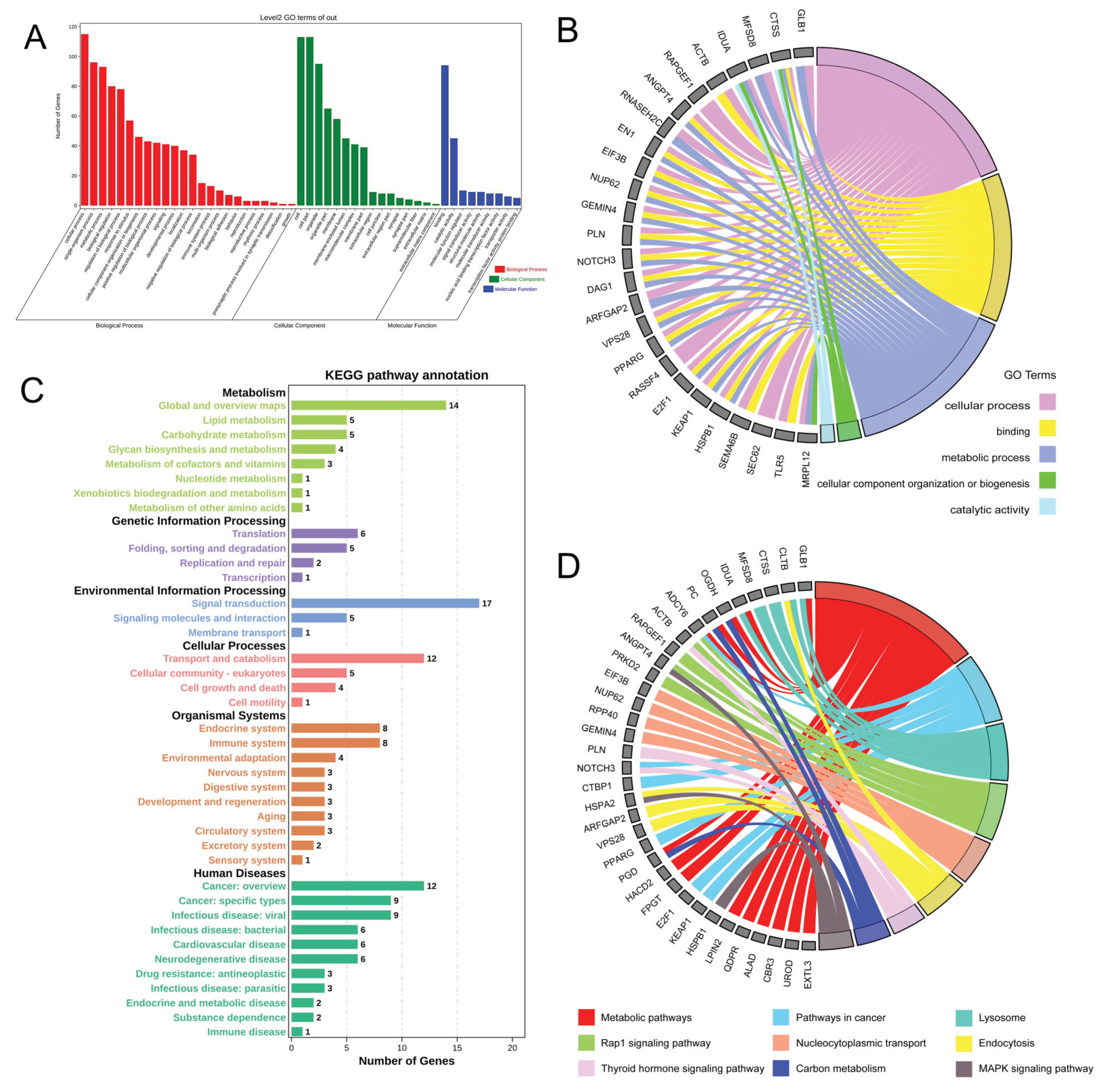
3.4. GO Annotation and KEGG Enrichment of DMGs
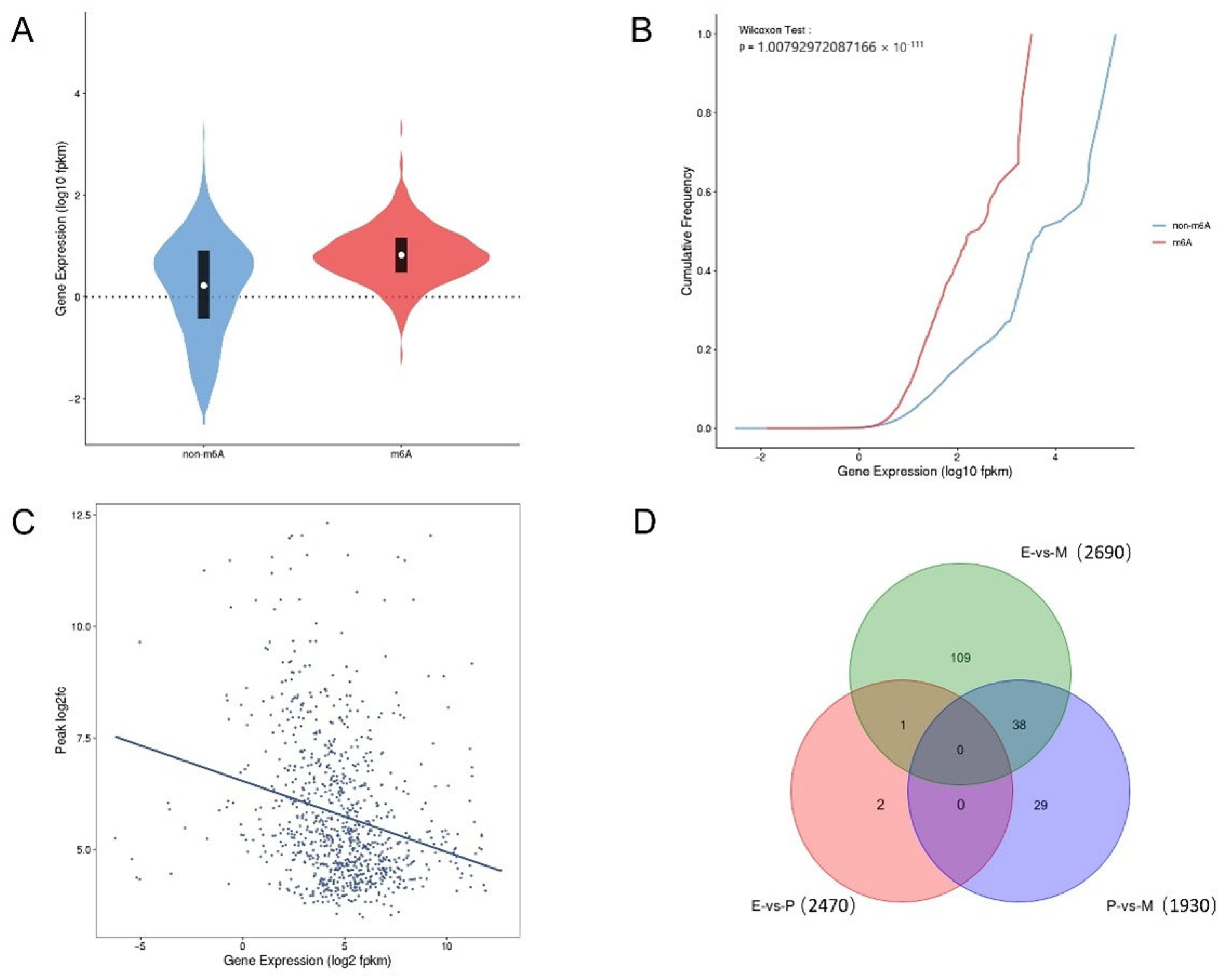
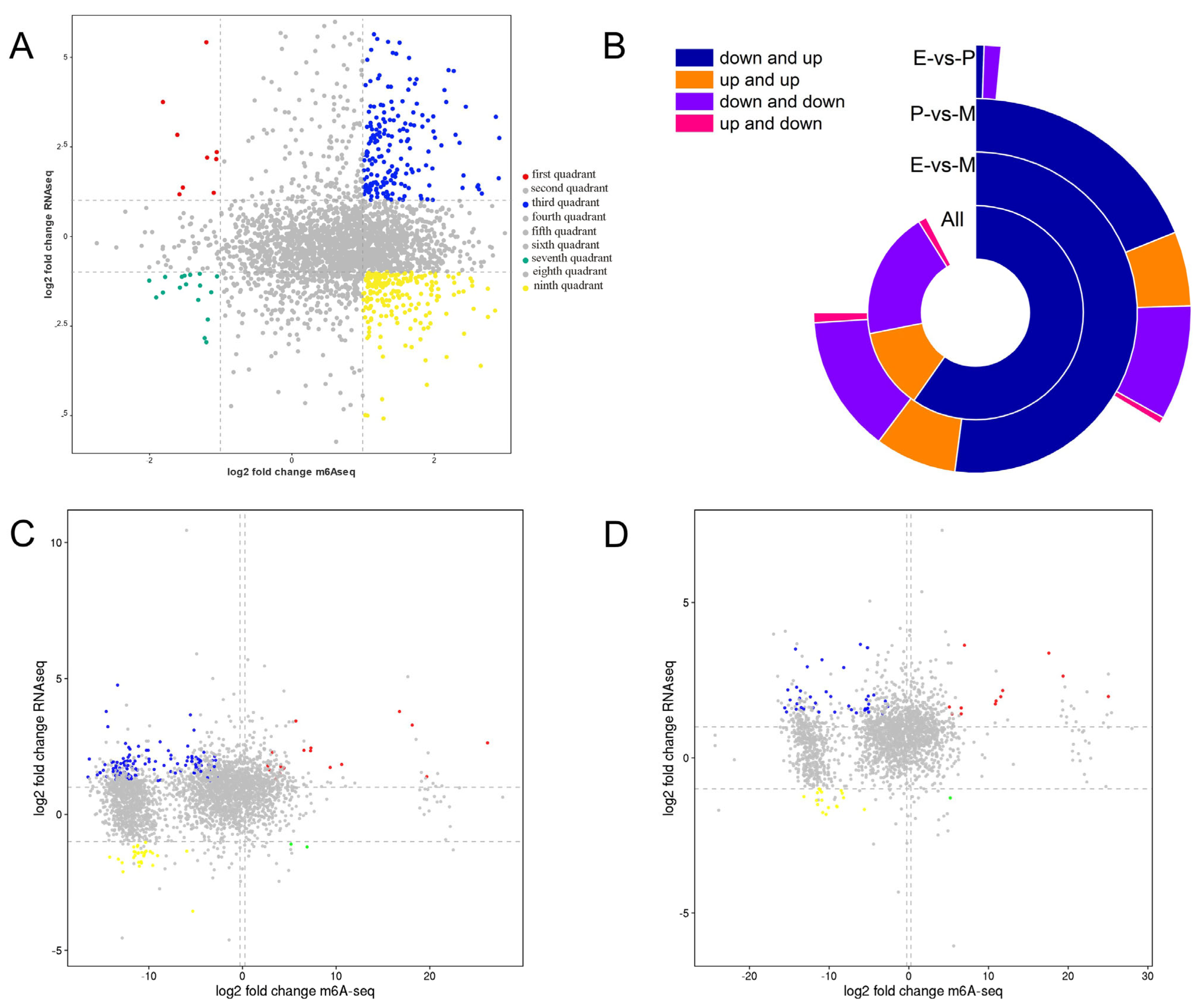
3.5. Correlation Analysis of MeRIP-seq and RNA-seq
3.6. Co-Regulation Relationship of Common DEGs in MeRIP-seq and RNA-seq
3.7. GO Annotation and KEGG of Common DEGs in MeRIP-seq and RNA-seq
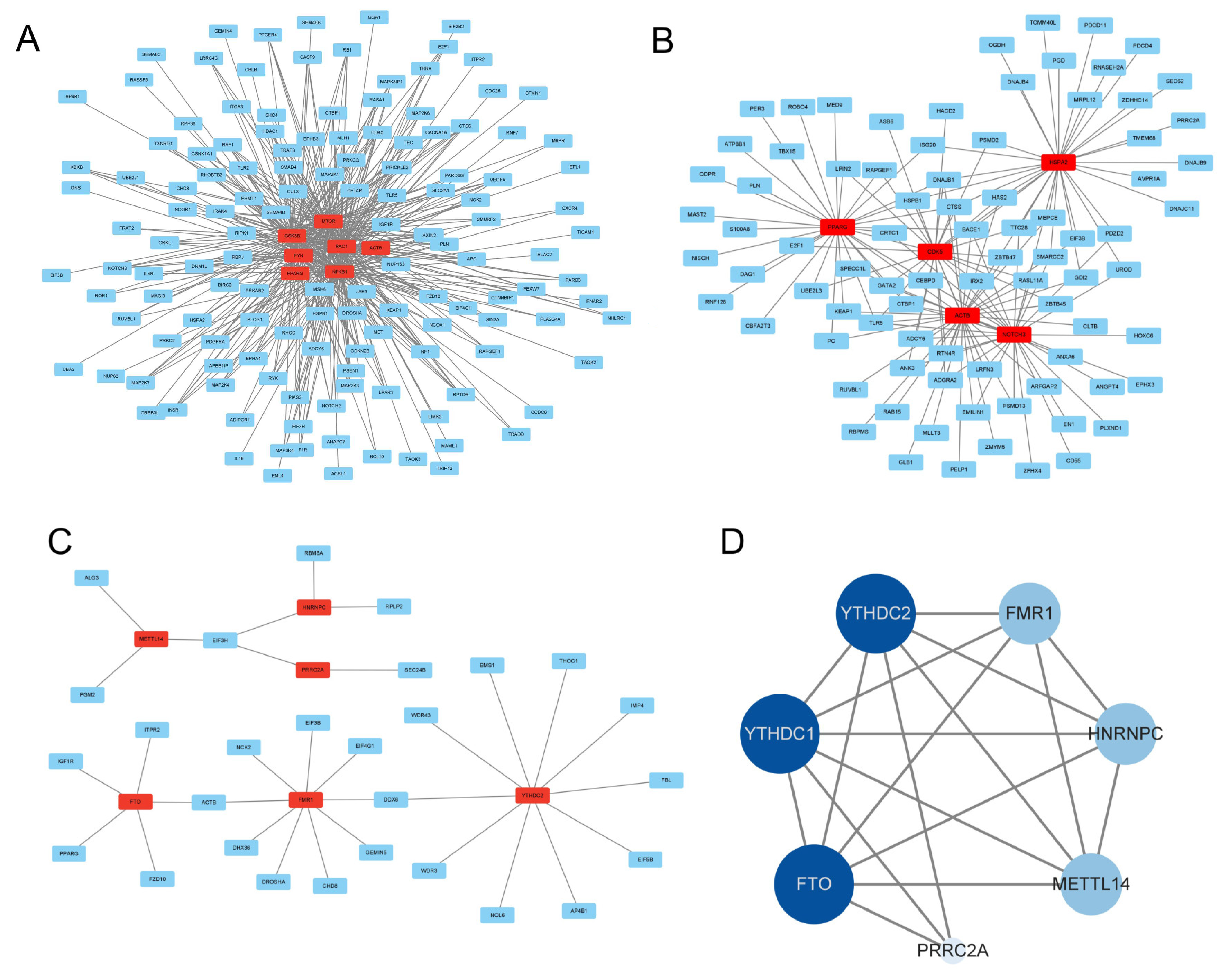
3.8. Regulatory Network Construction of DMGs in MeRIP-seq and RNA-seq
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ana, M.F.; Stine, L.B.; Emøke, B.; André, M.A. The mammary gland in domestic ruminants: A systems biology perspective. J. Proteom. 2013, 94, 110–123. [Google Scholar] [CrossRef] [PubMed]
- Akers, R.M. Lactation physiology: A ruminant animal perspective. Protoplasma 1990, 159, 96–111. [Google Scholar] [CrossRef]
- Macias, H.; Hinck, L. Mammary gland development. Wiley Interdiscip. Rev. Dev. Biol. 2012, 1, 533–557. [Google Scholar] [CrossRef] [PubMed]
- Hannan, F.M.; Elajnaf, T.; Vandenberg, L.N.; Kennedy, S.H.; Thakker, R.V. Hormonal regulation of mammary gland development and lactation. Nat. Rev. Endocrinol. 2023, 19, 46–61. [Google Scholar] [CrossRef]
- Slepicka, P.F.; Somasundara, A.V.H.; Dos Santos, C.O. The molecular basis of mammary gland development and epithelial differentiation. Semin. Cell Dev. Biol. 2021, 114, 93–112. [Google Scholar] [CrossRef]
- Song, P.S.; Tayier, Z.; Cai, Z.; Jia, G. RNA methylation in mammalian development and cancer. Cell Biol. Toxicol. 2021, 37, 811–831. [Google Scholar] [CrossRef]
- Sang, L.; Sun, L.; Wang, A.; Zhang, H.; Yuan, Y. The N6-Methyladenosine Features of mRNA and Aberrant Expression of m6A Modified Genes in Gastric Cancer and Their Potential Impact on the Risk and Prognosis. Front. Genet. 2020, 11, 561–566. [Google Scholar] [CrossRef]
- Liu, N.; Pan, T. N6-methyladenosine–encoded epitranscriptomics. Nat. Struct. Mol. Biol. 2016, 23, 98–102. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, B.S.; Roundtree, I.A.; Lu, Z.; Han, D.; Ma, H.; Weng, X.; Chen, K.; Shi, H.; He, C. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 2015, 161, 1388–1399. [Google Scholar] [CrossRef]
- Liu, J.; Dou, X.; Chen, C.; Chen, C.; Liu, C.; Xu, M.M.; Zhao, S.; Shen, B.; Gao, Y.; Han, D.; et al. N(6)-methyladenosine of chromosome-associated regulatory RNA regulates chromatin state and transcription. Science 2020, 367, 580–586. [Google Scholar] [CrossRef]
- Fu, Y.; Dominissini, D.; Rechavi, G.; He, C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat. Rev. Genet. 2014, 15, 293–306. [Google Scholar] [CrossRef]
- Xu, Y.; Song, M.; Hong, Z.; Chen, W.; Zhang, Q.; Zhou, J.; Yang, C.; He, Z.; Yu, J.; Peng, X.; et al. The N6-methyladenosine METTL3 regulates tumorigenesis and glycolysis by mediating m6A methylation of the tumor suppressor LATS1 in breast cancer. J. Exp. Clin. Cancer Res. 2023, 42, 10. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Xie, D.; Guan, W.; Li, Q.; He, J.; Li, Y.; Chen, J.; Gao, S.; Liu, Z. Exploring Site-Specific N6-Methyladenosine Modifications as New Cancer Biomarkers via SERS-Based Single-Cell Assay and Histopathological Imaging. Anal. Chem. 2025, 97, 16822–16831. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Qi, X.; Zhang, L.; Ning, W.; Gao, D.; Xu, T.; Ma, Y.; Knott, J.G.; Sathanawongs, A.; Cao, Z.; et al. Dynamic reprogramming and function of RNA N(6)-methyladenosine modification during porcine early embryonic development. Zygote 2021, 29, 417–426. [Google Scholar] [CrossRef]
- Li, D.; Liu, Z.; Deng, M.; Liu, L.; Lu, J.; Wang, F.; Wan, Y. The function of the m6A methyltransferase METTL3 in goat early embryo development under hypoxic and normoxic conditions. Theriogenology 2022, 177, 140–150. [Google Scholar] [CrossRef] [PubMed]
- Hui, T.; Zhu, Y.; Shen, J.; Bai, M.; Fan, Y.; Feng, S.; Wang, Z.; Zhao, J.; Zhang, Q.; Liu, X.; et al. Identification and Molecular Analysis of m(6)A-circRNAs from Cashmere Goat Reveal Their Integrated Regulatory Network and Putative Functions in Secondary Hair Follicle during Anagen Stage. Animals 2022, 12, 694. [Google Scholar] [CrossRef]
- Wang, J.; Li, X.; Qubi, W.; Li, Y.; Wang, Y.; Wang, Y.; Lin, Y. The Important Role of m6A-Modified circRNAs in the Differentiation of Intramuscular Adipocytes in Goats Based on MeRIP Sequencing Analysis. Int. J. Mol. Sci. 2023, 24, 4817. [Google Scholar] [CrossRef]
- Li, K.; Huang, W.; Wang, Z.; Nie, Q. m(6)A demethylase FTO regulate CTNNB1 to promote adipogenesis of chicken preadipocyte. J. Anim. Sci. Biotechnol. 2022, 13, 147. [Google Scholar] [CrossRef]
- Xu, T.; Xu, Z.; Lu, L.; Zeng, T.; Gu, L.; Huang, Y.; Zhang, S.; Yang, P.; Wen, Y.; Lin, D.; et al. Transcriptome-wide study revealed m6A regulation of embryonic muscle development in Dingan goose (Anser cygnoides orientalis). BMC Genom. 2021, 22, 270. [Google Scholar] [CrossRef]
- Yu, B.; Liu, J.; Cai, Z.; Wang, H.; Feng, X.; Zhang, T.; Ma, R.; Gu, Y.; Zhang, J. RNA N(6)-methyladenosine profiling reveals differentially methylated genes associated with intramuscular fat metabolism during breast muscle development in chicken. Poult. Sci. 2023, 102, 102793. [Google Scholar] [CrossRef]
- Zou, J.; Shen, Y.; Zou, J.; Yu, J.; Jiang, Y.; Huang, Y.; Jiang, Q. Transcriptome-Wide Study Revealed That N6-Methyladenosine Participates in Regulation Meat Production in Goats. Foods 2023, 12, 1159. [Google Scholar] [CrossRef]
- Wang, L.; Qi, H.; Li, D.; Liu, L.; Chen, D.; Gao, X. METTL3 is a key regulator of milk synthesis in mammary epithelial cells. Cell Biol. Int. 2022, 46, 359–369. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, X.; Wang, X.; Sun, C.; Zheng, J.; Li, J.; Yi, G.; Yang, N. The m6A methylation regulates gonadal sex differentiation in chicken embryo. J. Anim. Sci. Biotechnol. 2022, 13, 52. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wu, Y.N.; Yang, S.T.; Gao, R.; Lv, X.Y.; Yang, Z.; Jiao, P.; Zhang, N.; Loor, J.J.; Chen, Z. m6A Methylation Mediates the Function of the circRNA-08436/miR-195/ELOVL6 Axis in Regards to Lipid Metabolism in Dairy Goat Mammary Glands. Animals 2024, 14, 1715. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Zhang, Y.M.; Zhang, J.; Wang, J.; Li, Q.L. The alteration of N6-methyladenosine (m6A) modification at the transcriptome-wide level in response of heat stress in bovine mammary epithelial cells. BMC Genom. 2022, 23, 829. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Sirén, J.; Välimäki, N.; Mäkinen, V. Indexing Graphs for Path Queries with Applications in Genome Research. IEEE/ACM Trans. Comput. Biol. Bioinform. 2014, 11, 375–388. [Google Scholar]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, Y.; Wang, Z.; Liu, L.; Zhang, G.; Li, J.; Ren, Z.; Dong, Z.; Yu, Z. PRRC2A Promotes Hepatocellular Carcinoma Progression and Associates with Immune Infiltration. J. Hepatocell. Carcinoma 2021, 8, 1495–1511. [Google Scholar] [CrossRef]
- Wu, R.; Li, A.; Sun, B.; Sun, J.G.; Zhang, J.; Zhang, T.; Chen, Y.; Xiao, Y.; Gao, Y.; Zhang, Q.; et al. A novel m(6)A reader Prrc2a controls oligodendroglial specification and myelination. Cell Res. 2019, 29, 23–41. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Zheng, C.; Zhuang, Y.; Jin, P.; Wang, F. The m6A reader PRRC2A is essential for meiosis I completion during spermatogenesis. Nat. Commun. 2023, 14, 1636. [Google Scholar] [CrossRef] [PubMed]
- Merkestein, M.; Laber, S.; McMurray, F.; Andrew, D.; Sachse, G.; Sanderson, J.; Li, M.; Usher, S.; Sellayah, D.; Ashcroft, F.M.; et al. FTO influences adipogenesis by regulating mitotic clonal expansion. Nat. Commun. 2015, 6, 6792. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Song, C.; Wang, N.; Li, S.; Liu, Q.; Sun, Z.; Wang, K.; Yu, S.-C.; Yang, Q. NADP modulates RNA m(6)A methylation and adipogenesis via enhancing FTO activity. Nat. Chem. Biol. 2020, 16, 1394–1402. [Google Scholar] [CrossRef]
- Yang, Z.; Yu, G.L.; Zhu, X.; Peng, T.H.; Lv, Y.C. Critical roles of FTO-mediated mRNA m6A demethylation in regulating adipogenesis and lipid metabolism: Implications in lipid metabolic disorders. Genes 2022, 9, 51–61. [Google Scholar] [CrossRef]
- Fang, X.; Chen, J.; Meng, F.; Chen, F.; Chen, X.; Wang, Y.; Fang, X.; Zhang, C.; Song, C. Linc-smad7 is involved in the regulation of lipid synthesis in mouse mammary epithelial cells. Int. J. Biol. Macromol. 2024, 262, 129875. [Google Scholar] [CrossRef]
- Tanabe, A.; Nakayama, T.; Kashiyanagi, J.; Yamaga, H.; Hirohashi, Y.; Torigoe, T.; Satomi, F.; Shima, H.; Maeda, H.; Kutomi, G.; et al. YTHDC2 Promotes Malignant Phenotypes of Breast Cancer Cells. J. Oncol. 2022, 2022, 9188920. [Google Scholar] [CrossRef]
- Zhang, G.; Xu, Y.; Wang, X.; Zhu, Y.; Wang, L.; Zhang, W.; Wang, Y.; Gao, Y.; Wu, X.; Cheng, Y.; et al. Dynamic FMR1 granule phase switch instructed by m6A modification contributes to maternal RNA decay. Nat. Commun. 2022, 13, 859. [Google Scholar] [CrossRef]
- Yue, B.; Song, C.; Yang, L.; Cui, R.; Cheng, X.; Zhang, Z.; Zhao, G. METTL3-mediated N6-methyladenosine modification is critical for epithelial-mesenchymal transition and metastasis of gastric cancer. Mol. Cancer 2019, 18, 142. [Google Scholar] [CrossRef]
- Weng, H.; Huang, F.; Yu, Z.; Chen, Z.; Prince, E.; Kang, Y.; Zhou, K.; Li, W.; Hu, J.; Fu, C.; et al. The m6A reader IGF2BP2 regulates glutamine metabolism and represents a therapeutic target in acute myeloid leukemia. Cancer Cell 2022, 40, 1566–1582. [Google Scholar] [CrossRef]
- Yu, K.; Shi, H.; Luo, J.; Li, J.; Zhao, W.; Tian, H.; Shi, H. PPARG modulated lipid accumulation in dairy GMEC via regulation of ADRP gene. J. Cell. Biochem. 2015, 116, 192–201. [Google Scholar]
- Tian, H.; Luo, J.; Guo, P.; Li, C.; Zhang, X. C/EBPα promotes triacylglycerol synthesis via regulating PPARG promoter activity in goat mammary epithelial cells. J. Anim. Sci. 2023, 101, skac412. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.S.; Zhang, M.; Chen, P.; Xiong, X.F.; Liu, P.Q.; Wang, H.B.; Wang, J.J.; Shen, J. The m(6)A demethylase FTO promotes the osteogenesis of mesenchymal stem cells by downregulating PPARG. Acta Pharmacol. Sin. 2022, 43, 1311–1323. [Google Scholar] [CrossRef] [PubMed]
- Bouras, T.; Pal, B.; Vaillant, F.; Harburg, G.; Asselin-Labat, M.L.; Oakes, S.R.; Lindeman, G.J.; Visvader, J.E. Notch signaling regulates mammary stem cell function and luminal cell-fate commitment. Cell Stem Cell 2008, 3, 429–441. [Google Scholar] [CrossRef] [PubMed]
- Fortini, M.E. Notch signaling: The core pathway and its posttranslational regulation. Dev. Cell 2009, 16, 633–647. [Google Scholar] [CrossRef]
- Xie, Y.G.; Yu, Y.; Hou, L.K.; Wang, X.; Zhang, B.; Cao, X.C. FYN promotes breast cancer progression through epithelial-mesenchymal transition. Oncol. Rep. 2016, 36, 1000–1006. [Google Scholar] [CrossRef]
- Sun, S.; Liu, Y.; Zhou, M.; Wen, J.; Xue, L.; Han, S.; Liang, J.; Wang, Y.; Wei, Y.; Yu, J.; et al. PA2G4 promotes the metastasis of hepatocellular carcinoma by stabilizing FYN mRNA in a YTHDF2-dependent manner. Cell Biosci. 2022, 12, 55. [Google Scholar] [CrossRef]
- Wang, X.; Proud, C.G. The mTOR pathway in the control of protein synthesis. Physiology 2006, 21, 362–369. [Google Scholar] [CrossRef]
- Liu, S.; Cao, H.; Guo, D.; Jiang, Y.; Yin, H.; Zhu, J.; Duan, Q.; Seleh-Zo, E.D.M.; Li, G.; An, X.; et al. Pou2F3 silencing enhanced the proliferation of mammary epithelial cells in dairy goat via PI3K/AKT/mTOR signaling pathway. Anim. Biotechnol. 2022, 33, 321–329. [Google Scholar] [CrossRef]
- Pires, B.R.B.; Silva, R.; Ferreira, G.M.; Abdelhay, E. NF-kappaB: Two Sides of the Same Coin. Genes 2018, 9, 24. [Google Scholar] [CrossRef]
- Yu, M.; Qi, H.; Gao, X. Daidzein promotes milk synthesis and proliferation of mammary epithelial cells via the estrogen receptor α-dependent NFκB1 activation. Anim. Biotechnol. 2022, 33, 43–52. [Google Scholar] [CrossRef]
- Zhao, Q.; Zhao, Y.; Hu, W.; Zhang, Y.; Wu, X.; Lu, J.; Li, M.; Li, W.; Wu, W.; Wang, J.; et al. m(6)A RNA modification modulates PI3K/Akt/mTOR signal pathway in Gastrointestinal Cancer. Theranostics 2020, 10, 9528–9543. [Google Scholar] [CrossRef]
- Mironov, A.; Fisher, M.; Narayanan, P.; Elsayed, R.; Karabulutoglu, M.; Akhtar, N. Rac1 controls cell turnover and reversibility of the involution process in postpartum mammary glands. PLoS Biol. 2023, 21, e3001583. [Google Scholar] [CrossRef]
- Zhang, X.; Zhao, F.; Si, Y.; Huang, Y.; Yu, C.; Luo, C.; Zhang, N.; Li, Q.; Gao, X. GSK3β regulates milk synthesis in and proliferation of dairy cow mammary epithelial cells via the mTOR/S6K1 signaling pathway. Molecules 2014, 19, 9435–9452. [Google Scholar] [CrossRef]
- Zhu, L.; Shen, X.B.; Yuan, P.C.; Shao, T.L.; Wang, G.D.; Liu, X.P. Arctigenin inhibits proliferation of ER-positive breast cancer cells through cell cycle arrest mediated by GSK3-dependent cyclin D1 degradation. Life Sci. 2020, 256, 117983. [Google Scholar] [CrossRef] [PubMed]
- Bagci, H.; Laurin, M.; Huber, J.; Muller, W.J.; Côté, J.F. Impaired cell death and mammary gland involution in the absence of Dock1 and Rac1 signaling. Cell Death Dis. 2014, 5, e1375. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Li, X.; Gong, Z.; Wang, W.; Li, Y.; Nair, B.C.; Piao, H.; Yang, K.; Wu, G.; Chen, J. Proteomic analysis of the human cyclin-dependent kinase family reveals a novel CDK5 complex involved in cell growth and migration. Mol. Cell Proteom. 2014, 13, 2986–3000. [Google Scholar] [CrossRef]
- Shah, K.; Rossie, S. Tale of the Good and the Bad Cdk5, Remodeling of the Actin Cytoskeleton in the Brain. Mol. Neurobiol. 2018, 55, 3426–3438. [Google Scholar] [CrossRef] [PubMed]


| Sample | Raw Data | Clean Data and Base Quality | Reference Genome Mapping | ||||
|---|---|---|---|---|---|---|---|
| Clean Data | Q20 (%) | Q30 (%) | Unique_Mapped_Reads | Multiple_Mapped_Reads | All Mapped Reads | ||
| E1-IN | 49336232 | 48847722 (99.01%) | 6989937247 (97.26%) | 6613256954 (92.02%) | 41781542 (85.75%) | 3549962 (7.29%) | 45331504 (93.04%) |
| E1-IP | 47274568 | 45972812 (97.25%) | 2021227037 (92.21%) | 1871690851 (85.39%) | 25455216 (55.42%) | 9434574 (20.54%) | 34889790 (75.96%) |
| E2-IN | 52823280 | 52304560 (99.02%) | 7497513916 (97.32%) | 7095665407 (92.10%) | 45085065 (86.34%) | 4680842 (8.96%) | 49765907 (95.31%) |
| E2-IP | 50759632 | 49362212 (97.25%) | 2216090699 (92.50%) | 2054835901 (85.77%) | 30472303 (61.76%) | 8603775 (17.44%) | 39076078 (79.20%) |
| E3-IN | 64813138 | 64543396 (99.58%) | 9254754450 (97.39%) | 8792123099 (92.53%) | 55296223 (85.92%) | 6533475 (10.15%) | 61829698 (96.07%) |
| E3-IP | 58311116 | 57006772 (97.76%) | 2373500143 (93.15%) | 2216974301 (87.00%) | 36019943 (63.23%) | 10062207 (17.66%) | 46082150 (80.89%) |
| M1-IN | 45990328 | 45873914 (99.75%) | 6676884375 (97.26%) | 6360487594 (92.65%) | 41528790 (90.58%) | 2293689 (5.00%) | 43822479 (95.58%) |
| M1-IP | 60920318 | 59851776 (98.25%) | 4620013058 (98.50%) | 4493473372 (95.80%) | 46720976 (78.13%) | 6314407 (10.56%) | 53035383 (88.69%) |
| M2-IN | 45138384 | 45022574 (99.74%) | 6559955218 (97.40%) | 6261243385 (92.96%) | 38979574 (86.71%) | 1889772 (4.20%) | 40869346 (90.92%) |
| M2-IP | 59322628 | 56784348 (95.72%) | 3227223883 (98.05%) | 3115696572 (94.66%) | 31123475 (54.89%) | 7857644 (13.86%) | 38981119 (68.74%) |
| M3-IN | 52372706 | 52283468 (99.83%) | 7644172658 (97.65%) | 7296392557 (93.21%) | 47941023 (91.72%) | 2080862 (3.98%) | 50021885 (95.70%) |
| M3-IP | 62105882 | 60811202 (97.92%) | 4300057659 (98.25%) | 4167031814 (95.21%) | 47128789 (77.64%) | 8972234 (14.78%) | 56101023 (92.42%) |
| P1-IN | 50993434 | 50610932 (99.25%) | 7295972140 (97.32%) | 6901424197 (92.06%) | 43640296 (86.38%) | 4544886 (9.00%) | 48185182 (95.37%) |
| P1-IP | 44199300 | 43162816 (97.65%) | 1688812739 (93.05%) | 1577322072 (86.91%) | 27756879 (64.34%) | 6412478 (14.86%) | 34169357 (79.20%) |
| P2-IN | 49823380 | 49382116 (99.11%) | 7017856509 (97.28%) | 6651148714 (92.20%) | 43060635 (87.49%) | 3391325 (6.89%) | 46451960 (94.39%) |
| P2-IP | 49680806 | 48674586 (97.97%) | 2206924336 (93.49%) | 2056514036 (87.12%) | 29752521 (61.17%) | 9099850 (18.71%) | 38852371 (79.88%) |
| P3-IN | 59977268 | 59518652 (99.24%) | 8522800666 (97.60%) | 8108542674 (92.85%) | 50915336 (85.69%) | 6397019 (10.77%) | 57312355 (96.45%) |
| P3-IP | 46226638 | 45333466 (98.07%) | 2087434412 (93.51%) | 1947410259 (87.24%) | 31992607 (70.60%) | 5985301 (13.21%) | 37977908 (83.81%) |
| Period | DRACH | Motif | Motif Number | RRACH | Motif | Motif Number |
|---|---|---|---|---|---|---|
| E |  | AAACA | 1258 (9.05%) | 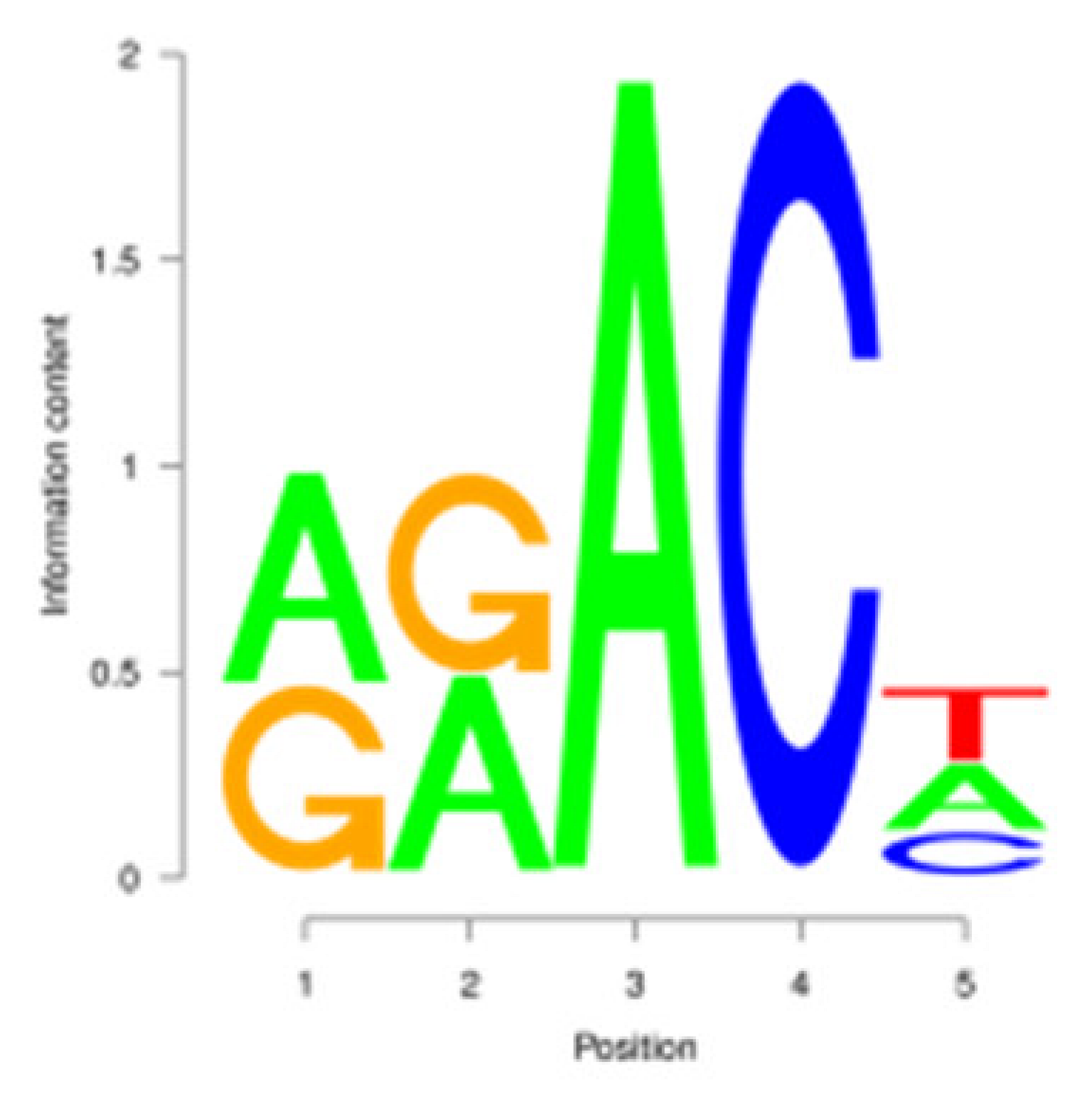 | AAACA | 1281 (11.93%) |
| P |  | GGACT | 709 (9.31%) | 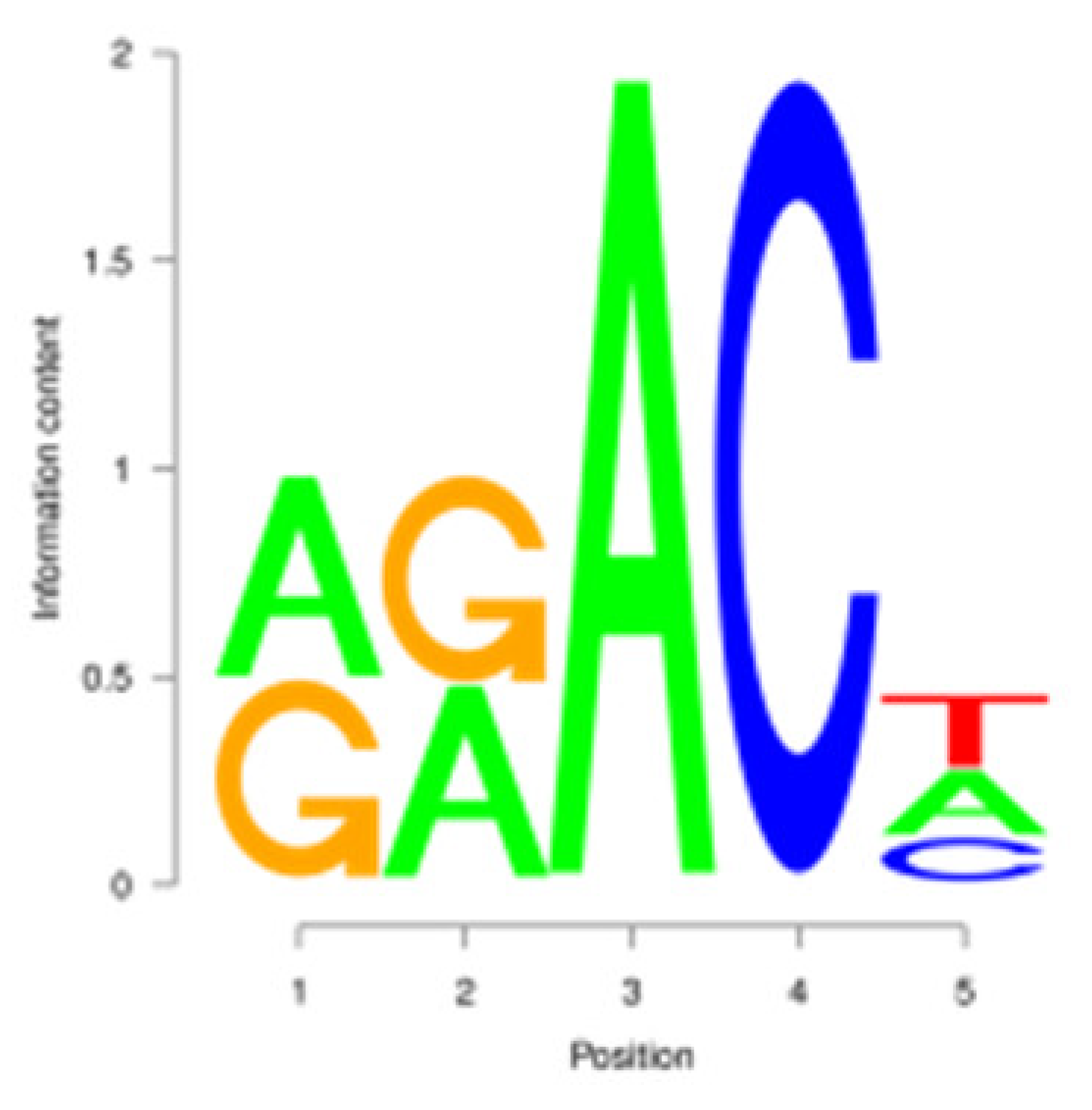 | GGACT | 732 (12.35%) |
| M | 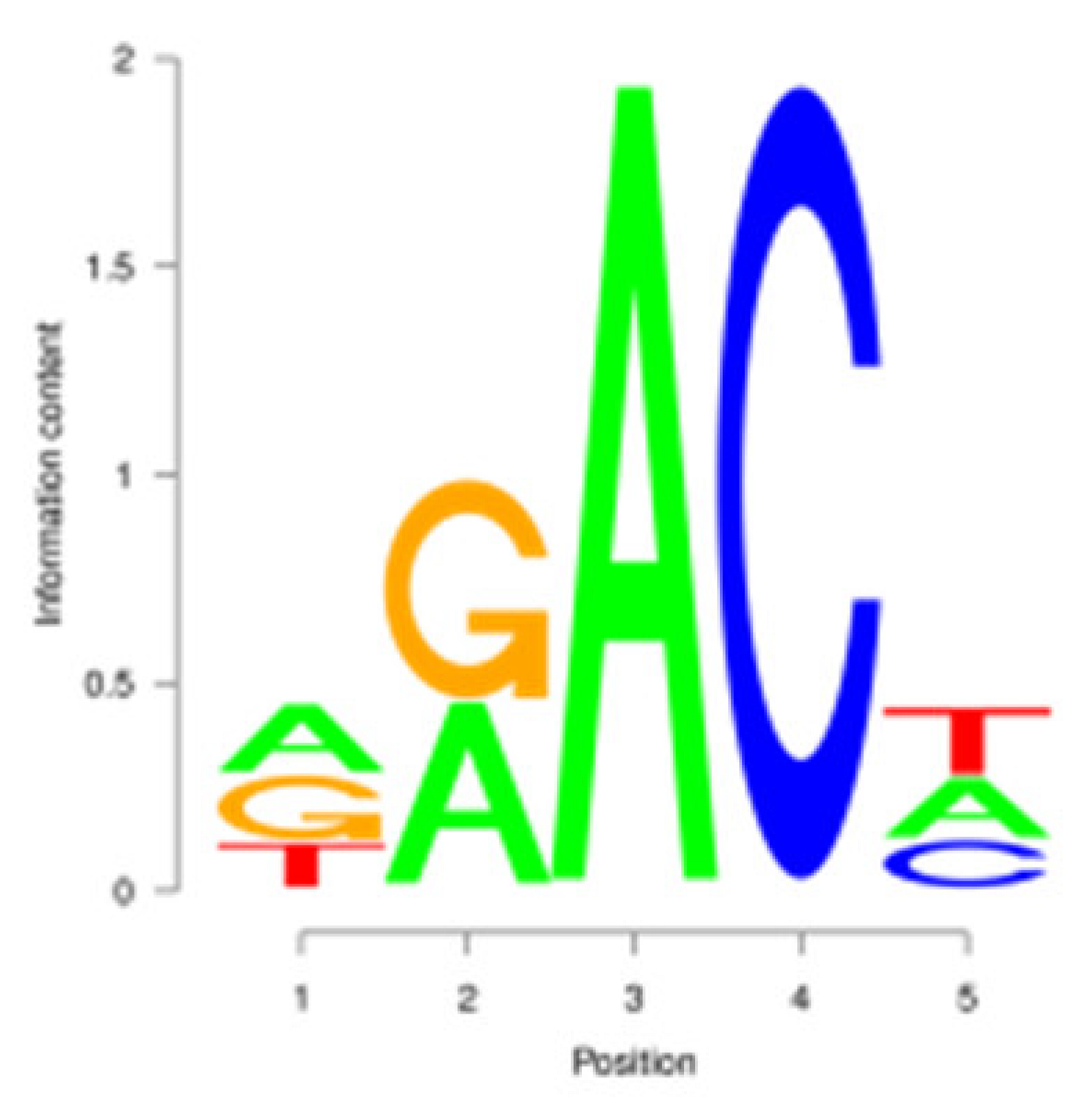 | AAACA | 218 (8.92%) | 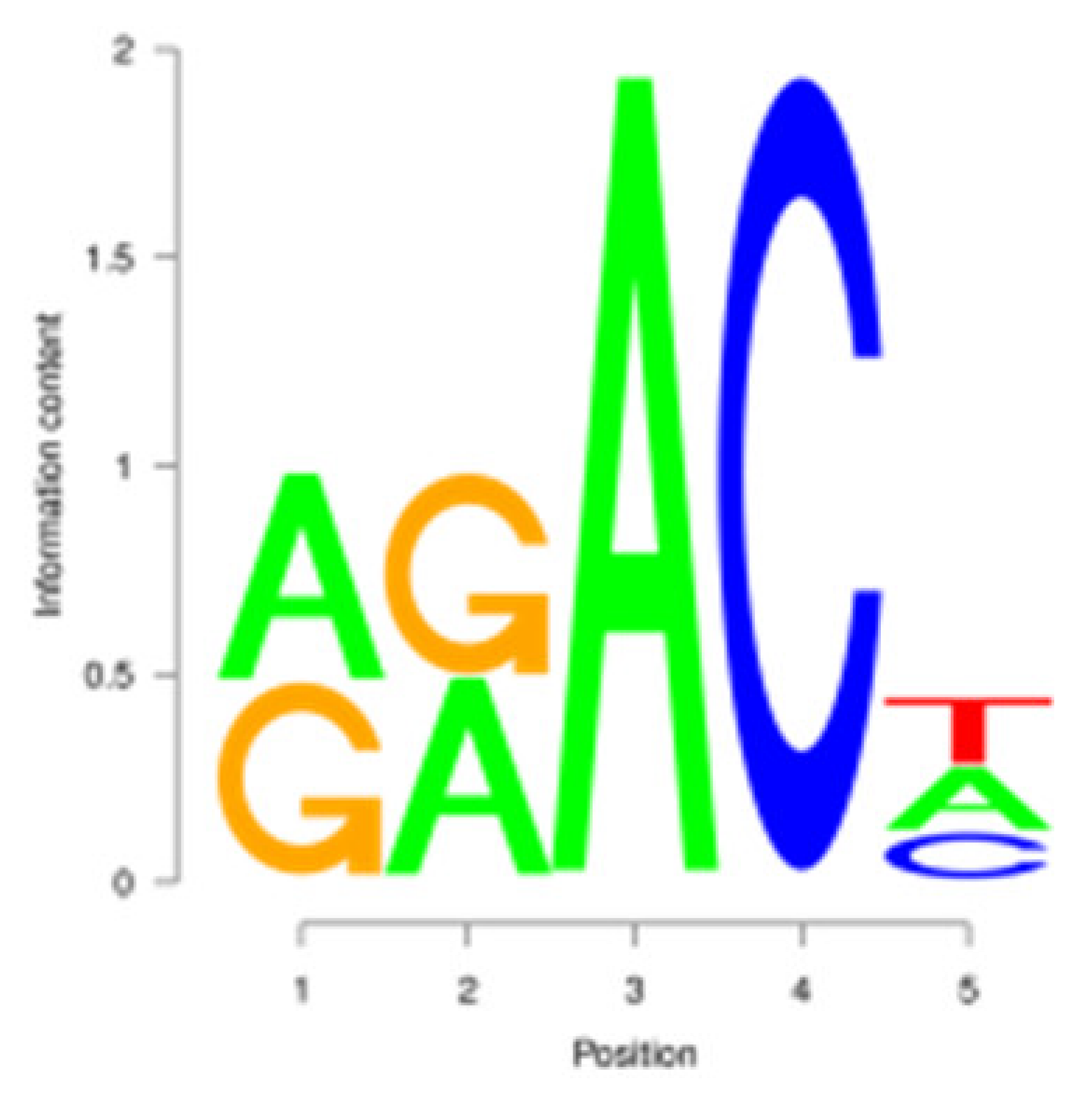 | AAACA | 222 (11.76%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Ji, Z.; Zhao, M.; Fu, J.; Meng, X. Epigenetic Effects and Potential Contributions of m6A Modification to Mammary Gland Development and Lactation of Dairy Goats Explored via MeRIP-seq. Animals 2025, 15, 2775. https://doi.org/10.3390/ani15192775
Zhang L, Ji Z, Zhao M, Fu J, Meng X. Epigenetic Effects and Potential Contributions of m6A Modification to Mammary Gland Development and Lactation of Dairy Goats Explored via MeRIP-seq. Animals. 2025; 15(19):2775. https://doi.org/10.3390/ani15192775
Chicago/Turabian StyleZhang, Lu, Zhibin Ji, Mingxin Zhao, Jianzhi Fu, and Xianglei Meng. 2025. "Epigenetic Effects and Potential Contributions of m6A Modification to Mammary Gland Development and Lactation of Dairy Goats Explored via MeRIP-seq" Animals 15, no. 19: 2775. https://doi.org/10.3390/ani15192775
APA StyleZhang, L., Ji, Z., Zhao, M., Fu, J., & Meng, X. (2025). Epigenetic Effects and Potential Contributions of m6A Modification to Mammary Gland Development and Lactation of Dairy Goats Explored via MeRIP-seq. Animals, 15(19), 2775. https://doi.org/10.3390/ani15192775





