A New Family-Based Approach for Detecting Allele-Specific Expression and for Mapping Possible eQTLs
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Library Preparation and Sequencing
2.2.1. Whole-Genome Sequencing
2.2.2. Quality Control (QC)
2.3. Generation of an Annotation File for OryCun3.0
2.4. Read Mapping
2.5. Read Counts for DEG Analysis
2.6. Variant Identification
2.7. ASE Analysis in the Entire Family
2.8. Pinpointing ASE Cases (Genes)
2.9. Identification of Relevant Variants in the Whole Genome and RNA Sequencing
2.10. Identifying Potential Causative Transcription Factor Binding Sites (TFBSs)
2.11. Haplotype Phasing
3. Results
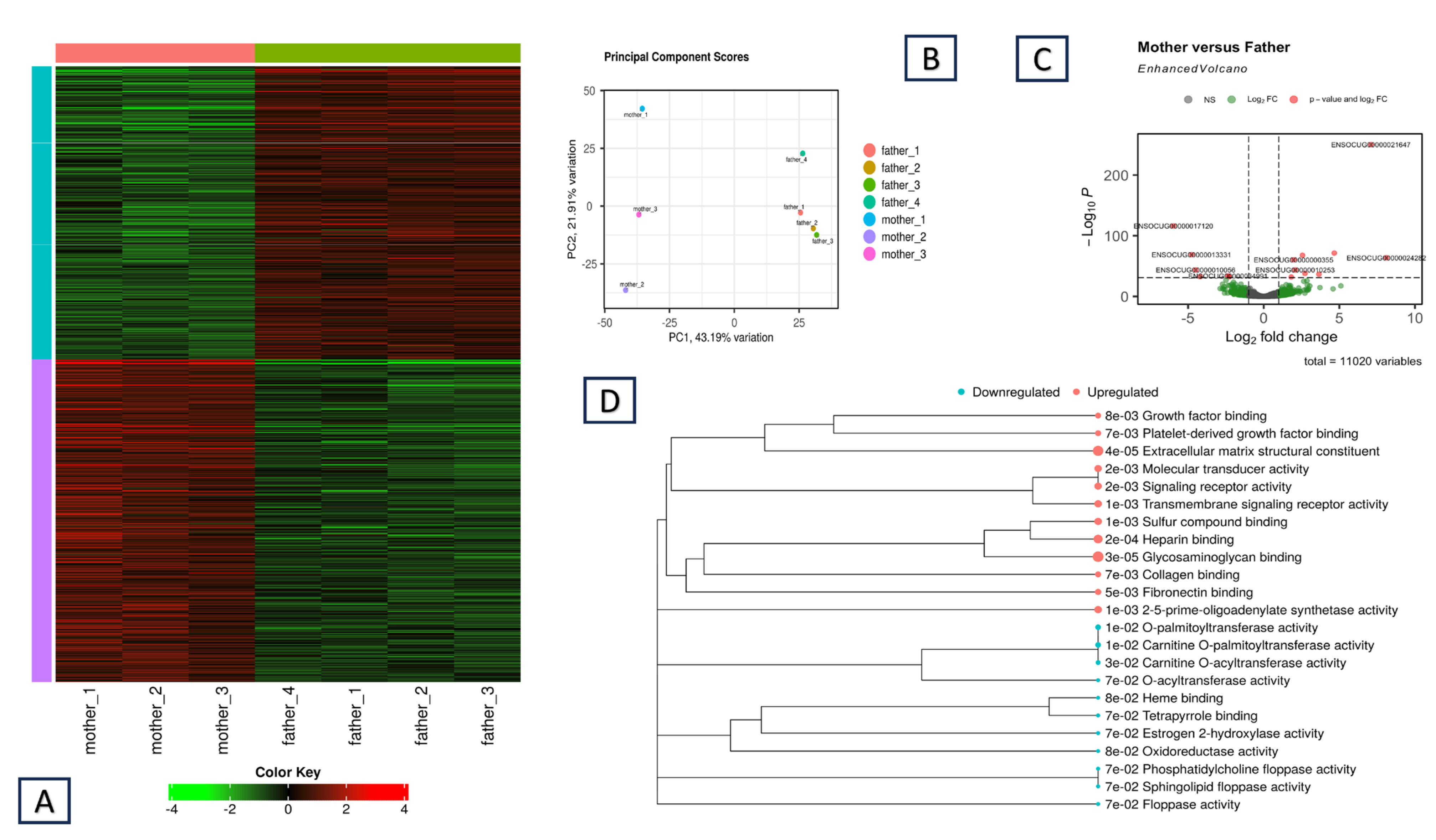
3.1. Differences in Gene Expression Between the Two Parents
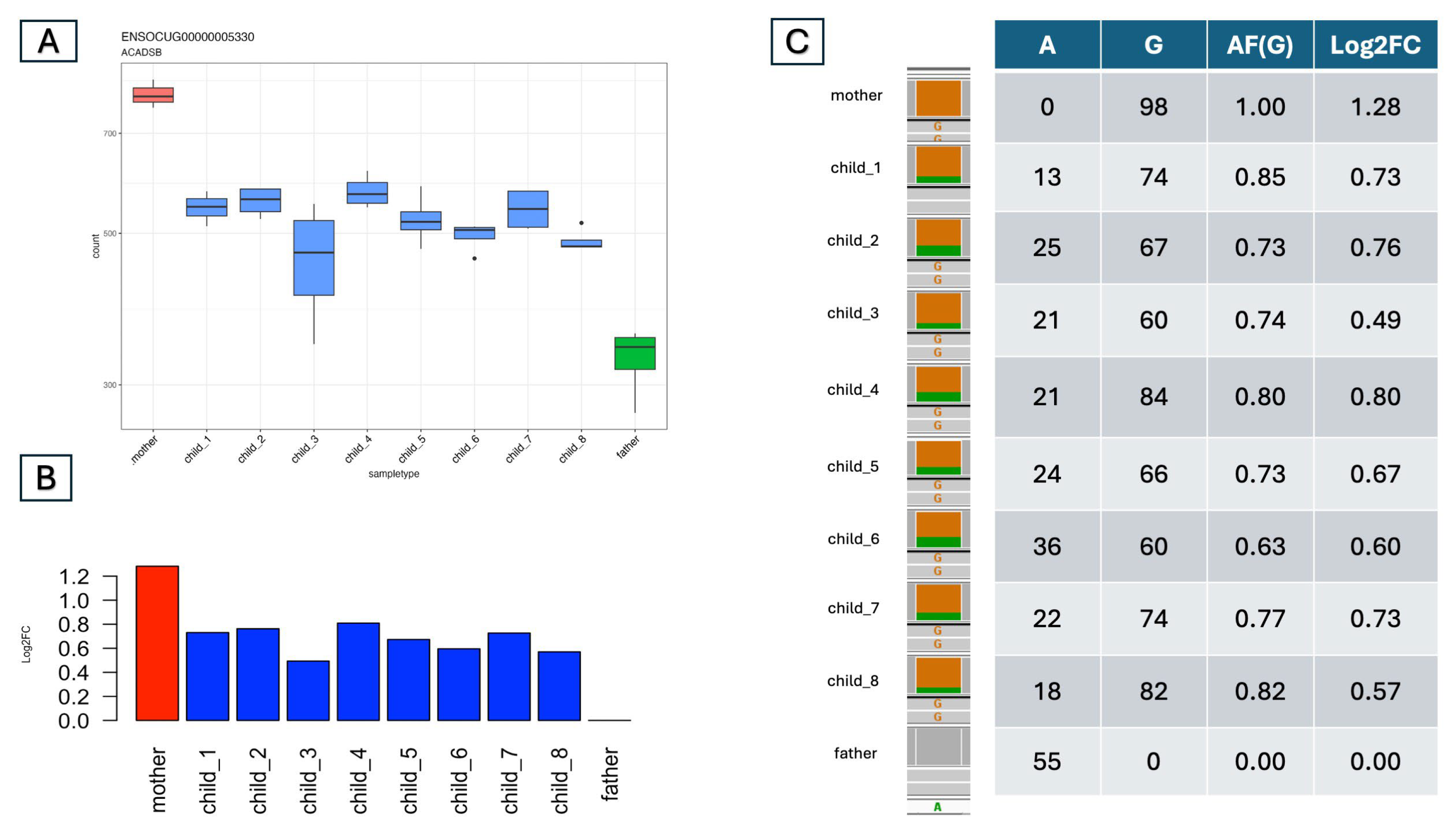
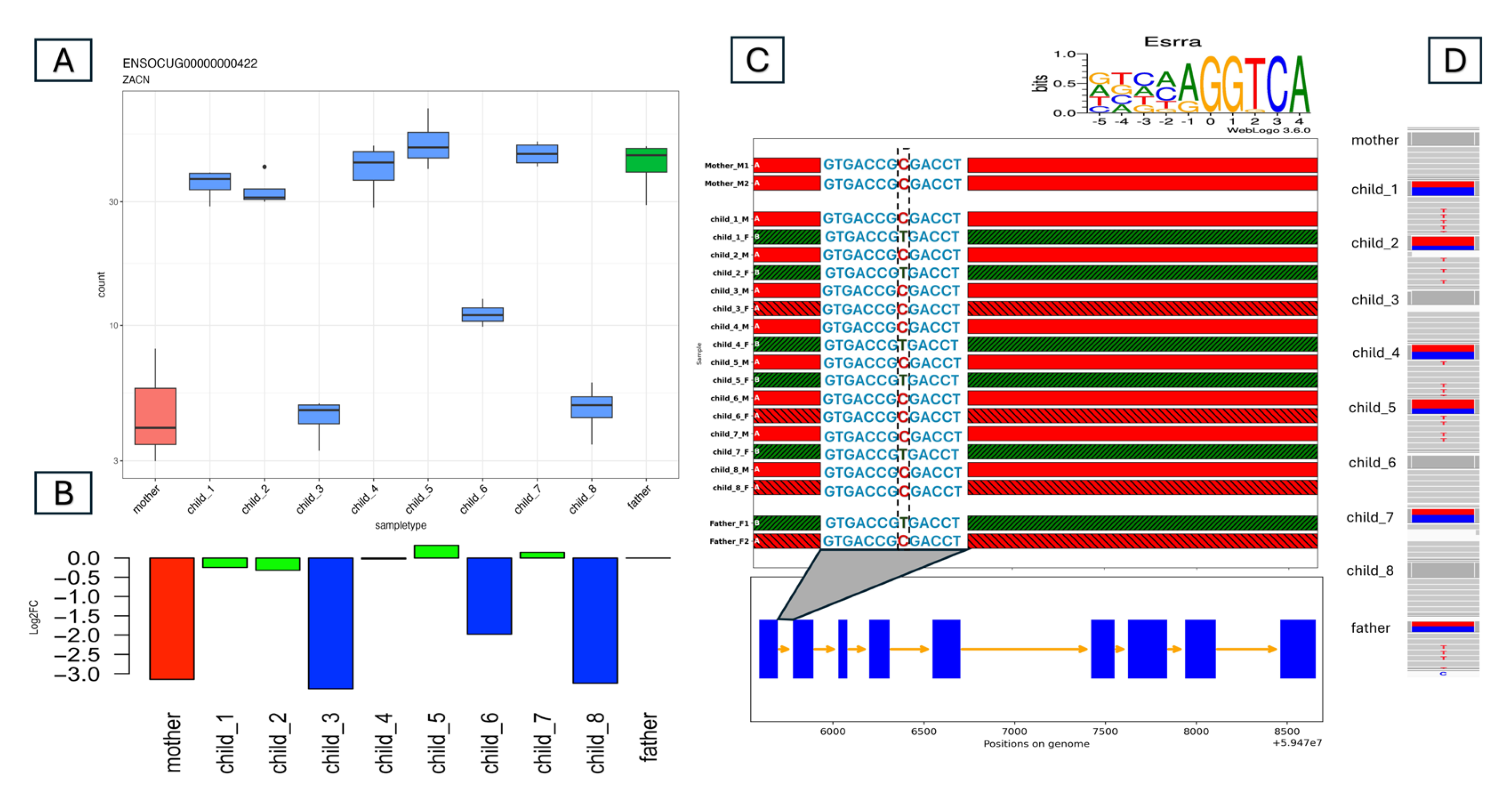
3.2. Detecting Allele-Specific Expression
- (1)
- The two parents have significantly different expression levels: |Log2FC| > 1 and p-adj value < 0.05. If all the offspring show expression levels intermediate (M) between those of the parents, then we can assign these genes to the H_L or L_H categories (Figure 2A,B) depending on which parent’s expression was higher (the first letter refers to the mother).
- (2)
- The parents have different expression levels; the individual offspring’s expression levels are distributed into two categories, which align with the parents’ two expression levels; and at least one individual has a |Log2FC| > 1 and a p-adj value < 0.05. In this case, we can assume that one of the parents is under heterozygous regulation (M). The (H_M, L_M) and (M_H, M_L) cases fall into this category (Figure 2C,D).
- (3)
- There is also a special case where both parents’ expression levels are similar (Log2FC in the range of [−0.4, 0.4]), but in accordance with the Mendelian law of segregation, the expression levels of the offspring fall into three categories (Figure 2E); some will be above (H), some will be below (L), and some will be similar (M) to the parent’s expression level. Although eight offspring are not a sufficient number for detailed statistical analysis, the ratios of the three cases should adhere roughly to Mendel’s law of segregation for the F2 generation (1:2:1). Here, the second category, with a twofold ratio, should correspond to the offspring whose expression level is similar to that of the parents.
- (4)
- In many cases, the parents’ expression levels will be similar and the expression levels in the offspring will not segregate. In these cases, we can assume that no ASE exists (Figure 2F).
3.3. Validation of the Predicted ASEs
3.4. Looking for Regulatory Variants
4. Discussion
Study Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ASE | Allele-Specific Expression |
| DEG | Differentially Expressed Gene |
| TFBS | Transcription Factor Binding Site |
| WGS | Whole-Genome Sequencing |
References
- Diaz, R.; Wang, Z.; Jeffrey, P.T. Chapter 5—Measurement and meaning in gene expression evolution. In Transcriptome Profiling; Mohammad, A.A., Joongku, L., Eds.; Academic Press: Cambridge, MA, USA, 2023; pp. 111–129. [Google Scholar]
- Blanco, A.; Gustavo, B. Chapter 23—Regulation of gene expression. In Medical Biochemistry, 2nd ed.; Antonio, B., Gustavo, B., Eds.; Academic Press: Cambridge, MA, USA, 2022; pp. 569–581. [Google Scholar]
- Istrail, S.; Pi, S. How Does the Regulatory Genome Work? J. Comput. Biol. 2019, 26, 685–695. [Google Scholar] [CrossRef]
- Rister, J.; Desplan, C. Deciphering the genome’s regulatory code: The many languages of DNA. Bioessays 2010, 32, 381–384. [Google Scholar] [CrossRef] [PubMed]
- Moutsopoulos, I.; Williams, E.C.; Mohorianu, I.I. bulkAnalyseR: An accessible, interactive pipeline for analysing and sharing bulk multi-modal sequencing data. Brief. Bioinform. 2023, 24, bbac591. [Google Scholar] [CrossRef] [PubMed]
- Cleary, S.; Seoighe, C. Perspectives on Allele-Specific Expression. Annu. Rev. Biomed. Data Sci. 2021, 4, 101–122. [Google Scholar] [CrossRef] [PubMed]
- Wittkopp, P.J.; Haerum, B.K.; Clark, A.G. Evolutionary changes in cis and trans gene regulation. Nature 2004, 430, 85–88. [Google Scholar] [CrossRef]
- Cavalli, M.; Baltzer, N.; Umer, H.M.; Grau, J.; Lemnian, I.; Pan, G.; Wallerman, O.; Spalinskas, R.; Sahlén, P.; Grosse, I.; et al. Allele specific chromatin signals, 3D interactions, and motif predictions for immune and B cell related diseases. Sci. Rep. 2019, 9, 2695. [Google Scholar] [CrossRef]
- Li, D.; Zand, M.S.; Dye, T.D.; Goniewicz, M.L.; Rahman, I.; Xie, Z. An evaluation of RNA-seq differential analysis methods. PLoS ONE 2022, 17, e0264246. [Google Scholar] [CrossRef]
- Ngwa, J.S.; Yanek, L.R.; Kammers, K.; Kanchan, K.; Taub, M.A.; Scharpf, R.B.; Faraday, N.; Becker, L.C.; Mathias, R.A.; Ruczinski, I. Secondary analyses for genome-wide association studies using expression quantitative trait loci. Genet. Epidemiol. 2022, 46, 170–181. [Google Scholar] [CrossRef]
- Little, P.; Liu, S.; Zhabotynsky, V.; Li, Y.; Lin, D.Y.; Sun, W. A computational method for cell type-specific expression quantitative trait loci mapping using bulk RNA-seq data. Nat. Commun. 2023, 14, 3030. [Google Scholar] [CrossRef]
- Bruscadin, J.J.; Cardoso, T.F.; da Silva Diniz, W.J.; Afonso, J.; de Souza, M.M.; Petrini, J.; Nascimento Andrade, B.G.; da Silva, V.H.; Ferraz, J.B.S.; Zerlotini, A.; et al. Allele-specific expression reveals functional SNPs affecting muscle-related genes in bovine. Biochim. Biophys. Acta Gene Regul. Mech. 2022, 1865, 194886. [Google Scholar] [CrossRef]
- Dong, X.; Luo, H.; Yao, J.; Guo, Q.; Yu, S.; Zhang, X.; Cheng, X.; Meng, D. Characterization of Genes That Exhibit Genotype-Dependent Allele-Specific Expression and Its Implications for the Development of Maize Kernel. Int. J. Mol. Sci. 2023, 24, 4766. [Google Scholar] [CrossRef]
- Her, L.; Shi, J.; Wang, X.; He, B.; Smith, L.S.; Jiang, H.; Zhu, H.J. Identification of regulatory variants of carboxylesterase 1 (CES1): A proof-of-concept study for the application of the Allele-Specific Protein Expression (ASPE) assay in identifying cis-acting regulatory genetic polymorphisms. Proteomics 2023, 23, e2200176. [Google Scholar] [CrossRef]
- Zou, L.S.; Cable, D.M.; Barrera-Lopez, I.A.; Zhao, T.; Murray, E.; Aryee, M.J.; Chen, F.; Irizarry, R.A. Detection of allele-specific expression in spatial transcriptomics with spASE. Genome Biol. 2024, 25, 180. [Google Scholar] [CrossRef]
- Van de Geijn, B.; McVicker, G.; Gilad, Y.; Pritchard, J.K. WASP: Allele-specific software for robust molecular quantitative trait locus discovery. Nat. Methods 2015, 12, 1061–1063. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC; A Quality Control Tool for High Throughput Sequence Data; Babraham Bioinformatics: Cambridge, UK, 2010. [Google Scholar]
- Chen, S. Ultrafast one-pass FASTQ data preprocessing, quality control, and deduplication using fastp. Imeta 2023, 2, e107. [Google Scholar] [CrossRef] [PubMed]
- Shumate, A.; Salzberg, S.L. Liftoff: Accurate mapping of gene annotations. Bioinformatics 2021, 37, 1639–1643. [Google Scholar] [CrossRef] [PubMed]
- Pertea, G.; Pertea, M. GFF Utilities: GffRead and GffCompare. F1000Research 2020, 9, 304. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Vasimuddin, M.; Misra, S.; Li, H.; Aluru, S. Efficient Architecture-Aware Acceleration of BWA-MEM for Multicore Systems. In Proceedings of the 2019 IEEE International Parallel and Distributed Processing Symposium (IPDPS), Rio de Janeiro, Brazil, 20–24 May 2019. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Ge, S.X.; Son, E.W.; Yao, R. iDEP: An integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinform. 2018, 19, 534. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Blighe, K.; Rana, S.; Lewis, M. EnhancedVolcano: Publication-Ready Volcano Plots with Enhanced Colouring and Labeling. 2018. Available online: https://bioconductor.org/packages/release/bioc/html/EnhancedVolcano.html (accessed on 15 June 2025).
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- DePristo, M.A.; Banks, E.; Poplin, R.; Garimella, K.V.; Maguire, J.R.; Hartl, C.; Philippakis, A.A.; del Angel, G.; Rivas, M.A.; Hanna, M.; et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011, 43, 491–498. [Google Scholar] [CrossRef]
- Van der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; Del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From FastQ data to high confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinform. 2013, 43, 11.10.1–11.10.33. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Czipa, E.; Schiller, M.; Nagy, T.; Kontra, L.; Steiner, L.; Koller, J.; Pálné-Szén, O.; Barta, E. ChIPSummitDB: A ChIP-seq-based database of human transcription factor binding sites and the topological arrangements of the proteins bound to them. Database 2020, 2020, baz141. [Google Scholar] [CrossRef] [PubMed]
- Fekete, Z.; Német, Z.; Ninausz, N.; Fehér, P.; Schiller, M.; Alnajjar, M.; Szenes, Á.; Nagy, T.; Stéger, V.; Kontra, L.; et al. Whole-Genome Sequencing-Based Population Genetic Analysis of Wild and Domestic Rabbit Breeds. Animals 2025, 15, 775. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, Y.; Zhang, B.; Zhong, H.; Lu, Y.; Zhang, H. Candidate gene screening for lipid deposition using combined transcriptomic and proteomic data from Nanyang black pigs. BMC Genom. 2021, 22, 441. [Google Scholar] [CrossRef]
- Nagy, G.; Nagy, L. Motif grammar: The basis of the language of gene expression. Comput. Struct. Biotechnol. J. 2020, 18, 2026–2032. [Google Scholar] [CrossRef]
- Lin, Y.; Li, J.; Chen, L.; Bai, J.; Zhang, J.; Wang, Y.; Liu, P.; Long, K.; Ge, L.; Jin, L.; et al. Allele-specific regulatory effects on the pig transcriptome. GigaScience 2022, 12, giad076. [Google Scholar] [CrossRef]
- Quan, J.; Yang, M.; Wang, X.; Cai, G.; Ding, R.; Zhuang, Z.; Zhou, S.; Tan, S.; Ruan, D.; Wu, J.; et al. Multi-omic characterization of allele-specific regulatory variation in hybrid pigs. Nat. Commun. 2024, 15, 5587. [Google Scholar] [CrossRef] [PubMed]
- Lek, M.; Karczewski, K.J.; Minikel, E.V.; Samocha, K.E.; Banks, E.; Fennell, T.; O’Donnell-Luria, A.H.; Ware, J.S.; Hill, A.J.; Cummings, B.B.; et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016, 536, 285–291. [Google Scholar] [CrossRef] [PubMed]
- Bruscadin, J.J.; de Souza, M.M.; de Oliveira, K.S.; Rocha, M.I.P.; Afonso, J.; Cardoso, T.F.; Zerlotini, A.; Coutinho, L.L.; Niciura, S.C.M.; de Almeida Regitano, L.C. Muscle allele-specific expression QTLs may affect meat quality traits in Bos indicus. Sci. Rep. 2021, 11, 7321. [Google Scholar] [CrossRef] [PubMed]
- Guillocheau, G.M.; El Hou, A.; Meersseman, C.; Esquerré, D.; Rebours, E.; Letaief, R.; Simao, M.; Hypolite, N.; Bourneuf, E.; Bruneau, N.; et al. Survey of allele specific expression in bovine muscle. Sci. Rep. 2019, 9, 4297. [Google Scholar] [CrossRef]
- Liu, D.; Zhang, H.; Yang, Y.; Liu, T.; Guo, Z.; Fan, W.; Wang, Z.; Yang, X.; Zhang, B.; Liu, H.; et al. Metabolome-Based Genome-Wide Association Study of Duck Meat Leads to Novel Genetic and Biochemical Insights. Adv. Sci. 2023, 10, e2300148. [Google Scholar] [CrossRef]
- Andrews, G.; Fan, K.; Pratt, H.E.; Phalke, N.; Consortium, Z.; Karlsson, E.K.; Lindblad-Toh, K.; Gazal, S.; Moore, J.E.; Weng, Z.; et al. Mammalian evolution of human cis-regulatory elements and transcription factor binding sites. Science 2023, 380, eabn7930. [Google Scholar] [CrossRef]
- Li, Y.; Chen, C.Y.; Kaye, A.M.; Wasserman, W.W. The identification of cis-regulatory elements: A review from a machine learning perspective. Biosystems 2015, 138, 6–17. [Google Scholar] [CrossRef]
- Crowley, J.J.; Zhabotynsky, V.; Sun, W.; Huang, S.; Pakatci, I.K.; Kim, Y.; Wang, J.R.; Morgan, A.P.; Calaway, J.D.; Aylor, D.L.; et al. Analyses of allele-specific gene expression in highly divergent mouse crosses identifies pervasive allelic imbalance. Nat. Genet. 2015, 47, 353–360. [Google Scholar] [CrossRef]
- Goncalves, A.; Leigh-Brown, S.; Thybert, D.; Stefflova, K.; Turro, E.; Flicek, P.; Brazma, A.; Odom, D.T.; Marioni, J.C. Extensive compensatory cis-trans regulation in the evolution of mouse gene expression. Genome Res. 2012, 22, 2376–2384. [Google Scholar] [CrossRef]

| Reference | A | A | A | A | A | A | A | A |
| Mother | AA | BB | BB | AB | AA | AB | BB | AB |
| Father | BB | AA | BB | AB | AB | AA | AB | BB |
| Var_Count | 1,612,167 | 1,693,817 | 13,833,792 | 2,896,419 | 4,198,928 | 3,657,972 | 2,765,210 | 2,439,608 |
| Cases | H_L | L_H | M_M | H_M or M_L | L_M or M_H | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. Genes | 97 | 104 | 469 | 110 | 133 | ||||||||||
| All | Matched | % | All | Matched | % | All | Matched | % | All | Matched | % | All | Matched | % | |
| RNA | 65 | 27 | 41.5 | 72 | 35 | 48.6 | 341 | 1 | 0.3 | 72 | 3 | 4.2 | 94 | 8 | 8.5 |
| DNA_EXON | 81 | 34 | 42 | 88 | 45 | 51.1 | 374 | 1 | 0.3 | 89 | 6 | 6.7 | 105 | 14 | 13.3 |
| DNA_INTRON | 84 | 42 | 50 | 97 | 60 | 61.9 | 423 | 16 | 3.8 | 103 | 14 | 13.6 | 116 | 24 | 20.7 |
| DNA_Outside | 97 | 49 | 50.5 | 103 | 61 | 59.2 | 462 | 13 | 2.8 | 110 | 17 | 15.5 | 132 | 21 | 15.9 |
| No. of Unique Genes | 97 | 55 | 56.7 | 104 | 65 | 62.5 | 467 | 25 | 7.3 | 110 | 21 | 29.2 | 132 | 31 | 33 |
| TFs | ||||||
| H_L | L_H | M_M | H_M/M_L | L_M/H_M | Sum | |
| Surrounding_regions | 24 | 43 | 11 | 5 | 14 | 97 |
| Introns | 33 | 83 | 0 | 2 | 7 | 125 |
| Genes | ||||||
| Surrounding_regions | 11 | 22 | 1 | 3 | 6 | 43 |
| Introns | 11 | 30 | 0 | 1 | 5 | 47 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alnajjar, M.; Fekete, Z.; Nagy, T.; Német, Z.; Sakif, A.; Ninausz, N.; Fehér, P.; Stéger, V.; Barta, E. A New Family-Based Approach for Detecting Allele-Specific Expression and for Mapping Possible eQTLs. Animals 2025, 15, 2766. https://doi.org/10.3390/ani15182766
Alnajjar M, Fekete Z, Nagy T, Német Z, Sakif A, Ninausz N, Fehér P, Stéger V, Barta E. A New Family-Based Approach for Detecting Allele-Specific Expression and for Mapping Possible eQTLs. Animals. 2025; 15(18):2766. https://doi.org/10.3390/ani15182766
Chicago/Turabian StyleAlnajjar, Maher, Zsófia Fekete, Tibor Nagy, Zoltán Német, Agshin Sakif, Nóra Ninausz, Péter Fehér, Viktor Stéger, and Endre Barta. 2025. "A New Family-Based Approach for Detecting Allele-Specific Expression and for Mapping Possible eQTLs" Animals 15, no. 18: 2766. https://doi.org/10.3390/ani15182766
APA StyleAlnajjar, M., Fekete, Z., Nagy, T., Német, Z., Sakif, A., Ninausz, N., Fehér, P., Stéger, V., & Barta, E. (2025). A New Family-Based Approach for Detecting Allele-Specific Expression and for Mapping Possible eQTLs. Animals, 15(18), 2766. https://doi.org/10.3390/ani15182766






