Genetic Diversity and Core Germplasm Identification in Penaeus japonicus Using Whole-Genome Resequencing
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
2.2. Growth Performance Analysis
2.3. Sequencing and Sequence Alignment
2.4. Mutation Detection and Structural Annotation
2.5. Analysis of Population Genetics
2.6. Core Germplasm Analysis
3. Results
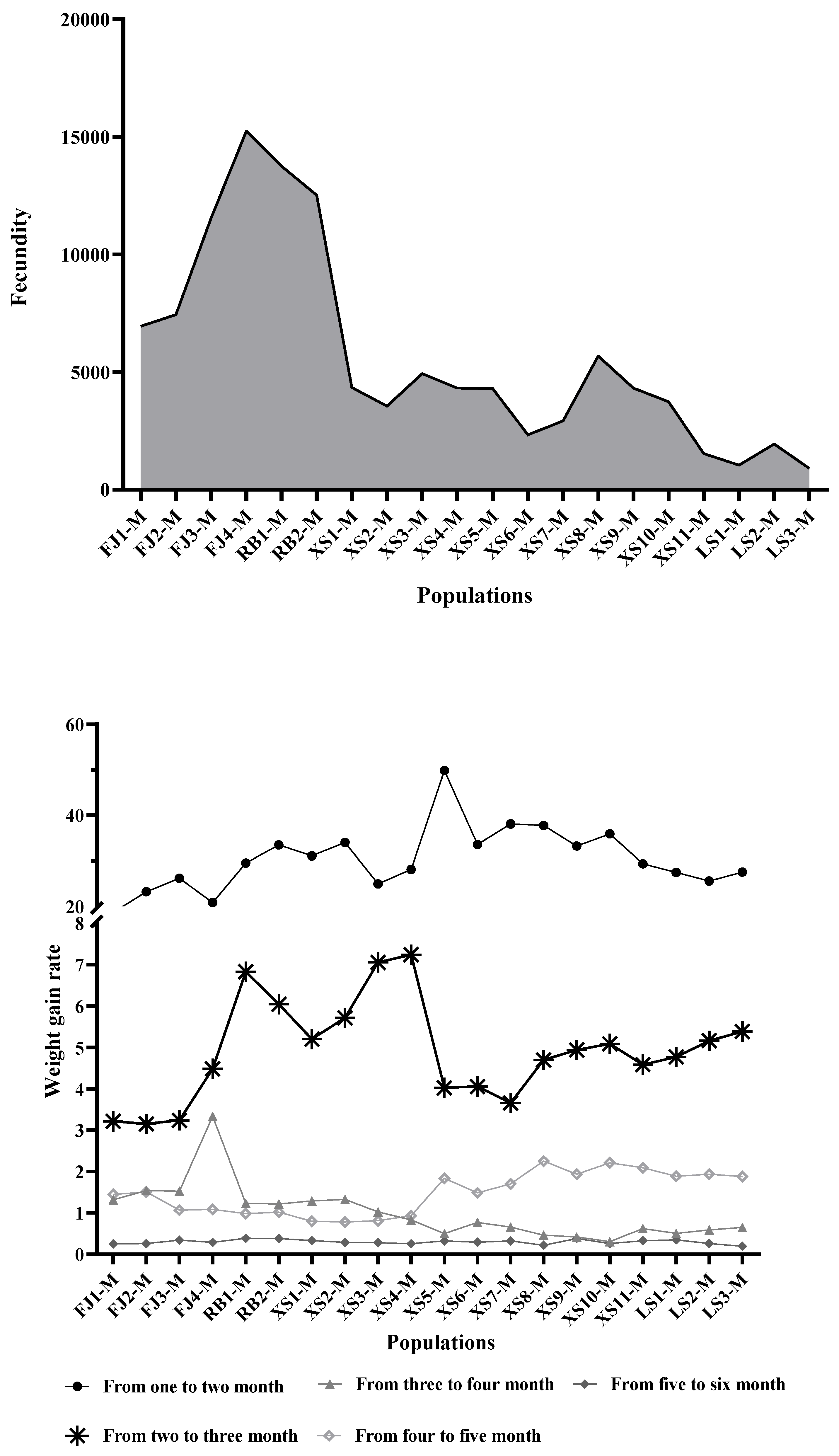
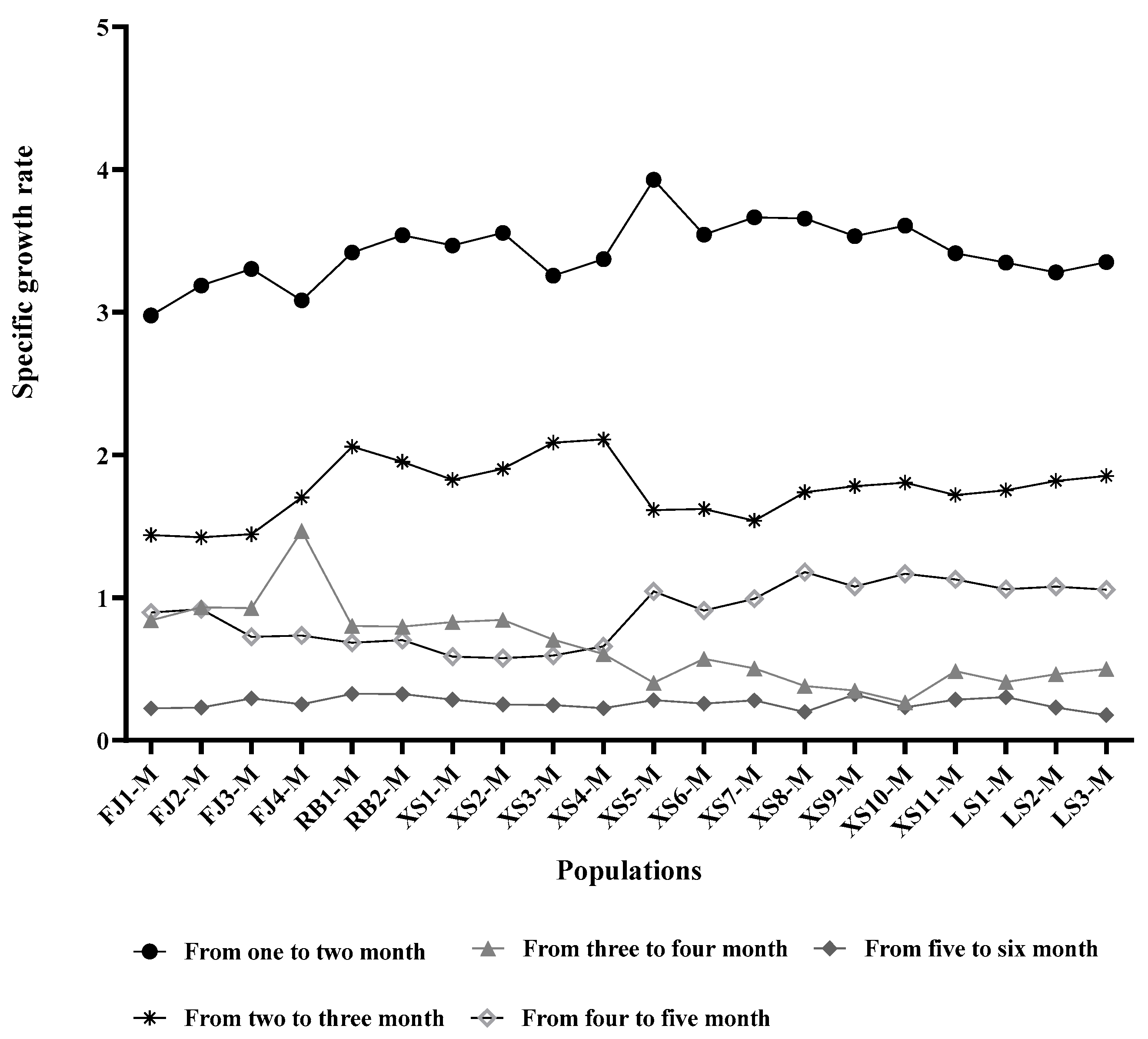
3.1. Analysis of Growth Performance Results
3.2. Quality Control and Evaluation of Data, and Comparison with Reference Genome
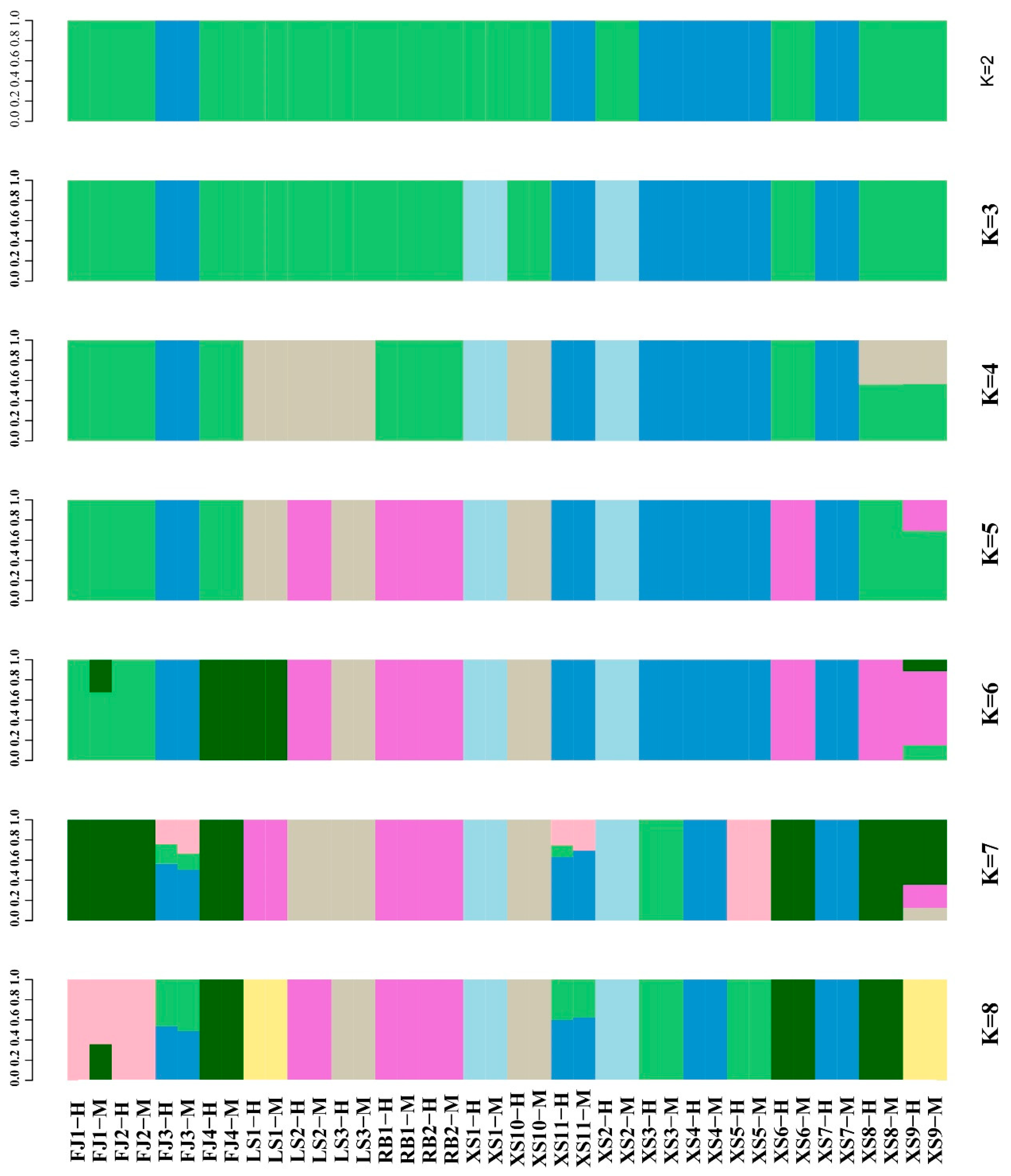
3.3. Genetic Structure
3.4. Localization Analysis, Detection, and Annotation of SNPs
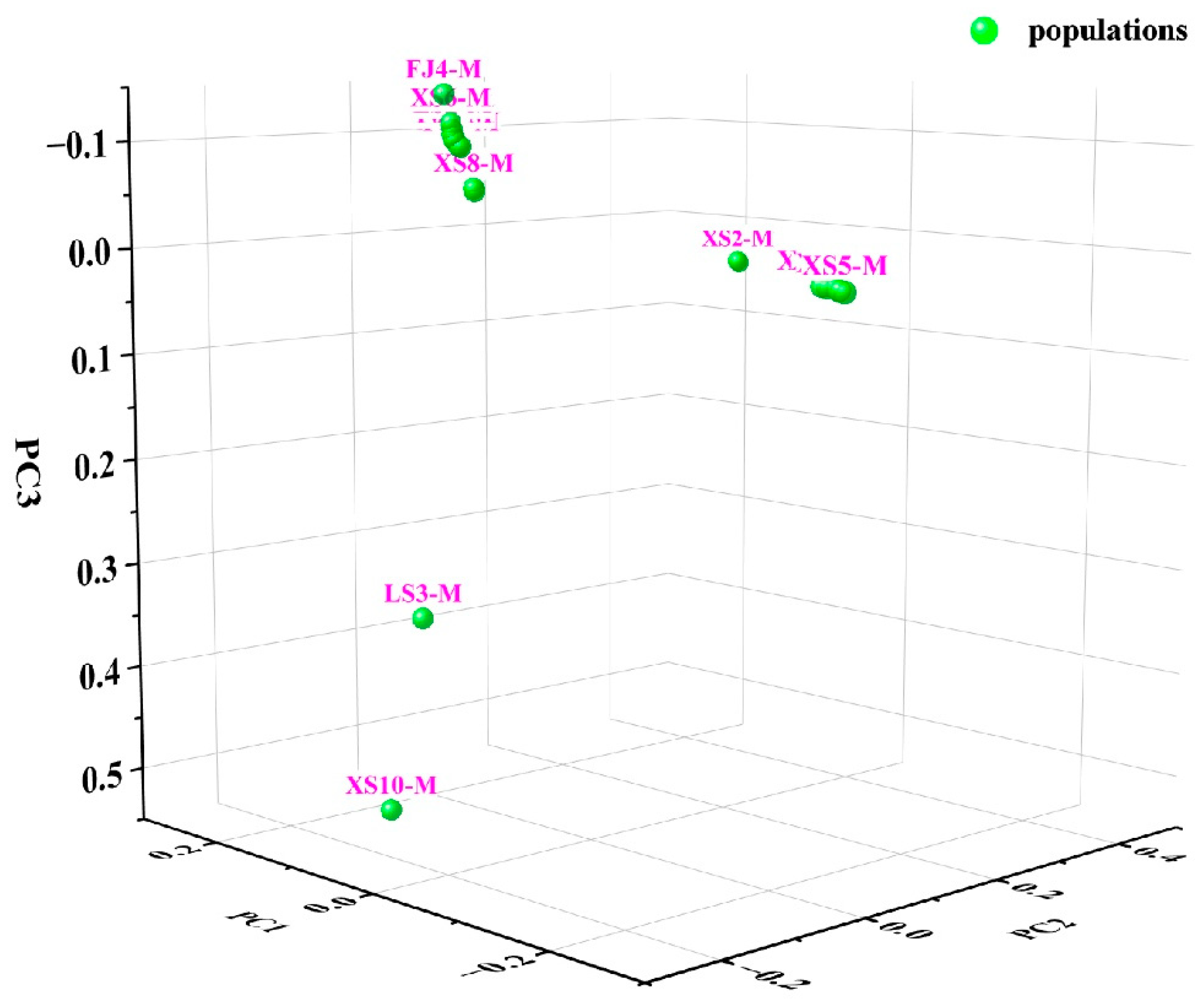
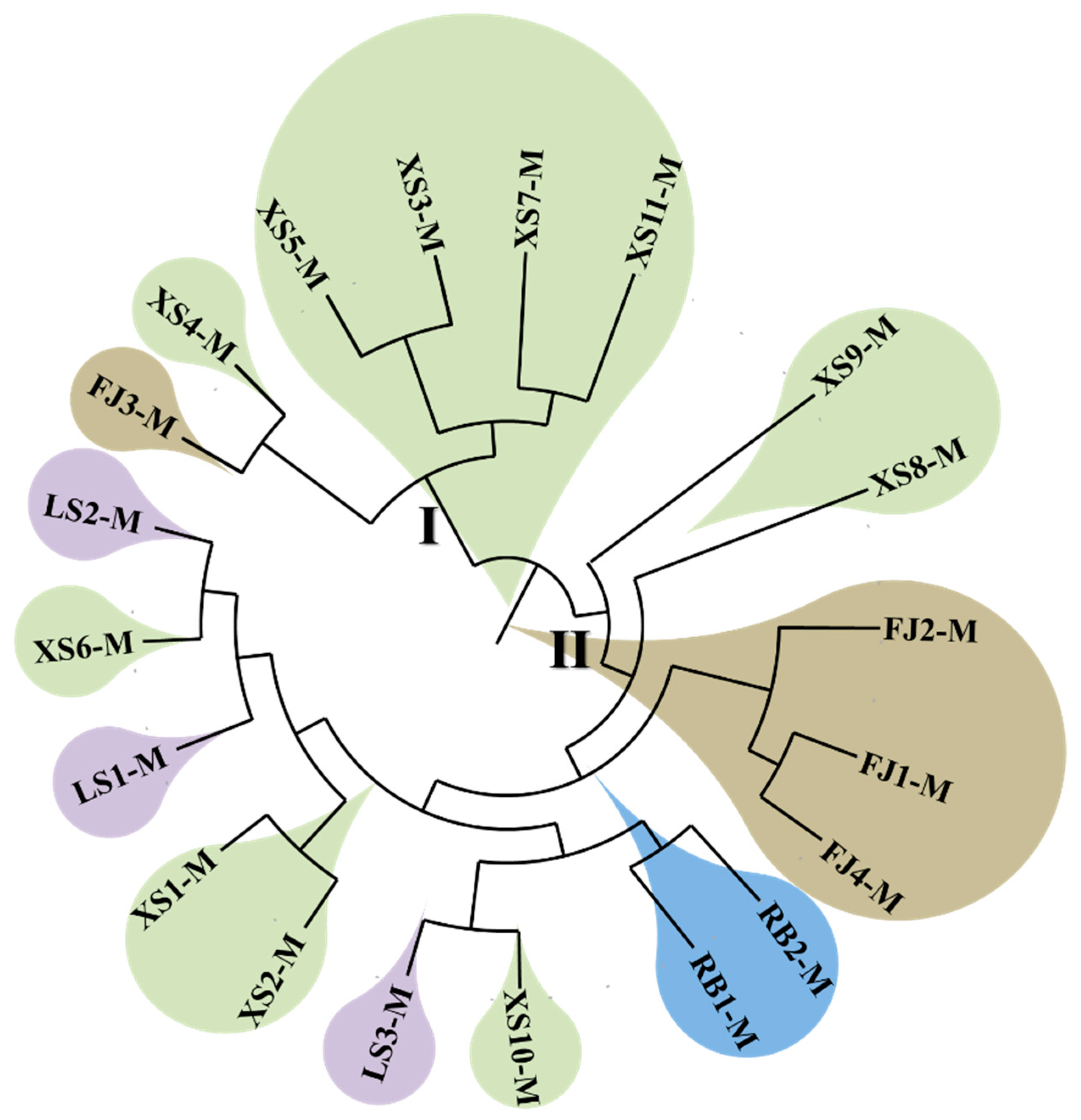
3.5. Construction of the Phylogenetic Tree
3.6. The Construction and Evaluation of Core Germplasm
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statements
Conflicts of Interest
References
- FAO. The State of World Fisheries and Aquaculture 2024; FAO: Rome, Italy, 2024; pp. 1–264. [Google Scholar]
- Zhang, X.; Jiao, L.; Li, M.; Ren, F.; Tao, X.; Jin, M.; Zhang, L.; Liu, W.; Zhou, Q. Effects of different dietary carbohydrate-to-protein ratios on the growth performance, antioxidant capacity and energy metabolism in Marsupenaeus japonicus. Aquac. Rep. 2023, 30, 101626. [Google Scholar] [CrossRef]
- Kaikkolante, N.; Katneni, V.K.; Palliyath, G.K.; Jangam, A.K.; Syamadayal, J.; Krishnan, K.; Prabhudas, S.K.; Shekhar, M.S. Computational insights into host-pathogen protein interactions: Unveiling penaeid shrimp and white spot syndrome virus interplay. Mol. Genet. Genom. 2025, 300, 35. [Google Scholar] [CrossRef]
- Huang, X.; Zhao, M.; Huang, J.; Liu, M.; Yang, L.; Wang, C. DRAM2 inhibits white spot syndrome virus infection via activating autophagy in Penaeus vannamei. Fish Shellfish Immunol. 2025, 160, 110240. [Google Scholar] [CrossRef]
- Shih, C.-H.; Haung, H.-L.; Chu, T.-J.; Lee, Y.-C.; Wang, C.-M.; Tzeng, T.-D. Genetic diversity and historical demography of kuruma shrimp (Penaeus japonicus) species complex off China based on mitochondrial DNA analysis. Afr. J. Biotechnol. 2011, 10, 1065–1072. [Google Scholar]
- Tsoi, K.H. Molecular Population Structure of the Kuruma Shrimp Panaeus japonicus in Western Pacific. Ph.D. Thesis, The Chinese University of Hong Kong, Hong Kong, China, 2006. [Google Scholar]
- Ellegren, H.; Galtier, N. Determinants of genetic diversity. Nat. Rev. Genet. 2016, 17, 422–433. [Google Scholar] [CrossRef] [PubMed]
- Frankham, R. Quantitative genetics in conservation biology. Genet. Res. 1999, 74, 237–244. [Google Scholar] [CrossRef]
- Vinay, T.N.; Raymond, J.A.J.; Katneni, V.K.; Aravind, R.; Balasubramanian, C.P.; Jayachandran, K.V.; Shekhar, M.S.; Vijayan, K.K. Mitochondrial DNA Study Reveals the Cryptic Species Penaeus japonicus (form-II) in Indian Waters. J. Coast. Res. 2019, 86, 149–155. [Google Scholar] [CrossRef]
- Zhang, M.; Zhao, Y.; Shan, B.; Liu, Y.; Yang, C.; Wang, L.; Liu, M.; Xie, Q.; Li, Y.; Zou, J.; et al. Microsatellite-Marker-Based Evaluation of Stock Enhancement for Kuruma Prawn Pernaeus japonicus in Beibu Gulf, South China Sea. Fishes 2023, 8, 568. [Google Scholar] [CrossRef]
- Soares, P.E.T.; Dantas, M.D.A.; Silva-Portela, R.d.C.B.; Agnez-Lima, L.F.; Lanza, D.C.F. Characterization of Penaeus vannamei mitogenome focusing on genetic diversity. PLoS ONE 2021, 16, e0255291. [Google Scholar] [CrossRef]
- Azad, M.; Yahyavi, M.; Bahri, A.; Jahromi, T.S.; Tala, M. Genetic identification of Penaeus vannamei broodstocks in Hormozgan and Bushehr Provinces using COI mitochondrial gene. Iran. J. Fish. Sci. 2021, 20, 209–217. [Google Scholar]
- Lu, Y.; Li, M.; Gao, Z.; Ma, H.; Chong, Y.; Hong, J.; Wu, J.; Wu, D.; Xi, D.; Deng, W. Advances in whole genome sequencing: Methods, tools, and applications in population genomics. Int. J. Mol. Sci. 2025, 26, 372. [Google Scholar] [CrossRef]
- Wang, H.; Teng, M.; Liu, P.; Zhao, M.; Wang, S.; Hu, J.; Bao, Z.; Zeng, Q. Selection Signatures of Pacific White Shrimp Litopenaeus vannamei Revealed by Whole-Genome Resequencing Analysis. Front. Mar. Sci. 2022, 9, 844597. [Google Scholar] [CrossRef]
- Bao, Z.; Yu, Y.; Lin, P.; Li, F. Identification of growth-related genes based on BSA in Pacific white shrimp Litopenaeus vannamei. Aquaculture 2025, 596, 741708. [Google Scholar] [CrossRef]
- Sui, J.; Sun, K.; Kong, J.; Tan, J.; Dai, P.; Cao, J.; Luo, K.; Luan, S.; Xing, Q.; Meng, X. Estimation of Genetic Parameters for Growth and WSSV Resistance Traits in Litopenaeus vannamei. Animals 2024, 14, 1817. [Google Scholar] [CrossRef]
- Ceng, F.R. Studies on Genetic Diversity of Marsupenaeus japonicus and Melocular Phylogenetics in Penaeidae. Master’s Thesis, Xiamen University, Xiamen, China, 2010. [Google Scholar]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Der-Auwera, G.A.V.; Carneiro, M.; Hartl, C.; Poplin, R.; Thibault, J. From FastQ data to high confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinform. 2013, 43, 11.10.1–11.10.33. [Google Scholar] [CrossRef]
- Abyzov, A.; Urban, A.; Snyder, M.; Gerstein, M. CNVnator: An approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 2011, 21, 974–984. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- Cingolani, P. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Vilella, A.J.; Severin, J.; Ureta-Vidal, A.; Heng, L.; Durbin, R.; Birney, E. EnsemblCompara GeneTrees: Complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 2009, 19, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Price, A.; Patterson, N.; Plenge, R.; Weinblatt, M.; Shadick, N.; Reich, D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006, 38, 904–909. [Google Scholar] [CrossRef]
- Yang, J.; Lee, S.; Goddard, M.; Visscher, P. GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.; Banks, E.; Depristo, M. The variant call format and vcftools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Chakraborty, R.; Weiss, K. Admixture as a tool for finding linked genes and detecting that difference from allelic association between loci. Proc. Natl. Acad. Sci. USA 1988, 85, 9119–9123. [Google Scholar] [CrossRef]
- Thachuk, C.; José, C.; Franco, J.; Dreisigacker, S.; Warburton, M.; Davenport, G. Core Hunter: An algorithm for sampling genetic resources based on multiple genetic measures. BMC Bioinform. 2009, 10, 203. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Zhuang, X.; Wu, L.; Lin, H.; Li, Y.; Wu, L.; Yao, J.; Liu, J.; Ding, S. Assessing the population genetic structure of yellow croaker in China: Insights into the ecological and genetic consequences of artificial breeding on natural populations. Aquaculture 2024, 590, 741026. [Google Scholar] [CrossRef]
- Gjedrem, T.; Baranski, M. Selective Breeding in Aquaculture: An Introduction; Springer: Dordrecht, The Netherlands, 2009; pp. 151–176. [Google Scholar]
- Jiraporn, R.; Ikuo, H.; Toshiaki, I.; Yukinori, T.; Takashi, A. Gene expression in haemocytes of kuruma prawn, Penaeus japonicus, in response to infection with WSSV by EST approach. Fish Shellfish Immunol. 2002, 13, 69–83. [Google Scholar]
- Ryosuke, O.; Teruyoshi, H.; Puttirungroj, P.; Kenichi, O.; Ikuo, H. Estimating the basic reproduction number and final epidemic size of white spot syndrome virus outbreak in Penaeus japonicus in aquaculture ponds. Aquaculture 2024, 582, 740548. [Google Scholar] [CrossRef]
- Cui, X.; Deng, J.; Zhang, Y.; Han, Y.; Ou, M.; Sun, Y. Investigation of genetic diversity in the loach Misgurnus anguillicaudatus revealed by whole-genome resequencing. BMC Genom. 2024, 25, 1126. [Google Scholar] [CrossRef]
- You, E.; Chiu, T.; Liu, K.; Tassanakajon, A.; Klinbunga, S.; Triwitayakorn, K.; de la Peña, L.D.; Li, Y.; Yu, H. Microsatellite and mitochondrial haplotype diversity reveals population differentiation in the tiger shrimp (Penaeus monodon) in the Indo-Pacific region. Anim. Genet. 2008, 39, 267–277. [Google Scholar] [CrossRef]
- Bo, Q.; Yu, Y.; Chen, C.; Zhang, Y.; Ma, C.; Wang, S.; Yu, J.; Lu, Y.; Hao, J.; Jiang, J. Genetic diversity losses in mantis shrimp Oratosquilla oratoria offspring created for supportive releases using an ecological breeding in earthen pond method. Mar. Ecol. 2024, 45, e12842. [Google Scholar] [CrossRef]
- Frankel, O. Genetic Perspectives of Germplasm Conservation; Cambridge University Press: Cambridge, UK, 1984; pp. 161–170. [Google Scholar]
- Zhao, J.; Tong, Y.; Ge, T.; Ge, J. Genetic diversity estimation and core collection construction of Sinojackia huangmeiensis based on novel microsatellite markers. Biochem. Syst. Ecol. 2016, 64, 74–80. [Google Scholar] [CrossRef]
- Mir, A.; Bhat, M.; Fayaz, H.; Wani, A.; Dar, S.; Maqbool, S.; Yasin, M.; Mir, J.; Khan, M.; Sofi, P. SSR markers in revealing extent of genetic diversity and phylogenetic relationships among chickpea core collection accessions for Western Himalayas. Mol. Biol. Rep. 2022, 49, 11469–11479. [Google Scholar] [CrossRef]
- Chabane, K.; Valkoun, J. Characterisation of genetic diversity in ICARDA core collection of cultivated barley. Czech J. Genet. Plant Breed. 2018, 40, 134–136. [Google Scholar] [CrossRef]
- Wu, S.; Zeng, Q.; Han, W.; Wang, M.; Ding, H.; Teng, M.; Wang, M.; Li, P.; Gao, X.; Bao, Z. Deciphering the population structure and genetic basis of growth traits from whole-genome resequencing of the leopard coral grouper (Plectropomus leopardus). Zool. Res. 2024, 45, 329–340. [Google Scholar] [CrossRef] [PubMed]
- Garcia, A.; Tsuruta, S.; Gao, G.; Palti, Y.; Lourenco, D.; Leeds, T. Genomic selection models substantially improve the accuracy of genetic merit predictions for fillet yield and body weight in rainbow trout using a multi-trait model and multi-generation progeny testing. Genet. Sel. Evol. 2023, 55, 11. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Dong, T.; Yan, X.; Wang, W.; Tian, Z.; Hu, H. Using Bayesian threshold model and machine learning method to improve the accuracy of genomic prediction for ordered categorical traits in fish. Agric. Commun. 2023, 1, 100005. [Google Scholar] [CrossRef]
- Garcia, B.F.; Caceres, P.A.; Marin-Nahuelpi, R.; Lopez, P.; Cichero, D.; Odegard, J.; Moen, T.; Yanez, J.M. Prioritized imputed sequence variants from multi-population GWAS improve prediction accuracy for sea lice count in Atlantic salmon (Salmo salar). Aquaculture 2024, 581, 740422. [Google Scholar] [CrossRef]
| Serial Number | Sample Name | Sample Source |
|---|---|---|
| 1 | FJ1-M | Fujian, China |
| 2 | FJ2-M | Fujian, China |
| 3 | FJ3-M | Fujian, China |
| 4 | FJ4-M | Fujian, China |
| 5 | RB1-M | Japan |
| 6 | RB2-M | Japan |
| 7 | XS1-M | Qifeng, Zhejiang, China |
| 8 | XS2-M | Qifeng, Zhejiang, China |
| 9 | XS3-M | Qifeng, Zhejiang, China |
| 10 | XS4-M | Qifeng, Zhejiang, China |
| 11 | XS5-M | Qifeng, Zhejiang, China |
| 12 | XS6-M | Qifeng, Zhejiang, China |
| 13 | XS7-M | Qifeng, Zhejiang, China |
| 14 | XS8-M | Qifeng, Zhejiang, China |
| 15 | XS9-M | Qifeng, Zhejiang, China |
| 16 | XS10-M | Qifeng, Zhejiang, China |
| 17 | XS11-M | Qifeng, Zhejiang, China |
| 18 | LS1-M | Shipu, Zhejiang, China |
| 19 | LS2-M | Shipu, Zhejiang, China |
| 20 | LS3-M | Shipu, Zhejiang, China |
| Category | Number of SNPs | |
|---|---|---|
| Upstream | 153,651 | |
| UTR3 | 122,233 | |
| UTR5 | 64,768 | |
| UTR5; UTR3 | 79 | |
| Exonic | Stop gain | 999 |
| Stop loss | 125 | |
| Synonymous | 306,354 | |
| Non-synonymous | 120,879 | |
| Unknown | 12,834 | |
| Intronic | 2,836,035 | |
| Splicing | 754 | |
| Downstream | 120,999 | |
| Upstream/downstream | 5744 | |
| Intergenic | 5,192,740 | |
| Other | 208,054 | |
| ts | 5,806,141 | |
| tv | 3,340,107 | |
| ts/tv | 1.738 | |
| Total | 9,146,248 | |
| Method | Core Collection | MR | MRmin | CE | CEmin | PN | CV |
|---|---|---|---|---|---|---|---|
| CoreHunter (20%) | 4 | 0.34 | 0.21 | 0.37 | 0.23 | 0.07 | 0.93 |
| Group Name | Na | Ne | PIC | Pi | HWB_Ratio | Ho | He | I |
|---|---|---|---|---|---|---|---|---|
| Core | 1.86 | 1.32 | 0.18 | 0.22 | 0.17 | 0.21 | 0.21 | 0.49 |
| ALL | 2 | 1.34 | 0.19 | 0.23 | 0.08 | 0.23 | 0.23 | 0.54 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, D.; Shentu, J.; Liu, W.; Wang, Y.; Zhu, M.; Yang, Z.; Si, L. Genetic Diversity and Core Germplasm Identification in Penaeus japonicus Using Whole-Genome Resequencing. Animals 2025, 15, 2759. https://doi.org/10.3390/ani15182759
Zhang D, Shentu J, Liu W, Wang Y, Zhu M, Yang Z, Si L. Genetic Diversity and Core Germplasm Identification in Penaeus japonicus Using Whole-Genome Resequencing. Animals. 2025; 15(18):2759. https://doi.org/10.3390/ani15182759
Chicago/Turabian StyleZhang, Dingyuan, Jikang Shentu, Weijian Liu, Yanxia Wang, Minjun Zhu, Zhiming Yang, and Liegang Si. 2025. "Genetic Diversity and Core Germplasm Identification in Penaeus japonicus Using Whole-Genome Resequencing" Animals 15, no. 18: 2759. https://doi.org/10.3390/ani15182759
APA StyleZhang, D., Shentu, J., Liu, W., Wang, Y., Zhu, M., Yang, Z., & Si, L. (2025). Genetic Diversity and Core Germplasm Identification in Penaeus japonicus Using Whole-Genome Resequencing. Animals, 15(18), 2759. https://doi.org/10.3390/ani15182759




