Development of a 5K Liquid-Phase Genome-Wide Breeding Chip for Xinglong Buffalo
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection and SNP Calling
2.2. Site Selection
2.3. Design and Synthesis of Probe
2.4. Functional Analysis of Liquid-Phase Chip Loci
2.5. DNA Extraction and Sequencing Library Construction
2.6. Verification of Liquid-Phase Chip
2.7. Validation of Non-Synonymous Mutation Sites
2.8. Analysis of Breed and Kinship
3. Results
3.1. Identification of Functional and Specific Sites
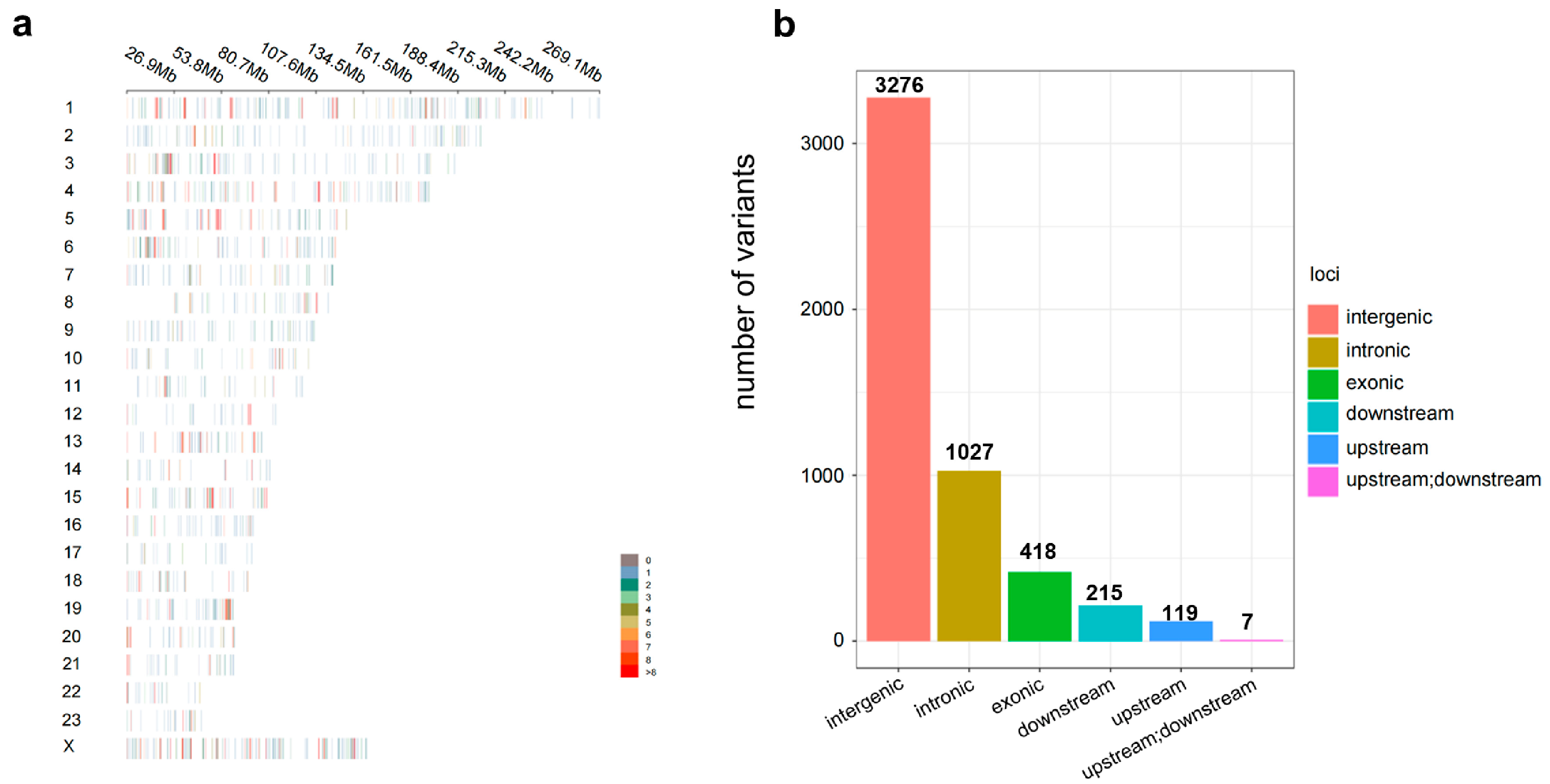
3.2. Analysis of SNP Loci on the 5K Liquid-Phase Chip
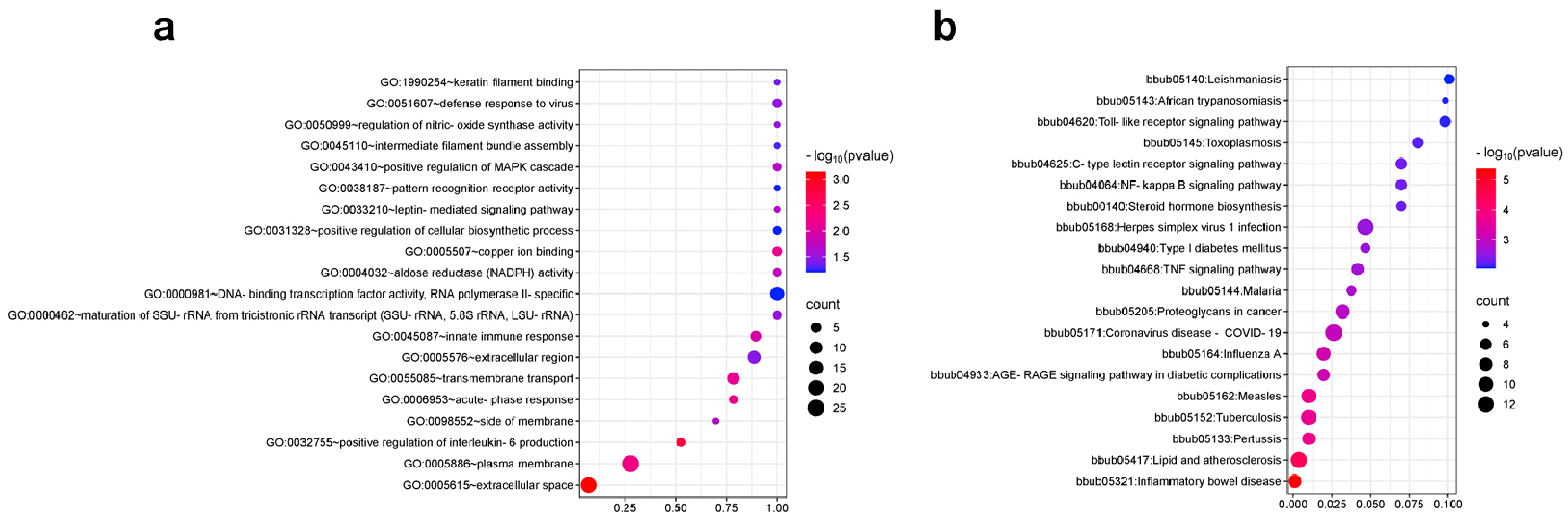
3.3. Functional Analysis of 5K Liquid-Phase Chip
3.4. Verification of 5K Liquid-Phase Chip
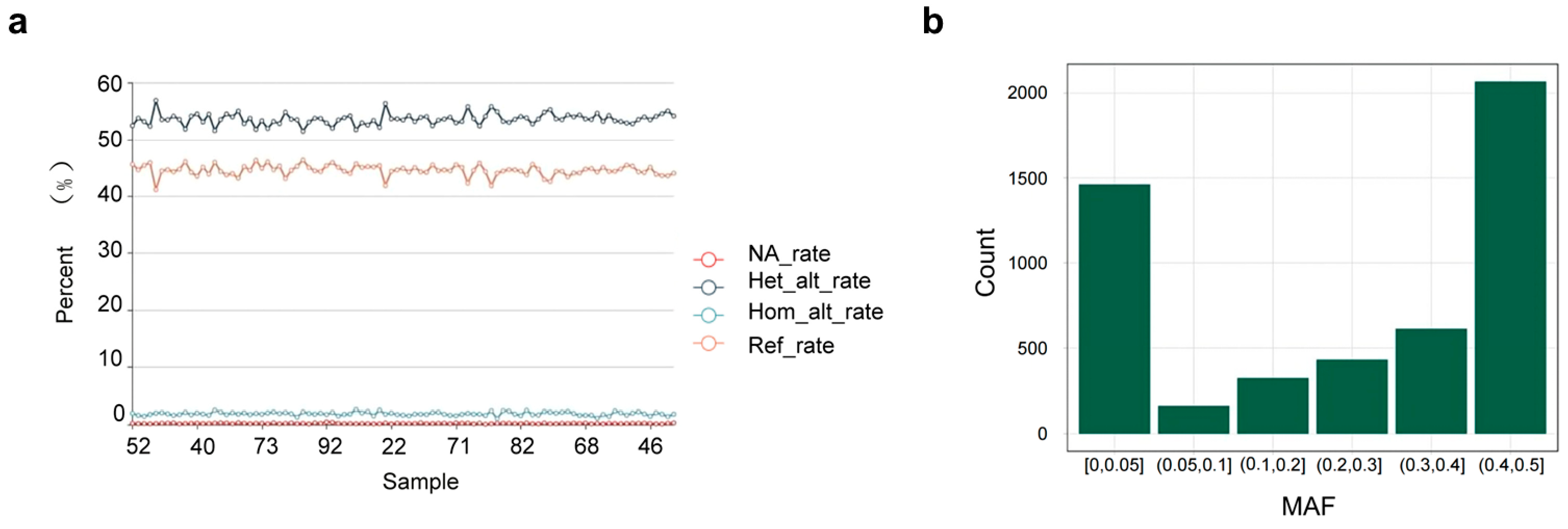
3.5. Consistency and Repeatability Analysis of the Chip
3.6. Validation of Non-Synonymous Mutation Sites of Liquid-Phase Chip
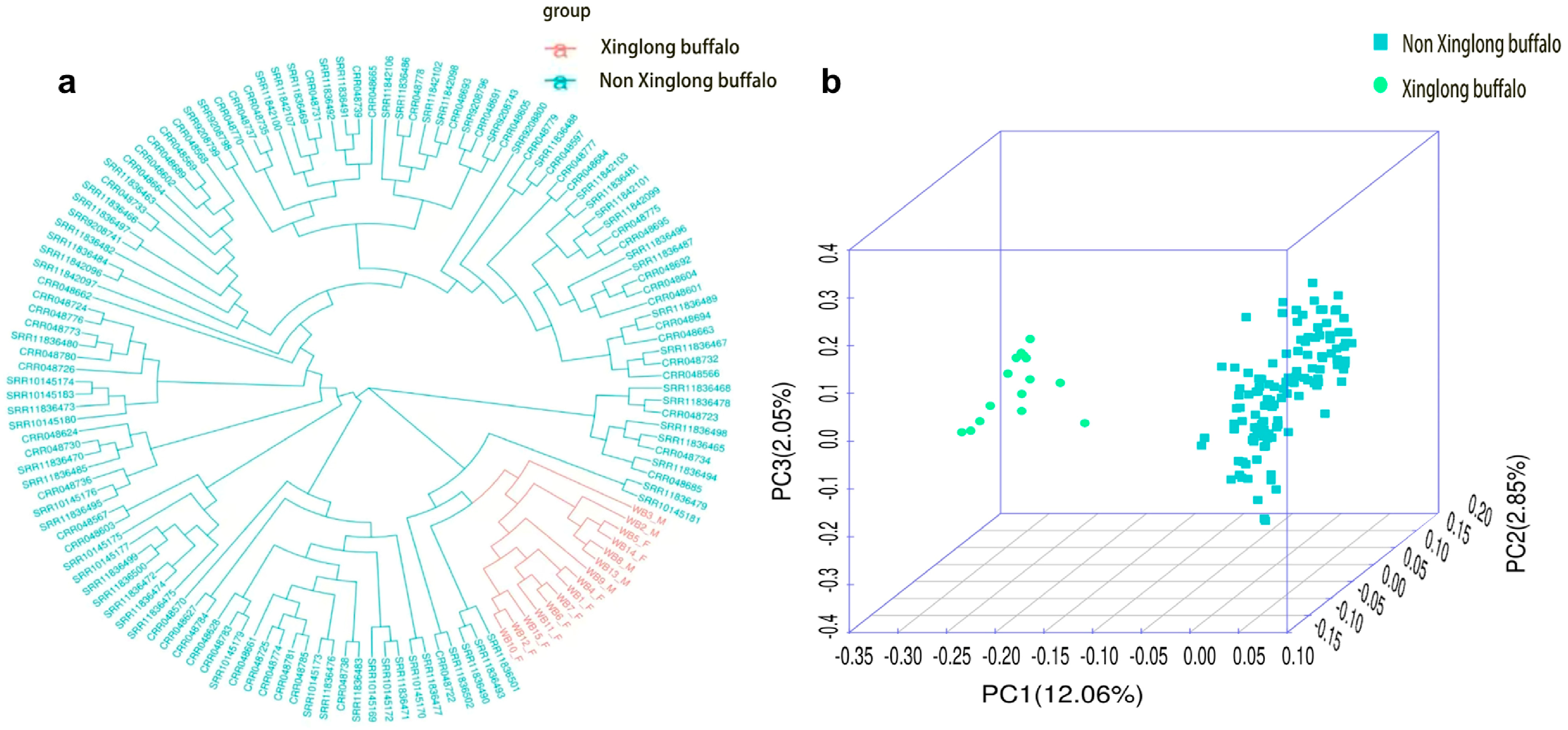
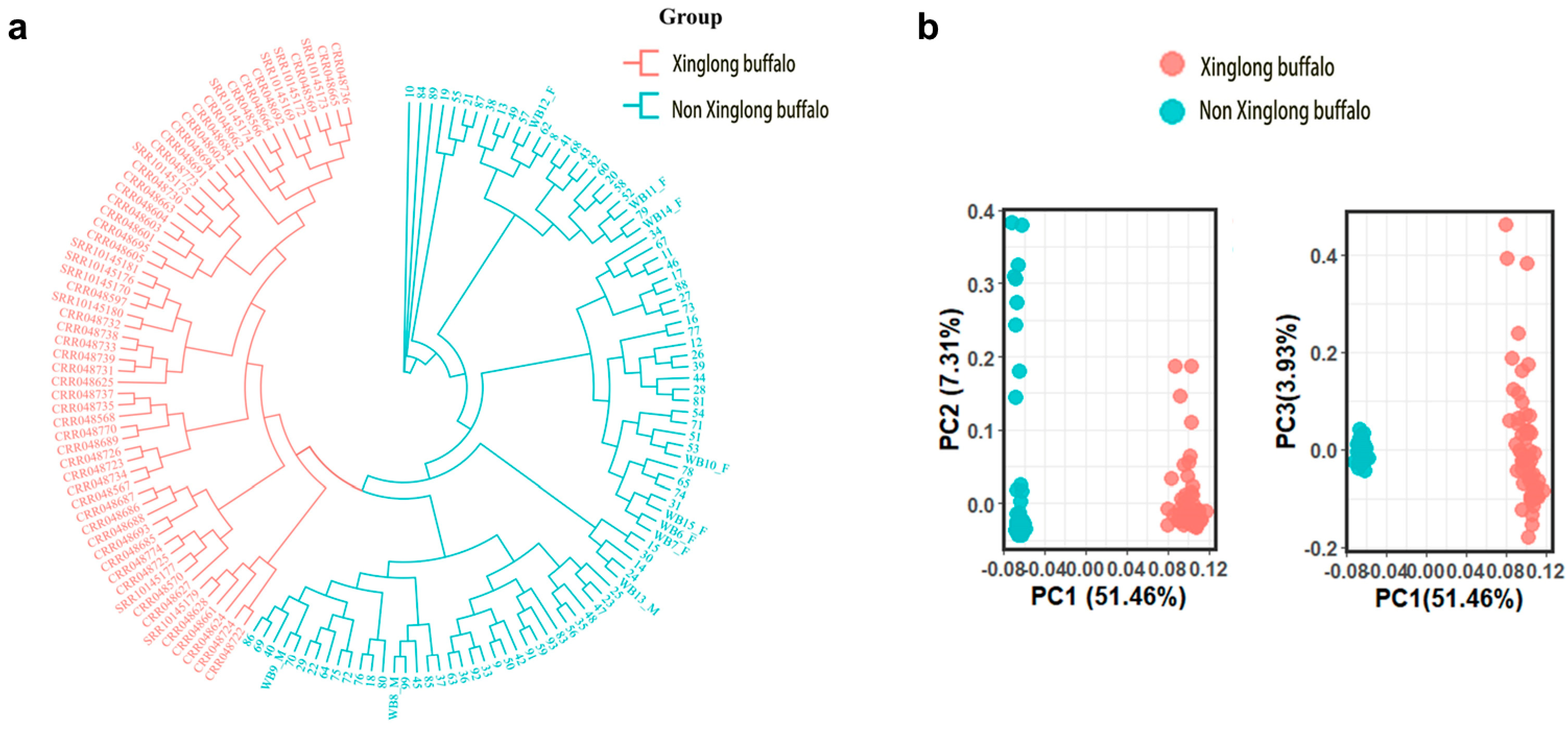
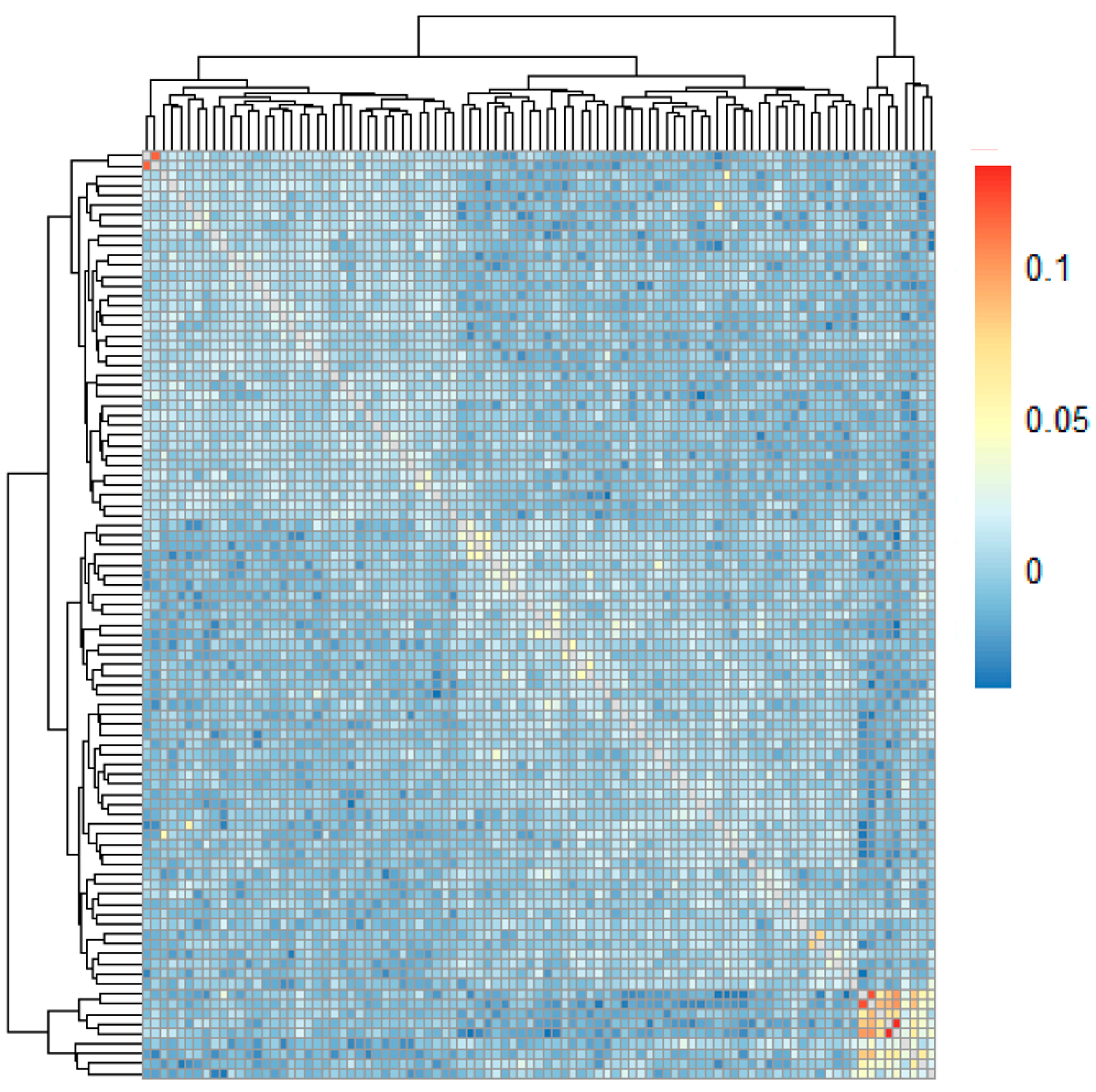
3.7. Analysis of Breed and Kinship Based on the Chip Data
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| SNP | Single nucleotide polymorphism |
| GBTS | Genotyping by target sequencing |
| MAF | Minor allele frequency |
| GO | Gene ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| PCA | Principal component analysis |
| PCR | Polymerase chain reaction |
References
- Zhang, Y.; Colli, L.; Barker, J.S.F. Asian water buffalo: Domestication, history and genetics. Anim. Genet. 2020, 51, 177–191. [Google Scholar] [CrossRef]
- Minervino, A.H.H.; Zava, M.; Vecchio, D.; Borghese, A. Bubalus bubalis: A Short Story. Front. Vet. Sci. 2020, 7, 570413. [Google Scholar] [CrossRef]
- Chai, Y.; Li, S.; Wu, H.; Meng, Y.; Fu, Y.; Li, H.; Wu, G.; Jiang, J.; Chen, T.; Jiao, Y.; et al. The genome landscape of the Xinglong buffalo. BMC Genom. 2024, 25, 1054. [Google Scholar] [CrossRef]
- Wu, G.; Qiu, X.; Jiao, Z.; Yang, W.; Pan, H.; Li, H.; Bian, Z.; Geng, Q.; Wu, H.; Jiang, J.; et al. Integrated Analysis of Transcriptome and Metabolome Profiles in the Longissimus Dorsi Muscle of Buffalo and Cattle. Curr. Issues. Mol. Biol. 2023, 45, 9723–9736. [Google Scholar] [CrossRef]
- Vignal, A.; Milan, D.; SanCristobal, M.; Eggen, A. A review on SNP and other types of molecular markers and their use in animal genetics. Genet. Sel. Evol. 2002, 34, 275–305. [Google Scholar] [CrossRef]
- Tosser-Klopp, G.; Bardou, P.; Bouchez, O.; Cabau, C.; Crooijmans, R.; Dong, Y.; Donnadieu-Tonon, C.; Eggen, A.; Heuven, H.C.; Jamli, S.; et al. Design and characterization of a 52K SNP chip for goats. PLoS ONE 2014, 9, e86227. [Google Scholar] [CrossRef]
- Zhang, T.; Guo, L.; Shi, M.; Xu, L.; Chen, Y.; Zhang, L.; Gao, H.; Li, J.; Gao, X. Selection and effectiveness of informative SNPs for paternity in Chinese Simmental cattle based on a high-density SNP array. Gene 2018, 673, 211–216. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Tang, D.; Ni, J.; Li, P.; Wang, L.; Zhou, J.; Li, C.; Lan, H.; Li, L.; Liu, J. Development of genic KASP SNP markers from RNA-Seq data for map-based cloning and marker-assisted selection in maize. BMC Plant Biol. 2021, 21, 157. [Google Scholar] [CrossRef] [PubMed]
- Druet, T.; Schrooten, C.; de Roos, A.P. Imputation of genotypes from different single nucleotide polymorphism panels in dairy cattle. J. Dairy Sci. 2010, 93, 5443–5454. [Google Scholar] [CrossRef]
- Xu, Y.; Yang, Q.; Zheng, H.; Xu, Y.; Sang, Z.; Guo, Z.; Peng, H.; Zhang, C.; Lan, H.; Wang, Y.; et al. Genotyping by target sequencing (GBTS) and its applications. Sci. Agric. Sin. 2020, 53, 2983–3004. [Google Scholar]
- Wu, H.C.; Chiu, Y.T.; Wu, I.C.; Liou, C.H.; Cheng, H.W.; Kuo, S.C.; Lauderdale, T.L.; Sytwu, H.K.; Liao, Y.C.; Chen, F.J. Streamlining whole genome sequencing for clinical diagnostics with ONT technology. Sci. Rep. 2025, 15, 6270. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Xiang, M.; Wang, X.; Li, J.; Cheng, X.; Li, H.; Singh, R.P.; Bhavani, S.; Huang, S.; Zheng, W.; et al. Development and application of the GenoBaits WheatSNP16K array to accelerate wheat genetic research and breeding. Plant Commun. 2025, 6, 101138. [Google Scholar] [CrossRef]
- Guan, S.; Li, W.; Jin, H.; Zhang, L.; Liu, G. Development and Validation of a 54K Genome-Wide Liquid SNP Chip Panel by Target Sequencing for Dairy Goat. Genes 2023, 14, 1122. [Google Scholar] [CrossRef]
- Guo, Y.; Bai, F.; Wang, J.; Fu, S.; Zhang, Y.; Liu, X.; Zhang, Z.; Shao, J.; Li, R.; Wang, F.; et al. Design and characterization of a high-resolution multiple-SNP capture array by target sequencing for sheep. J. Anim. Sci. 2023, 101, skac383. [Google Scholar] [CrossRef]
- Wang, H.; Wu, H.; Zhang, W.; Jiang, J.; Qian, H.; Man, C.; Gao, H.; Chen, Q.; Du, L.; Chen, S.; et al. Development and validation of a 5K low-density SNP chip for Hainan cattle. BMC Genom. 2024, 25, 873. [Google Scholar] [CrossRef]
- Han, J. Efficient Excavation of Functional Change Sites in Water Buffalo and Development of SNP Breeding Chips. Master’s Thesis, Huazhong Agricultural University, Wuhan, China, 2023. [Google Scholar]
- de Camargo, G.M.; Aspilcueta-Borquis, R.R.; Fortes, M.R.; Porto-Neto, R.; Cardoso, D.F.; Santos, D.J.; Lehnert, S.A.; Reverter, A.; Moore, S.S.; Tonhati, H. Prospecting major genes in dairy buffaloes. BMC Genom. 2015, 16, 872. [Google Scholar] [CrossRef]
- Iamartino, D.; Nicolazzi, E.L.; Van Tassell, C.P.; Reecy, J.M.; Fritz-Waters, E.R.; Koltes, J.E.; Biffani, S.; Sonstegard, T.S.; Schroeder, S.G.; Ajmone-Marsan, P.; et al. Design and validation of a 90K SNP genotyping assay for the water buffalo (Bubalus bubalis). PLoS ONE 2017, 12, e0185220. [Google Scholar] [CrossRef]
- Luo, X.; Zhou, Y.; Zhang, B.; Zhang, Y.; Wang, X.; Feng, T.; Li, Z.; Cui, K.; Wang, Z.; Luo, C.; et al. Understanding divergent domestication traits from the whole-genome sequencing of swamp- and river-buffalo populations. Natl. Sci. Rev. 2020, 7, 686–701. [Google Scholar] [CrossRef] [PubMed]
- Liang, D.; Zhao, P.; Si, J.; Fang, L.; Pairo-Castineira, E.; Hu, X.; Xu, Q.; Hou, Y.; Gong, Y.; Liang, Z.; et al. Genomic Analysis Revealed a Convergent Evolution of LINE-1 in Coat Color: A Case Study in Water Buffaloes (Bubalus bubalis). Mol. Biol. Evol. 2021, 38, 1122–1136. [Google Scholar] [CrossRef]
- Sun, T.; Huang, G.Y.; Wang, Z.H.; Teng, S.H.; Cao, Y.H.; Sun, J.L.; Hanif, Q.; Chen, N.B.; Lei, C.Z.; Liao, Y.Y. Selection signatures of Fuzhong Buffalo based on whole-genome sequences. BMC Genom. 2020, 21, 674. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Shen, J.; Achilli, A.; Chen, N.; Chen, Q.; Dang, R.; Zheng, Z.; Zhang, H.; Zhang, X.; Wang, S.; et al. Genomic analyses reveal distinct genetic architectures and selective pressures in buffaloes. Gigascience 2020, 9, giz166. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Vasimuddin, M.; Misra, S.; Li, H.; Aluru, S. Efficient Architecture-Aware Acceleration of BWA-MEM for Multicore Systems. In Proceedings of the 2019 IEEE International Parallel and Distributed Processing Symposium (IPDPS), Rio de Janeiro, Brazil, 20–24 May 2019; pp. 314–324. [Google Scholar]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic. Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Greene, S.A. Protocols for anesthesia of cattle. Vet. Clin. N. Am. Food. Anim. Pract. 2003, 19, 679–693, vii. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Wang, T.; Li, Y.; Liu, H.; Dong, Y.; Zhang, R.; Wang, H.; Shang, L.; Xing, X. Development and validation of a 1 K sika deer (Cervus nippon) SNP Chip. BMC Genom. Data 2021, 22, 35. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Patterson, N.; Price, A.L.; Reich, D. Population structure and eigenanalysis. PLoS Genet. 2006, 2, e190. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Ginestet, C. ggplot2: Elegant Graphics for Data Analysis: Book Reviews. J. R. Stat. Soc. Ser. A (Stat. Soc.) 2013, 174, 245–256. [Google Scholar] [CrossRef]
- Xie, J.; Chen, Y.; Cai, G.; Cai, R.; Hu, Z.; Wang, H. Tree Visualization by One Table (tvBOT): A web application for visualizing, modifying and annotating phylogenetic trees. Nucleic. Acids Res. 2023, 51, W587–W592. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Gellatly, S.A.; Kalujnaia, S.; Cramb, G. Cloning, tissue distribution and sub-cellular localisation of phospholipase C X-domain containing protein (PLCXD) isoforms. Biochem. Biophys. Res. Commun. 2012, 424, 651–656. [Google Scholar] [CrossRef] [PubMed]
- Ji, M.; Zhao, Z.; Li, Y.; Xu, P.; Shi, J.; Li, Z.; Wang, K.; Huang, X.; Ji, J.; Liu, W.; et al. FBXO16-mediated hnRNPL ubiquitination and degradation plays a tumor suppressor role in ovarian cancer. Cell Death Dis. 2021, 12, 758. [Google Scholar] [CrossRef]
- Khan, M.; Muzumdar, D.; Shiras, A. Attenuation of Tumor Suppressive Function of FBXO16 Ubiquitin Ligase Activates Wnt Signaling in Glioblastoma. Neoplasia 2019, 21, 106–116. [Google Scholar] [CrossRef]
- Paul, D.; Islam, S.; Manne, R.K.; Dinesh, U.S.; Malonia, S.K.; Maity, B.; Boppana, R.; Rapole, S.; Shetty, P.K.; Santra, M.K. F-box protein FBXO16 functions as a tumor suppressor by attenuating nuclear β-catenin function. J. Pathol. 2019, 248, 266–279. [Google Scholar] [CrossRef] [PubMed]
- Geraldes, A.; Difazio, S.P.; Slavov, G.T.; Ranjan, P.; Muchero, W.; Hannemann, J.; Gunter, L.E.; Wymore, A.M.; Grassa, C.J.; Farzaneh, N.; et al. A 34K SNP genotyping array for Populus trichocarpa: Design, application to the study of natural populations and transferability to other Populus species. Mol. Ecol. Resour. 2013, 13, 306–323. [Google Scholar] [CrossRef]
- Liu, R.; Xing, S.; Wang, J.; Zheng, M.; Cui, H.; Crooijmans, R.; Li, Q.; Zhao, G.; Wen, J. A new chicken 55K SNP genotyping array. BMC Genom. 2019, 20, 410. [Google Scholar] [CrossRef]
- Bernard, M.; Dehaullon, A.; Gao, G.; Paul, K.; Lagarde, H.; Charles, M.; Prchal, M.; Danon, J.; Jaffrelo, L.; Poncet, C.; et al. Development of a High-Density 665 K SNP Array for Rainbow Trout Genome-Wide Genotyping. Front. Genet. 2022, 13, 941340. [Google Scholar] [CrossRef]
- Wang, J.; Xing, S.; Zhou, Y.; Liu, J.; Sun, Y.; Lei, Q.; Han, H.; Liu, W.; Li, D.; Li, F.; et al. Research note: A low-density SNP genotyping panel for Chinese native chickens. Poult. Sci. 2025, 104, 104609. [Google Scholar] [CrossRef]
- Matukumalli, L.K.; Lawley, C.T.; Schnabel, R.D.; Taylor, J.F.; Allan, M.F.; Heaton, M.P.; O’Connell, J.; Moore, S.S.; Smith, T.P.; Sonstegard, T.S.; et al. Development and characterization of a high density SNP genotyping assay for cattle. PLoS ONE 2009, 4, e5350. [Google Scholar] [CrossRef]
- Berner, D. Allele Frequency Difference AFD—An Intuitive Alternative to F(ST) for Quantifying Genetic Population Differentiation. Genes 2019, 10, 308. [Google Scholar] [CrossRef]
- Ramos, E.; Doumatey, A.; Elkahloun, A.G.; Shriner, D.; Huang, H.; Chen, G.; Zhou, J.; McLeod, H.; Adeyemo, A.; Rotimi, C.N. Pharmacogenomics, ancestry and clinical decision making for global populations. Pharmacogenom. J. 2014, 14, 217–222. [Google Scholar] [CrossRef]
- Neumann, G.B.; Korkuć, P.; Arends, D.; Wolf, M.J.; May, K.; Reißmann, M.; Elzaki, S.; König, S.; Brockmann, G.A. Design and performance of a bovine 200 k SNP chip developed for endangered German Black Pied cattle (DSN). BMC Genom. 2021, 22, 905. [Google Scholar] [CrossRef]
- Guo, Z.; Yang, Q.; Huang, F.; Zheng, H.; Sang, Z.; Xu, Y.; Zhang, C.; Wu, K.; Tao, J.; Prasanna, B.M.; et al. Development of high-resolution multiple-SNP arrays for genetic analyses and molecular breeding through genotyping by target sequencing and liquid chip. Plant Commun. 2021, 2, 100230. [Google Scholar] [CrossRef]
- Stothard, P.; Choi, J.W.; Basu, U.; Sumner-Thomson, J.M.; Meng, Y.; Liao, X.; Moore, S.S. Whole genome resequencing of black Angus and Holstein cattle for SNP and CNV discovery. BMC Genom. 2011, 12, 559. [Google Scholar] [CrossRef] [PubMed]
- Gaggero, S.; Martinez-Fabregas, J.; Cozzani, A.; Fyfe, P.K.; Leprohon, M.; Yang, J.; Thomasen, F.E.; Winkelmann, H.; Magnez, R.; Conti, A.G.; et al. IL-2 is inactivated by the acidic pH environment of tumors enabling engineering of a pH-selective mutein. Sci. Immunol. 2022, 7, eade5686. [Google Scholar] [CrossRef] [PubMed]
- Esposito, S.; Caminiti, C.; Giordano, R.; Argentiero, A.; Ramundo, G.; Principi, N. Risks of SARS-CoV-2 Infection and Immune Response to COVID-19 Vaccines in Patients with Inflammatory Bowel Disease: Current Evidence. Front. Immunol. 2022, 13, 933774. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, S.; Jagannadham, J.; Kumari, J.; Iquebal, M.A.; Gurjar, A.K.S.; Nayan, V.; Angadi, U.B.; Kumar, S.; Kumar, R.; Datta, T.K.; et al. Genome Wide Prediction, Mapping and Development of Genomic Resources of Mastitis Associated Genes in Water Buffalo. Front. Vet. Sci. 2021, 8, 593871. [Google Scholar] [CrossRef]
- Roldan-Montes, V.; Cardoso, D.F.; Hurtado-Lugo, N.A.; do Nascimento, A.V.; Santos, D.J.A.; Scalez, D.C.B.; de Freitas, A.C.; Herrera, A.C.; Albuquerque, L.G.; de Camargo, G.M.F.; et al. Polymorphisms in TLR4 Gene Associated with Somatic Cell Score in Water Buffaloes (Bubalus bubalis). Front. Vet. Sci. 2020, 7, 568249. [Google Scholar] [CrossRef]
- Kumar, B.; Sahoo, A.K.; Dayal, S.; Das, A.K.; Taraphder, S.; Batabyal, S.; Ray, P.K.; Kumari, R. Genetic profiling of Hsp70 gene in Murrah buffalo (Bubalus bubalis) under sub-tropical climate of India. Cell Stress Chaperones 2019, 24, 1187–1195. [Google Scholar] [CrossRef]
- Qiao, X.; Su, R.; Wang, Y.; Wang, R.; Yang, T.; Li, X.; Chen, W.; He, S.; Jiang, Y.; Xu, Q.; et al. Genome-wide Target Enrichment-aided Chip Design: A 66 K SNP Chip for Cashmere Goat. Sci. Rep. 2017, 7, 8621. [Google Scholar] [CrossRef]
- Wei, K.; Wang, X.; Hao, X.; Qian, Y.; Li, X.; Xu, L.; Ruan, L.; Wang, Y.; Zhang, Y.; Bai, P.; et al. Development of a genome-wide 200K SNP array and its application for high-density genetic mapping and origin analysis of Camellia sinensis. Plant Biotechnol. J. 2022, 20, 414–416. [Google Scholar] [CrossRef]
- Fukami, K.; Inanobe, S.; Kanemaru, K.; Nakamura, Y. Phospholipase C is a key enzyme regulating intracellular calcium and modulating the phosphoinositide balance. Prog. Lipid. Res. 2010, 49, 429–437. [Google Scholar] [CrossRef]
- Gupta, Y.; Goicoechea, S.; Pearce, C.M.; Mathur, R.; Romero, J.G.; Kwofie, S.K.; Weyenberg, M.C.; Daravath, B.; Sharma, N.; Poonam Akala, H.M.; et al. The emerging paradigm of calcium homeostasis as a new therapeutic target for protozoan parasites. Med. Res. Rev. 2022, 42, 56–82. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Tan, H.; Gu, Z.; Liu, Z.; Geng, Y.; Liu, Y.; Tong, H.; Tang, Y.; Qiu, J.; Su, L. Heat stress induces apoptosis through a Ca2+-mediated mitochondrial apoptotic pathway in human umbilical vein endothelial cells. PLoS ONE 2014, 9, e111083. [Google Scholar] [CrossRef] [PubMed]
- Terrell, K.; Choi, S.; Choi, S. Calcium’s Role and Signaling in Aging Muscle, Cellular Senescence, and Mineral Interactions. Int. J. Mol. Sci. 2023, 24, 17034. [Google Scholar] [CrossRef]
- Schink, K.O.; Tan, K.W.; Stenmark, H. Phosphoinositides in Control of Membrane Dynamics. Annu. Rev. Cell Dev. Biol. 2016, 32, 143–171. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto-Ishige, A.; Jodo, A.; Tanaka, T. Fbxo16 mediates degradation of NF-κB p65 subunit and inhibits inflammatory response in dendritic cells. Front. Immunol. 2025, 16, 1524110. [Google Scholar] [CrossRef] [PubMed]
- Samuel, B.; Dadi, H.; Dejene, G.; Kang, M.; Park, C.; Dinka, H. Single nucleotide polymorphisms within exon four of the prolactin gene and their effect on milk traits in cattle populations of Ethiopia. Anim. Biotechnol. 2023, 34, 4634–4644. [Google Scholar] [CrossRef] [PubMed]
- Marcuzzi, O.; Calcaterra, F.; Loza Vega, A.; Ortega Masagué, M.F.; Armstrong, E.; Pereira Rico, J.A.; Jara, E.; Olivera, L.H.; Peral García, P.; Giovambattista, G. Genomic analysis of inbreeding level, kinship and breed relationships in Creole cattle from South America. Anim. Genet. 2024, 55, 527–539. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Wang, W.; Huang, J.; Xu, M.; Wang, B.; Wu, Y.; Xie, Y.; Jian, J. Kinship analysis and pedigree reconstruction by RAD sequencing in cattle. GigaByte 2024, 2024, 1–15. [Google Scholar] [CrossRef]
- Zhao, J.; Wang, Y.; Kamalibieke, J.; Gong, P.; Zhang, F.; Shi, H.; Wang, W.; Luo, J. Design and verification of a 25 K multiple-SNP liquid-capture chip by target sequencing for dairy goat. BMC Genom. 2025, 26, 377. [Google Scholar] [CrossRef]
| Sample ID | Number of Discordant SNPs | Concordance Rate |
|---|---|---|
| buffalo-5 and buffalo-5-re | 5 | 99.90% |
| buffalo-6 and buffalo-6-re | 3 | 99.94% |
| buffalo-12 and buffalo-12-re | 3 | 99.94% |
| buffalo-14 and buffalo-14-re | 2 | 99.96% |
| Gene | Location | SNP * | Attribute | Amino Acid |
|---|---|---|---|---|
| PLCXD1 | chrX:136183705 | c.464C>T | non-synonymous | p.155A>V |
| FBXO16 | chr4:71852182 | c.421T>G | non-synonymous | p.141F>V |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiao, Y.; Jiang, J.; Li, S.; Chen, T.; Qiu, X.; Cui, K.; Li, B.; Chen, S.; Chen, Q.; Du, L.; et al. Development of a 5K Liquid-Phase Genome-Wide Breeding Chip for Xinglong Buffalo. Animals 2025, 15, 2702. https://doi.org/10.3390/ani15182702
Jiao Y, Jiang J, Li S, Chen T, Qiu X, Cui K, Li B, Chen S, Chen Q, Du L, et al. Development of a 5K Liquid-Phase Genome-Wide Breeding Chip for Xinglong Buffalo. Animals. 2025; 15(18):2702. https://doi.org/10.3390/ani15182702
Chicago/Turabian StyleJiao, Yuqing, Junming Jiang, Shiyuan Li, Taoyu Chen, Xinjun Qiu, Ke Cui, Boling Li, Si Chen, Qiaoling Chen, Li Du, and et al. 2025. "Development of a 5K Liquid-Phase Genome-Wide Breeding Chip for Xinglong Buffalo" Animals 15, no. 18: 2702. https://doi.org/10.3390/ani15182702
APA StyleJiao, Y., Jiang, J., Li, S., Chen, T., Qiu, X., Cui, K., Li, B., Chen, S., Chen, Q., Du, L., Man, C., Li, L., Wang, F., & Gao, H. (2025). Development of a 5K Liquid-Phase Genome-Wide Breeding Chip for Xinglong Buffalo. Animals, 15(18), 2702. https://doi.org/10.3390/ani15182702







