Uncovering Genetic Diversity and Adaptive Candidate Genes in the Mugalzhar Horse Breed Using Whole-Genome Sequencing Data
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Sample Collection and DNA Extraction
2.2. WGS, Reads Preprocessing and Mapping
2.3. Variants Calling and Annotation
2.4. Genetic Diversity Analysis and Population Structure
- FGRM, the inbreeding coefficient driven from the genomic relationship matrix (GRM) and calculated as the deviation of the diagonal elements from unity:where Z is the standardized and centralized genotype file, p is the minor allele frequency, is the inbreeding coefficient for ith sample, and is the ith diagonal element of GRM corresponding to ith sample.
- FHOM, the Wright’s inbreeding coefficient based on the proportion of the loci with higher observed homozygosity than expected homozygosity:where , and are numbers of observed homozygous, expected homozygous, and non-missing loci, respectively.
- FUNI, the Wright’s inbreeding coefficient based on the correlation between alleles in uniting gametes:where is the number of copies of the reference allele for the ith SNP, n is the number of versions of the reference allele, and is the minor allele frequency for the ith SNP.
2.5. Segregating Variants from Online Mendelian Inheritance in Animals
3. Results
3.1. WGS, Reads Preprocessing, Mapping, Variant Calling, and Annotation
3.2. Genetic Diversity Analysis and Population Stratification
3.3. Adaptation Footprints and Candidate Genes
3.4. OMIA Variants Segregating Analysis
4. Discussion
4.1. WGS Outcome and Variant Characterization in the Mugalzhar Breed
4.2. Genomic Diversity, Inbreeding, and Population Stratification
4.3. PCGs
4.4. OMIA Variants
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schubert, M.; Jónsson, H.; Chang, D.; Der Sarkissian, C.; Ermini, L.; Ginolhac, A.; Albrechtsen, A.; Dupanloup, I.; Foucal, A.; Petersen, B.; et al. Prehistoric genomes reveal the genetic foundation and cost of horse domestication. Proc. Natl. Acad. Sci. USA 2014, 111, E5661–E5669. [Google Scholar] [CrossRef]
- Orlando, L. Ancient genomes reveal unexpected horse domestication and management dynamics. BioEssays 2020, 42, 1900164. [Google Scholar] [CrossRef]
- Orlando, L. The evolutionary and historical foundation of the modern horse: Lessons from ancient genomics. Annu. Rev. Genet. 2020, 54, 563–581. [Google Scholar] [CrossRef]
- Lawrence, E.A. Horses in society. Anthrozoös 1988, 1, 223–231. [Google Scholar] [CrossRef]
- Klecel, W.; Martyniuk, E. From the Eurasian steppes to the Roman circuses: A Review of early development of horse breeding and management. Animals 2021, 11, 1859. [Google Scholar] [CrossRef]
- Librado, P.; Khan, N.; Fages, A.; Kusliy, M.A.; Suchan, T.; Tonasso-Calvière, L.; Schiavinato, S.; Alioglu, D.; Fromentier, A.; Perdereau, A.; et al. The origins and spread of domestic horses from the Western Eurasian steppes. Nature 2021, 598, 634–640. [Google Scholar] [CrossRef]
- Kelekna, P. The Horse in Human History; Cambridge University Press: Cambridge, UK, 2009; Available online: https://www.cambridge.org/us/universitypress/subjects/archaeology/archaeology-europe-and-near-and-middle-east/horse-human-history (accessed on 5 July 2025).
- Librado, P.; Fages, A.; Gaunitz, C.; Leonardi, M.; Wagner, S.; Khan, N.; Hanghøj, K.; Alquraishi, S.A.; Alfarhan, A.H.; Al-Rasheid, K.A. The evolutionary origin and genetic makeup of domestic horses. Genetics 2016, 204, 423–434. [Google Scholar] [CrossRef] [PubMed]
- Outram, A.K.; Stear, N.A.; Bendrey, R.; Olsen, S.; Kasparov, A.; Zaibert, V.; Thorpe, N.; Evershed, R.P. The earliest horse harnessing and milking. Science 2009, 323, 1332–1335. [Google Scholar] [CrossRef]
- Outram, A.; Bendrey, R.; Evershed, R.P.; Orlando, L.; Zaibert, V.F. Rebuttal of Taylor and Barrón-Ortiz 2021 Rethinking the evidence for early horse domestication at Botai. Dataset Zenodo 2021. [Google Scholar] [CrossRef]
- Kosharov, A.N.; Pern, E.M.; Rozhdestvenskaya, G.A. Horses. In Animal Genetic Resources of the USSR; FAO Animal Production and Health Paper; Dmitriev, N.G., Ernst, L.K., Eds.; Food and Agriculture Organization of the United Nations: Rome, Italy, 1989; Volume 65, pp. 272–343. Available online: https://www.fao.org/4/ah759e/AH759E14.htm (accessed on 5 July 2025).
- Kabylbekova, D.; Assanbayev, T.S.; Kassymbekova, S.; Kantanen, J. Genetic studies and breed diversity of Kazakh native horses: A comprehensive review. Adv. Life Sci. 2024, 11, 18–27. Available online: https://www.als-journal.com/1113-24/ (accessed on 5 July 2025). [CrossRef]
- Sansyzbayev, B.; Sydykov, D.; Kozhanov, Z.; Akhmetov, U.; Zhenishbekov, A. Organization and analysis of production efficiency horse breeding products by breed and region of the Republic of Kazakhstan. [Oš Mamlekettik Univ. Žarčysy. Ajyl Čarba Agron. Vet. Žana Zootehniâ.] J. Osh State Univ. Agric. Agron. Vet. Zootech. 2024, 2, 219–226. [Google Scholar] [CrossRef]
- Rzabayev, S.S. Mugalzhar Horse Breed; LLP Information and Printing Center—Kokzhiyek: Aktobe, Kazakhstan, 2007. [Google Scholar]
- Satybaldin, A.A. Current State of Horse Breeding and Horse Sports in Kazakhstan. In Proceedings of the First International Conference; n.p.: Kostanay, Kazakhstan, 2002; Available online: https://scholar.google.com/scholar?q=Current+State+of+Horse+Breeding+and+Horse+Sports+in+Kazakhstan (accessed on 5 July 2025).
- Iskhan, K.Z.; Kalashnikov, V.V.; Akimbekov, A.R.; Mongush, S.D.; Demin, V.A.; Rzabayev, T.S.; Nesipbaeva, A.K.; Zhilkybaeva, M.M.; Zhikishev, Y.K. Zootechnic characteristics of modern populations of Mugalzhar horse breed. Bull. Natl. Acad. Sci. Rep. Kazakhstan 2019, 6, 75–82. [Google Scholar] [CrossRef]
- Baimukanov, D.A.; Iskhan, K.Z. Steppe Horse Breeds—Lecture 7.1; Agriexpert.ru: Moscow, Russia, 2022; Available online: https://agriexpert.ru/articles/555/stepnye-porody-losadei-lekciya-71 (accessed on 5 July 2025). (In Russian)
- Salkova, N.; Moldagaliev, B. Mugalzhar Horse Breed is 25 Years Old; TV channel 24KZ: Astana, Kazakhstan, 2023; Available online: https://24.kz/ru/news/social/620379-mugalzharskoj-porode-loshadej-25-let (accessed on 5 July 2025). (In Russian)
- Kalzhanov, A. Mugalzhar Horse Breed is 25 Years Old; Aktyubinskiy Vestnik: Aktobe, Kazakhstan, 2023; Available online: https://avestnik.kz/mugalzharskoj-porode-loshadej-25-let/ (accessed on 5 July 2025). (In Russian)
- Boss Agro. Mugalzhar Horse; Boss Agro: Ust-Kamenogorsk, Kazakhstan, 2023; Available online: https://bossagro.kz/glossary/mugalzharskaya-loshad/ (accessed on 5 July 2025). (In Russian)
- Baibolsyn. Mugalzhar Horse; Baibolsyn: Almaty, Kazakhstan, 2023; Available online: https://baibolsyn.kz/ru/zhivotnye/mugalzharskaya-loshad/ (accessed on 5 July 2025). (In Russian)
- Stachurska, A.M. Inheritance of primitive markings in horses. J. Anim. Breed. Genet. 1999, 116, 29–38. [Google Scholar] [CrossRef]
- Seleuova, L.A.; Naimanov, D.K.; Jaworski, Z.; Aubakirov, M.Z.H.; Mustafin, M.K.; Mustafin, B.M.; Safronova, O.S.; Baktybaev, G.T.; Turabaev, A.T.; Domatski, V.N. Population genetic characteristic of horses of Mugalzhar breed by STR-markers. Biomed. Res. 2018, 29, 3508–3511. [Google Scholar] [CrossRef]
- Akhmetov, U.A.; Kozhanov, Z.Y.; Sydykov, D.A. Optimal Timing of Slaughtering Horses of Different Breeds in the Southern Region of Kazakhstan. In Seifullin Readings—18(2): Science of the 21st Century—The Era of Transformation, Proceedings of the International Scientific-Practical Conference, Astana, Kazakhstan, 7 October 2022; Saken Seifullin Kazakh State Agrotechnical University: Astana, Kazakhstan, 2022; Volume I, Chapter II; pp. 169–172. Available online: https://kazatu.edu.kz/webroot/js/kcfinder/upload/files/наука/СЧ-18(2)/Akhmetov%20U.A..pdf (accessed on 5 July 2025).
- Shamshidin, A.S.; Beishova, I.S.; Alikhanov, O.; Aubakirov, K.A.; Shamekova, M.K.; Kargaeva, M.T.; Karibayeva, D.K.; Baimukanov, D.A. Productive longevity of Mugalzhar mares. [Ġylym Žἄne Bìlìm.] Sci. Educ. 2025, 2, 279–287. [Google Scholar] [CrossRef]
- Dyussegaliyev, M.Z. The genotypes of herd horses of the West Region of Kazakhstan. [Sel’skoe Hozâjstvo Èkosistemy Sovremennom Mire Reg. Mežstranovye Issledovaniâ.] Agric. Ecosyst. Mod. World Reg. Inter Countries Res. 2022, 1, 33–43, (In Russian with English summary). [Google Scholar]
- Nurushev, M. A Unique Breed of Horses Has Been Developed; Kazakhstanskaya Pravda: Astana, Kazakhstan, 2013; Available online: https://kazpravda.kz/n/vyvedena-unikalnaya-poroda-loshadey/ (accessed on 5 July 2025). (In Russian)
- Orazymbetova, Z.; Ualiyeva, D.; Dossybayev, K.; Torekhanov, A.; Sydykov, D.; Mussayeva, A.; Baktybayev, G. Genetic diversity of Kazakhstani Equus caballus (Linnaeus, 1758) horse breeds inferred from microsatellite markers. Vet. Sci. 2023, 10, 598. [Google Scholar] [CrossRef] [PubMed]
- Iskhan, K.; Uskenov, R.; Akimbekov, A.; Baymukanov, D.; Yuldashbayev, Y.; Orynaliev, K. The Irtysh factory type of the Mugalzhar breed and the line Zamana, Bakay. [Ìzdenìster Nἄtiželer.] Res. Results 2024, 4, 16–24. [Google Scholar] [CrossRef]
- Akim of the Abay Region. Presentation of the Mugalzhar Horse Breed “Irtysh” Took Place; Websites of Government Bodies, gov.kz: Semey City, Republic of Kazakhstan, 2024. Available online: https://www.gov.kz/memleket/entities/abay/press/news/details/825473?lang=ru (accessed on 5 July 2025)(In Russian and Kazakh).
- Orazymbetova, Z.S.; Sydykov, D.A.; Dossybayev, K.Z.; Razak, A.B. Analysis of the Population Structure of the Kozhamberdin Type Inside Mugalzhar Horse Breed Using Microsatellite Markers. In Scientific Support for Animal Husbandry in Siberia, Proceedings of the VII International Scientific and Practical Conference, Krasnoyarsk, Russia, 18–19 May 2023; Efimova, L.V., Tereshchenko, V.A., Eds.; KrasNIISKh FRC KSC SB RAS: Krasnoyarsk, Russia, 2023; pp. 177–182. Available online: https://sh.krasn.ru/upload/iblock/f5d/bmna0ji3o1516z19ezzz21wq46pdq2ca.pdf#page=178 (accessed on 5 July 2025).
- Kassymbekova, S.N.; Iskhan, K.Z.; Rzabaev, S.S.; Bimenova, Z.Z.; Kabylbekova, D.I.; Tursunkulov, S.A. Assessment of genetic diversity using microsatellite markers and milk productivity of Mugalzhar horses. [S Sejfullin Atyndaġy K̦az. Agroteh. Univ. Ġylym Žaršysy.] Her. Sci. S Seifullin Kazakh Agro Techn. Res. Univ. Vet. Sci. 2024, 3, 29–36. [Google Scholar] [CrossRef]
- Pozharskiy, A.; Abdrakhmanova, A.; Beishova, I.; Shamshidin, A.; Nametov, A.; Ulyanova, T.; Bekova, G.; Kikebayev, N.; Kovalchuk, A.; Ulyanov, V. Genetic structure and genome-wide association study of the traditional Kazakh horses. Animal 2023, 17, 100926. [Google Scholar] [CrossRef]
- Pozharskiy, A.; Beishova, I.; Nametov, A.; Shamshidin, A.; Ulyanova, T.; Kovalchuk, A.; Ulyanov, V.; Shamekova, M.; Bekova, G.; Gritsenko, D. Genetic composition of Kazakh horses of Zhabe type evaluated by SNP genotyping. Heliyon 2025, 11, e41173. [Google Scholar] [CrossRef]
- Dementeva, N.V.; Romanov, M.N.; Kudinov, A.A.; Mitrofanova, O.V.; Stanishevskaya, O.I.; Terletsky, V.P.; Fedorova, E.S.; Nikitkina, E.V.; Plemyashov, K.V. Studying the structure of a gene pool population of the Russian White chicken breed by genome-wide SNP scan. Sel’skokhozyaistvennaya Biol. (Agric. Biol.) 2017, 52, 1166–1174. [Google Scholar] [CrossRef]
- Romanov, M.N.; Dementyeva, N.V.; Terletsky, V.P.; Plemyashov, K.V.; Stanishevskaya, O.I.; Kudinov, A.A.; Perinek, O.Y.; Fedorova, E.S.; Larkina, T.A.; Pleshanov, N.V. Applying SNP Array Technology to Assess Genetic Diversity in Russian Gene Pool of Chickens. In Proceedings of the International Plant and Animal Genome XXV Conference, San Diego, CA, USA, 14–18 January 2017; Scherago International: San Diego, CA, USA, 2017. Abstract P0115. Available online: https://pag.confex.com/pag/xxv/webprogram/Paper23948.html (accessed on 5 July 2025).
- Dementieva, N.V.; Shcherbakov, Y.S.; Tyshchenko, V.I.; Terletsky, V.P.; Vakhrameev, A.B.; Nikolaeva, O.A.; Ryabova, A.E.; Azovtseva, A.I.; Mitrofanova, O.V.; Peglivanyan, G.K.; et al. Comparative analysis of molecular RFLP and SNP markers in assessing and understanding the genetic diversity of various chicken breeds. Genes 2022, 13, 1876. [Google Scholar] [CrossRef] [PubMed]
- Volkova, N.A.; German, N.Y.; Larionova, P.V.; Vetokh, A.N.; Romanov, M.N.; Zinovieva, N.A. Identification of SNPs and candidate genes associated with abdominal fat deposition in quails (Coturnix japonica). Sel’skokhozyaistvennaya Biol. (Agric. Biol.) 2023, 58, 1079–1087. [Google Scholar] [CrossRef]
- Deniskova, T.E.; Dotsev, A.V.; Koshkina, O.A.; Solovieva, A.D.; Churbakova, N.A.; Petrov, S.N.; Frolov, A.N.; Platonov, S.A.; Abdelmanova, A.S.; Vladimirov, M.A.; et al. Examination of runs of homozygosity distribution patterns and relevant candidate genes of potential economic interest in Russian goat breeds using whole-genome sequencing. Genes 2025, 16, 631. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-H.; Wang, L.; Xu, T.; Guo, X.; Li, Y.; Yin, T.-T.; Yang, H.-C.; Hu, Y.; Adeola, A.C.; Sanke, O.J.; et al. Whole-genome sequencing of African dogs provides insights into adaptations against tropical parasites. Mol. Biol. Evol. 2018, 35, 287–298. [Google Scholar] [CrossRef]
- Tuttle, E.; Korody, M.; Lear, T.; Gonser, R.; Houck, M.; Ryder, O.; Romanov, M.; Balakrishnan, C.; Bergland, A.; Warren, W. Whole Genome Sequence of the Behaviorally Polymorphic White-throated Sparrow. 1: Mapping Genes for Sociogenomics. In Proceedings of the Evolution 2014 Conference, Raleigh, NC, USA, 20–24 June 2014; Society for the Study of Evolution (SSE); Society of Systematic Biologists (SSB). American Society of Naturalists (ASN): Raleigh, NC, USA, 2014. Abstract 628. p. 183. Available online: https://kar.kent.ac.uk/id/eprint/46698 (accessed on 5 July 2025).
- Ryder, O.; Chemnick, L.G.; Thomas, S.; Martin, J.; Romanov, M.; Ralls, K.; Ballou, J.D.; Mace, M.; Ratan, A.; Miller, W.; et al. Supporting California Condor Conservation Management through Analysis of Species-wide Whole Genome Sequence Variation. In Proceedings of the International Plant and Animal Genome XXII Conference, San Diego, CA, USA, 11–15 January 2014; Scherago International: San Diego, CA, USA, 2014. Abstract W635. Available online: https://pag.confex.com/pag/xxii/webprogram/Paper11851.html (accessed on 5 July 2025).
- Ryder, O.; Miller, W.; Ralls, K.; Ballou, J.D.; Steiner, C.C.; Mitelberg, A.; Romanov, M.; Chemnick, L.G.; Mace, M.; Schuster, S. Whole Genome Sequencing of California Condors Is Now Utilized for Guiding Genetic Management. In Proceedings of the International Plant and Animal Genome XXIV Conference, San Diego, CA, USA, 8–13 January 2016; Scherago International: San Diego, CA, USA, 2016. Abstract W741. Available online: http://kar.kent.ac.uk/61072 (accessed on 5 July 2025).
- Rehman, S.U.; Hassan, F.-U.; Luo, X.; Li, Z.; Liu, Q. Whole-genome sequencing and characterization of buffalo genetic resources: Recent advances and future challenges. Animals 2021, 11, 904. [Google Scholar] [CrossRef]
- Sun, J.; Chen, T.; Zhu, M.; Wang, R.; Huang, Y.; Wei, Q.; Yang, M.; Liao, Y. Whole-genome sequencing revealed genetic diversity and selection of Guangxi indigenous chickens. PLoS ONE 2022, 17, e0250392. [Google Scholar] [CrossRef]
- Chen, N.; Xia, X.; Hanif, Q.; Zhang, F.; Dang, R.; Huang, B.; Lyu, Y.; Luo, X.; Zhang, H.; Yan, H.; et al. Global genetic diversity, introgression, and evolutionary adaptation of indicine cattle revealed by whole genome sequencing. Nat. Commun. 2023, 14, 7803. [Google Scholar] [CrossRef]
- Alemu, S.W.; Kadri, N.K.; Harland, C.; Faux, P.; Charlier, C.; Caballero, A.; Druet, T. An evaluation of inbreeding measures using a whole-genome sequenced cattle pedigree. Heredity 2021, 126, 410–423. [Google Scholar] [CrossRef]
- Huo, J.L.; Zhang, L.Q.; Zhang, X.; Wu, X.W.; Ye, X.H.; Sun, Y.H.; Cheng, W.M.; Yang, K.; Pan, W.R.; Zeng, Y.Z. Genome-wide single nucleotide polymorphism array and whole-genome sequencing reveal the inbreeding progression of Banna minipig inbred line. Anim. Genet. 2022, 53, 146–151. [Google Scholar] [CrossRef]
- Chen, H.M.; Zhao, H.; Zhu, Q.Y.; Yan, C.; Liu, Y.Q.; Si, S.; Jamal, M.A.; Xu, K.X.; Jiao, D.L.; Lv, M.J.; et al. Genomic consequences of intensive inbreeding in miniature inbred pigs. BMC Genom. 2025, 26, 154. [Google Scholar] [CrossRef]
- Lou, R.N.; Jacobs, A.; Wilder, A.P.; Therkildsen, N.O. A beginner’s guide to low-coverage whole genome sequencing for population genomics. Mol. Ecol. 2021, 30, 5966–5993. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Lan, T.; Li, H.; Sahu, S.K.; Shi, M.; Zhu, Y.; Han, L.; Yang, S.; Li, Q.; Zhang, L.; et al. Whole-genome resequencing of Chinese pangolins reveals a population structure and provides insights into their conservation. Commun. Biol. 2022, 5, 821. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Zhang, L.; Xin, Q.; Li, L.; Miao, Z.; Huang, Q.; Zheng, N. Whole-genome sequencing revealed the population structure of Fujian chicken breeds. Czech J. Anim. Sci. 2024, 69, 323–330. [Google Scholar] [CrossRef]
- Shi, H.; Li, T.; Su, M.; Wang, H.; Li, Q.; Lang, X.; Ma, Y. Whole genome sequencing revealed genetic diversity, population structure, and selective signature of Panou Tibetan sheep. BMC Genom. 2023, 24, 50. [Google Scholar] [CrossRef] [PubMed]
- Gebreselase, H.B.; Nigussie, H.; Wang, C.; Luo, C. Genetic diversity, population structure and selection signature in Begait goats revealed by whole-genome sequencing. Animals 2024, 14, 307. [Google Scholar] [CrossRef]
- Zhang, Y.; Wei, Z.; Zhang, M.; Wang, S.; Gao, T.; Huang, H.; Zhang, T.; Cai, H.; Liu, X.; Fu, T.; et al. Population structure and selection signal analysis of Nanyang cattle based on whole-genome sequencing data. Genes 2024, 15, 351. [Google Scholar] [CrossRef]
- Onogi, A.; Shirai, K.; Amano, T. Investigation of genetic diversity and inbreeding in a Japanese native horse breed for suggestions on its conservation. Anim. Sci. J. 2017, 88, 1902–1910. [Google Scholar] [CrossRef]
- Pokharel, K.; Weldenegodguad, M.; Reilas, T.; Kantanen, J. EquCab_Finn: A new reference genome assembly for the domestic horse, Finnhorse. Anim. Genet. 2024, 55, 766–771. [Google Scholar] [CrossRef]
- Tang, X.; Zhu, B.; Ren, R.; Chen, B.; Li, S.; Gu, J. Genome-wide copy number variation detection in a large cohort of diverse horse breeds by whole-genome sequencing. Front. Vet. Sci. 2023, 10, 1296213. [Google Scholar] [CrossRef]
- Chen, C.; Zhu, B.; Tang, X.; Chen, B.; Liu, M.; Gao, N.; Li, S.; Gu, J. Genome-wide assessment of runs of homozygosity by whole-genome sequencing in diverse horse breeds worldwide. Genes 2023, 14, 1211. [Google Scholar] [CrossRef]
- Maniatis, T.; Fritsch, E.F.; Sambrook, J. Molecular Cloning. A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratories: Cold Spring Harbor, NY, USA, 1982. [Google Scholar]
- NCBI. Equus caballus Genome Assembly EquCab3.0; NCBI RefSeq assembly GCF_002863925.1; National Center for Biotechnology Information; National Library of Medicine: Bethesda, MD, USA, 2018. Available online: https://www.ncbi.nlm.nih.gov/datasets/genome/GCF_002863925.1/ (accessed on 5 July 2025).
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Narasimhan, V.; Danecek, P.; Scally, A.; Xue, Y.; Tyler-Smith, C.; Durbin, R. BCFtools/RoH: A hidden Markov model approach for detecting autozygosity from next-generation sequencing data. Bioinformatics 2016, 32, 1749–1751. [Google Scholar] [CrossRef] [PubMed]
- Poplin, R.; Ruano-Rubio, V.; DePristo, M.A.; Fennell, T.J.; Carneiro, M.O.; Van der Auwera, G.A.; Kling, D.E.; Gauthier, L.D.; Levy-Moonshine, A.; Roazen, D. Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv 2017, 201178. [Google Scholar] [CrossRef]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.S.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Dyer, S.C.; Austine-Orimoloye, O.; Azov, A.G.; Barba, M.; Barnes, I.; Barrera-Enriquez, V.P.; Becker, A.; Bennett, R.; Beracochea, M.; Berry, A.; et al. Ensembl 2025. Nucleic Acids Res. 2025, 53, D948–D957. [Google Scholar] [CrossRef]
- Mushtaq, A. Ensembl 114 Has Been Released! Ensembl Blog. 2025. Available online: https://www.ensembl.info/2025/05/07/ensembl-114-has-been-released/ (accessed on 5 July 2025).
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Yu, N.; Jensen-Seaman, M.I.; Chemnick, L.; Ryder, O.; Li, W.-H. Nucleotide diversity in gorillas. Genetics 2004, 166, 1375–1383. [Google Scholar] [CrossRef]
- Yang, J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2009; ISBN 978-0-387-98141-3. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018; Available online: https://www.r-project.org/ (accessed on 5 July 2025).
- R Core Team. R-4; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://cran.r-project.org/src/base/R-4/ (accessed on 5 July 2025).
- Nicholas, F.; Tammen, I.; Sydney Informatics Hub. Online Mendelian Inheritance in Animals (OMIA); The University of Sydney: Sydney, Australia, 1995. [Google Scholar] [CrossRef]
- Yi, W.; Hu, M.; Shi, L.; Li, T.; Bai, C.; Sun, F.; Ma, H.; Zhao, Z.; Yan, S. Whole genome sequencing identified genomic diversity and candidated genes associated with economic traits in Northeasern Merino in China. Front. Genet. 2024, 15, 1302222. [Google Scholar] [CrossRef]
- Scherf, B.D. (Ed.) World Watch List for Domestic Animal Diversity, 2nd ed.; FAO; UNEP: Rome, Italy, 1995; Available online: https://web.archive.org/web/20151126034309/http://www.fao.org/ag/againfo/programmes/en/lead/toolbox/Indust/wwl.pdf (accessed on 26 November 2015).
- Nikiforov, A.A.; Moiseeva, I.G.; Zakharov, I.A. Mesto russkikh porod kur v raznoobrazii porod Yevrazii. Position of Russian chicken breeds in the diversity of Eurasian breeds. Genetika 1998, 34, 850–851. Available online: https://pubmed.ncbi.nlm.nih.gov/9719931/ (accessed on 5 July 2025). (In Russian with English summary).
- Romanov, M.N.; Weigend, S. Genetic Diversity in Chicken Populations Based on Microsatellite Markers. In Proceedings of the Conference “From Jay Lush to Genomics: Visions for Animal Breeding and Genetics”, Ames, IA, USA, 16–18 May 1999; Dekkers, J.C.M., Lamont, S.J., Rothschild, M.F., Eds.; Iowa State University, Department of Animal Science: Ames, IA, USA, 1999; p. 174. Available online: https://web.archive.org/web/20050314091227/http://www.agbiotechnet.com/proceedings/jaylush.asp#34 (accessed on 14 March 2025).
- Rzabayev, T.; Rzabayev, S.; Rzabayev, K. A new intra-breed type, “Mamyr-Aktobe,” of the Kushum breed of horses of the Aktobe population. Arch. Razi Inst. 2022, 77, 2273. [Google Scholar] [CrossRef]
- Boettcher, P.; Martin, J.F.; Gandini, G.; Joshi, B.K.; Oldenbroek, J.K. In Vivo Conservation of Animal Genetic Resources. In FAO Animal Production and Health Guidelines; Commission on Genetic Resources for Food and Agriculture, FAO: Rome, Italy, 2013; Volume 14, Available online: https://www.fao.org/4/i3327e/i3327e.pdf (accessed on 5 July 2025).
- Woolliams, J.A.; Oldenbroek, J.K. Genetic diversity issues in animal populations in the genomic era. In Genomic Management of Animal Genetic Diversity; Oldenbroek, J.K., Ed.; Wageningen Academic Publishers: Wageningen, The Netherlands, 2017; pp. 13–47. [Google Scholar] [CrossRef]
- Eusebi, P.G.; Martinez, A.; Cortes, O. Genomic tools for effective conservation of livestock breed diversity. Diversity 2019, 12, 8. [Google Scholar] [CrossRef]
- Moiseeva, I.G. Vliyaniye inbridinga na kachestvo kurinykh yaits. The effect of inbreeding on the quality of fowl eggs. Genetika 1970, 6, 99–107, (In Russian with English Summary). [Google Scholar]
- Bondarenko, Y.V.; Popsuy, V.V. Vykorystannya inbrydynhu v suchasnomu svynarstvi. In The Use of Inbreeding in Modern Pig Farming; LLC Ahrar Mediyen Ukrayina: Kyiv, Ukraine, 2019; Available online: https://repo.snau.edu.ua/bitstream/123456789/6994/1/Бoндapeнкo%20Ю.%20Bикopистaння%20iнбpидингy.pdf (accessed on 5 July 2025). (In Ukrainian)
- Bailey, E.; Finno, C.J.; Cullen, J.N.; Kalbfleisch, T.; Petersen, J.L. Analyses of whole-genome sequences from 185 North American Thoroughbred horses, spanning 5 generations. Sci. Rep. 2024, 14, 22930. [Google Scholar] [CrossRef]
- Tozaki, T.; Ohnuma, A.; Kikuchi, M.; Ishige, T.; Kakoi, H.; Hirota, K.; Kusano, K.; Nagata, S. Rare and common variant discovery by whole-genome sequencing of 101 Thoroughbred racehorses. Sci. Rep. 2021, 11, 16057. [Google Scholar] [CrossRef]
- Al Abri, M.A.; Holl, H.M.; Kalla, S.E.; Sutter, N.B.; Brooks, S.A. Whole genome detection of sequence and structural polymorphism in six diverse horses. PLoS ONE 2020, 15, e0230899. [Google Scholar] [CrossRef] [PubMed]
- Brooks, S.A.; Gabreski, N.; Miller, D.; Brisbin, A.; Brown, H.E.; Streeter, C.; Mezey, J.; Cook, D.; Antczak, D.F. Whole-genome SNP association in the horse: Identification of a deletion in myosin Va responsible for Lavender Foal Syndrome. PLoS Genet. 2010, 6, e1000909. [Google Scholar] [CrossRef]
- Piro, M.; Benjouad, A.; Tligui, N.S.; Allali, K.E.; Kohen, M.E.; Nabich, A.; Ouragh, L. Frequency of the severe combined immunodeficiency disease gene among horses in Morocco. Equine Vet. J. 2008, 40, 590–591. [Google Scholar] [CrossRef]
- Wade, C.; Giulotto, E.; Sigurdsson, S.; Zoli, M.; Gnerre, S.; Imsland, F.; Lear, T.; Adelson, D.; Bailey, E.; Bellone, R.; et al. Genome sequence, comparative analysis, and population genetics of the domestic horse. Science 2009, 326, 865–867. [Google Scholar] [CrossRef]
- Zhang, Q.; Calus, M.P.; Guldbrandtsen, B.; Lund, M.S.; Sahana, G. Estimation of inbreeding using pedigree, 50k SNP chip genotypes and full sequence data in three cattle breeds. BMC Genet. 2015, 16, 88. [Google Scholar] [CrossRef]
- Najmabadi, H.; Hu, H.; Garshasbi, M.; Zemojtel, T.; Abedini, S.S.; Chen, W.; Hosseini, M.; Behjati, F.; Haas, S.; Jamali, P. Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature 2011, 478, 57–63. [Google Scholar] [CrossRef]
- Carss, K.J.; Arno, G.; Erwood, M.; Stephens, J.; Sanchis-Juan, A.; Hull, S.; Megy, K.; Grozeva, D.; Dewhurst, E.; Malka, S. Comprehensive rare variant analysis via whole-genome sequencing to determine the molecular pathology of inherited retinal disease. Am. J. Hum. Genet. 2017, 100, 75–90. [Google Scholar] [CrossRef]
- Tatour, Y.; Bar-Joseph, H.; Shalgi, R.; Ben-Yosef, T. Male sterility and reduced female fertility in SCAPER-deficient mice. Hum. Mol. Genet. 2020, 29, 2240–2249. [Google Scholar] [CrossRef] [PubMed]
- Ghoreishifar, M.; Vahedi, S.M.; Salek Ardestani, S.; Khansefid, M.; Pryce, J.E. Genome-wide assessment and mapping of inbreeding depression identifies candidate genes associated with semen traits in Holstein bulls. BMC Genom. 2023, 24, 230. [Google Scholar] [CrossRef]
- Terefe, M.T. Identification of Adaptive Signatures in the Cattle Genome. Ph.D. Thesis, Seoul National University, Seoul, Republic of Korea, 2018. Available online: https://s-space.snu.ac.kr/handle/10371/140789 (accessed on 5 July 2025).
- Gao, J.; Lyu, Y.; Zhang, D.; Reddi, K.K.; Sun, F.; Yi, J.; Liu, C.; Li, H.; Yao, H.; Dai, J.; et al. Genomic characteristics and selection signatures in indigenous Chongming white goat (Capra hircus). Front. Genet. 2020, 11, 901. [Google Scholar] [CrossRef] [PubMed]
- Shang, P.; Li, W.; Tan, Z.; Zhang, J.; Dong, S.; Wang, K.; Chamba, Y. Population genetic analysis of ten geographically isolated Tibetan pig populations. Animals 2020, 10, 1297. [Google Scholar] [CrossRef]
- Yahyaoui, G.; Jemaa, S.B.; Kdidi, S.; Gaouar, S.B.S.; Yahyaoui, M.H. Identification of selection signatures in Algero-Tunisian sheep breeds using medium-density SNP chips. Genet. Biodiv. J. 2024, 8, 59–75. [Google Scholar]
- Elkjaer, M.L.; Frisch, T.; Reynolds, R.; Kacprowski, T.; Burton, M.; Kruse, T.A.; Thomassen, M.; Baumbach, J.; Illes, Z. Molecular signature of different lesion types in the brain white matter of patients with progressive multiple sclerosis. Acta Neuropathol. Commun. 2019, 7, 205. [Google Scholar] [CrossRef] [PubMed]
- van Kruistum, H.; van den Heuvel, J.; Travis, J.; Kraaijeveld, K.; Zwaan, B.J.; Groenen, M.A.M.; Megens, H.-J.; Pollux, B.J.A. The genome of the live-bearing fish Heterandria formosa implicates a role of conserved vertebrate genes in the evolution of placental fish. BMC Evol. Biol. 2019, 19, 156. [Google Scholar] [CrossRef]
- Dierks, C. Molecular Genetic Analysis of Quantitative Trait Loci (QTL) for Osteochondrosis in Hanoverian Warmblood Horses. Ph.D. Thesis, Institut für Tierzucht und Vererbungsforschung der Tierärztlichen Hochschule Hannover, Hannover, Germany, 2006. Available online: https://elib.tiho-hannover.de/receive/etd_mods_00002149 (accessed on 5 July 2025).
- Waddell, L.A.; Lefevre, L.; Bush, S.J.; Raper, A.; Young, R.; Lisowski, Z.M.; McCulloch, M.E.B.; Muriuki, C.; Sauter, K.A.; Clark, E.L.; et al. ADGRE1 (EMR1, F4/80) is a rapidly-evolving gene expressed in mammalian monocyte-macrophages. Front. Immunol. 2018, 9, 2246. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.-H.; Faunce, D.E.; Stacey, M.; Terajewicz, A.; Nakamura, T.; Zhang-Hoover, J.; Kerley, M.; Mucenski, M.L.; Gordon, S.; Stein-Streilein, J. The macrophage F4/80 receptor is required for the induction of antigen-specific efferent regulatory T cells in peripheral tolerance. J. Exp. Med. 2005, 201, 1615–1625. [Google Scholar] [CrossRef]
- Al-Quraishy, S.; Dkhil, M.A.; Abdel-Baki, A.A.S.; Delic, D.; Santourlidis, S.; Wunderlich, F. Genome-wide screening identifies Plasmodium chabaudi-induced modifications of DNA methylation status of Tlr1 and Tlr6 gene promoters in liver, but not spleen, of female C57BL/6 mice. Parasitol. Res. 2013, 112, 3757–3770. [Google Scholar] [CrossRef]
- Dander, E.; Vinci, P.; Vetrano, S.; Recordati, C.; Piazza, R.; Fazio, G.; Bardelli, D.; Bugatti, M.; Sozio, F.; Piontini, A.; et al. The chemerin/CMKLR1 axis regulates intestinal graft-versus-host disease. JCI Insight. 2023, 8, e154440. [Google Scholar] [CrossRef]
- de Camargo, G.M.F.; Aspilcueta-Borquis, R.R.; Fortes, M.R.S.; Porto-Neto, R.; Cardoso, D.F.; Santos, D.J.A.; Lehnert, S.A.; Reverter, A.; Moore, S.S.; Tonhati, H. Prospecting major genes in dairy buffaloes. BMC Genom. 2015, 16, 872. [Google Scholar] [CrossRef]
- Bozaoglu, K.; Bolton, K.; McMillan, J.; Zimmet, P.; Jowett, J.; Collier, G.; Walder, K.; Segal, D. Chemerin is a novel adipokine associated with obesity and metabolic syndrome. Endocrinology 2007, 148, 4687–4694. [Google Scholar] [CrossRef]
- Sell, H.; Divoux, A.; Poitou, C.; Basdevant, A.; Bouillot, J.-L.; Bedossa, P.; Tordjman, J.; Eckel, J.; Clement, K. Chemerin correlates with markers for fatty liver in morbidly obese patients and strongly decreases after weight loss induced by bariatric surgery. J. Clin. Endocrinol. Metab. 2010, 95, 2892–2896. [Google Scholar] [CrossRef]
- Adhikari, M.; Kantar, M.B.; Longman, R.J.; Lee, C.N.; Oshiro, M.; Caires, K.; He, Y. Genome-wide association study for carcass weight in pasture-finished beef cattle in Hawai’i. Front. Genet. 2023, 14, 1168150. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.Z.; Haque, M.A.; Iqbal, A.; Lee, Y.-M.; Ha, J.-J.; Jin, S.; Park, B.; Kim, N.-Y.; Won, J.I.; Kim, J.-J. Genome-wide association study to identify QTL for carcass traits in Korean Hanwoo cattle. Animals 2023, 13, 2737. [Google Scholar] [CrossRef]
- Atashi, H.; Chen, Y.; Wilmot, H.; Vanderick, S.; Hubin, X.; Soyeurt, H.; Gengler, N. Single-step genome-wide association for selected milk fatty acids in Dual-Purpose Belgian Blue cows. J. Dairy Sci. 2023, 106, 6299–6315. [Google Scholar] [CrossRef]
- de las Heras-Saldana, S.; Clark, S.A.; Duijvesteijn, N.; Gondro, C.; van der Werf, J.H.J.; Chen, Y. Combining information from genome-wide association and multi-tissue gene expression studies to elucidate factors underlying genetic variation for residual feed intake in Australian Angus cattle. BMC Genom. 2019, 20, 939. [Google Scholar] [CrossRef]
- Lindholm-Perry, A.K.; Freetly, H.C.; Oliver, W.T.; Rempel, L.A.; Keel, B.N. Genes associated with body weight gain and feed intake identified by meta-analysis of the mesenteric fat from crossbred beef steers. PLoS ONE 2020, 15, e0227154. [Google Scholar] [CrossRef]
- Ablondi, M.; Dadousis, C.; Vasini, M.; Eriksson, S.; Mikko, S.; Sabbioni, A. Genetic diversity and signatures of selection in a native Italian horse breed based on SNP data. Animals 2020, 10, 1005. [Google Scholar] [CrossRef] [PubMed]
- Sievers, J.; Distl, O. Genomic patterns of homozygosity and genetic diversity in the Rhenish German draught horse. Genes 2025, 16, 327. [Google Scholar] [CrossRef] [PubMed]
- Vahedi, S.M.; Salek Ardestani, S.; Karimi, K.; Banabazi, M.H. Weighted single-step GWAS for body mass index and scans for recent signatures of selection in Yorkshire pigs. J. Hered. 2022, 113, 325–335. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Zha, Z.; He, Y.; Li, J.; Zhong, Z.; Xiao, Q.; Tan, Z. Genome-wide re-sequencing data reveals the population structure and selection signatures of Tunchang pigs in China. Animals 2023, 13, 1835. [Google Scholar] [CrossRef]
- Murgiano, L.; Becker, D.; Spector, C.; Carlin, K.; Santana, E.; Niggel, J.K.; Jagannathan, V.; Leeb, T.; Pearce-Kelling, S.; Aguirre, G.D.; et al. CCDC66 frameshift variant associated with a new form of early-onset progressive retinal atrophy in Portuguese Water Dogs. Sci. Rep. 2020, 10, 21162. [Google Scholar] [CrossRef]
- Dekomien, G.; Vollrath, C.; Petrasch-Parwez, E.; Boevé, M.H.; Akkad, D.A.; Gerding, W.M.; Epplen, J.T. Progressive retinal atrophy in Schapendoes dogs: Mutation of the newly identified CCDC66 gene. Neurogenetics 2010, 11, 163–174. [Google Scholar] [CrossRef]
- Volkova, N.A.; Kotova, T.O.; Vetokh, A.N.; Larionova, P.V.; Volkova, L.A.; Romanov, M.N.; Zinovieva, N.A. Genome-wide association study of testes development indicators in roosters (Gallus gallus L.). Sel’skokhozyaistvennaya Biol. (Agric. Biol.) 2024, 59, 649–657. [Google Scholar] [CrossRef]
- Lu, D.; Willard, D.; Patel, I.R.; Kadwell, S.; Overton, L.; Kost, T.; Luther, M.; Chen, W.; Woychik, R.P.; Wilkison, W.O. Agouti protein is an antagonist of the melanocyte-stimulating-hormone receptor. Nature 1994, 371, 799–802. [Google Scholar] [CrossRef]
- Cosso, G.; Carcangiu, V.; Luridiana, S.; Fiori, S.; Columbano, N.; Masala, G.; Careddu, G.M.; Sanna Passino, E.; Mura, M.C. Characterization of the Sarcidano Horse coat color genes. Animals 2022, 12, 2677. [Google Scholar] [CrossRef] [PubMed]
- Haase, B.; Brooks, S.A.; Schlumbaum, A.; Azor, P.J.; Bailey, E.; Alaeddine, F.; Mevissen, M.; Burger, D.; Poncet, P.-A.; Rieder, S.; et al. Allelic heterogeneity at the equine KIT locus in dominant white (W) horses. PLoS Genet. 2007, 3, e195. [Google Scholar] [CrossRef]
- Rönnstrand, L. Signal transduction via the stem cell factor receptor/c-Kit. Cell. Mol. Life Sci. 2004, 61, 2535–2548. [Google Scholar] [CrossRef] [PubMed]
- Henkel, J.; Lafayette, C.; Brooks, S.A.; Martin, K.; Patterson-Rosa, L.; Cook, D.; Jagannathan, V.; Leeb, T. Whole-genome sequencing reveals a large deletion in the MITF gene in horses with white spotted coat colour and increased risk of deafness. Anim. Genet. 2019, 50, 172–174. [Google Scholar] [CrossRef] [PubMed]
- Magdesian, K.G.; Tanaka, J.; Bellone, R.R. A de novo MITF deletion explains a novel splashed white phenotype in an American Paint Horse. J. Hered. 2020, 111, 287–293. [Google Scholar] [CrossRef]
- Hauswirth, R.; Haase, B.; Blatter, M.; Brooks, S.A.; Burger, D.; Drögemüller, C.; Gerber, V.; Henke, D.; Janda, J.; Jude, R.; et al. Mutations in MITF and PAX3 cause “splashed white” and other white spotting phenotypes in horses. PLoS Genet. 2012, 8, e1002653. [Google Scholar] [CrossRef]
- McFadden, A.; Martin, K.; Foster, G.; Vierra, M.; Lundquist, E.W.; Everts, R.E.; Martin, E.; Volz, E.; McLoone, K.; Brooks, S.A.; et al. Two novel variants in MITF and PAX3 associated with splashed white phenotypes in horses. J. Equine Vet. Sci. 2023, 128, 104875. [Google Scholar] [CrossRef]
- Bellone, R.R.; Tanaka, J.; Esdaile, E.; Sutton, R.B.; Payette, F.; Leduc, L.; Till, B.J.; Abdel-Ghaffar, A.K.; Hammond, M.; Magdesian, K.G. A de novo 2.3 kb structural variant in MITF explains a novel splashed white phenotype in a Thoroughbred family. Anim. Genet. 2023, 54, 752–762. [Google Scholar] [CrossRef]
- Negro, S.; Imsland, F.; Valera, M.; Molina, A.; Solé, M.; Andersson, L. Association analysis of KIT, MITF, and PAX3 variants with white markings in Spanish horses. Anim. Genet. 2017, 48, 349–352. [Google Scholar] [CrossRef]
- Promerová, M.; Andersson, L.S.; Juras, R.; Penedo, M.C.T.; Reissmann, M.; Tozaki, T.; Bellone, R.; Dunner, S.; Hořín, P.; Imsland, F.; et al. Worldwide frequency distribution of the ‘Gait keeper’ mutation in the DMRT3 gene. Anim. Genet. 2014, 45, 274–282. [Google Scholar] [CrossRef] [PubMed]
- Moazemi, I.; Mohammadabadi, M.R.; Mostafavi, A.; Esmailizadeh, A.K.; Babenko, O.I.; Bushtruk, M.V.; Tkachenko, S.V.; Stavetska, R.V.; Klopenko, N.I. Polymorphism of DMRT3 Gene and its association with body measurements in horse breeds. Russ. J. Genet. 2020, 56, 1232–1240. [Google Scholar] [CrossRef]
- Sonali; Bhardwaj, A.; Unnati; Nayan, V.; Legha, R.A.; Bhattacharya, T.K.; Pal, Y.; Giri, S.K. Identification and characterization of single nucleotide polymorphisms in DMRT3 gene in Indian horse (Equus caballus) and donkey (Equus asinus) populations. Anim. Biotechnol. 2023, 34, 4910–4920. [Google Scholar] [CrossRef] [PubMed]
- Kolberg, L.; Raudvere, U.; Kuzmin, I.; Adler, P.; Vilo, J.; Peterson, H. g:Profiler—Interoperable web service for functional enrichment analysis and gene identifier mapping (2023 update). Nucleic Acids Res. 2023, 51, W207–W212. [Google Scholar] [CrossRef]
- Nurmakhanbetov, D.M.; Sydykov, D.A.; Nasyrkhanova, B.K.; Kozhanov, Z.E. Selection and breeding work with Kazakh horse type Zhabe in peasant farms of Kazakhstan. [S Sejfullin Atyndaġy K̦az. Agroteh. Univ. Ġylym Žaršysy] Her. Sci. S Seifullin Kazakh Agro Techn. Res. Univ. Multidiscip. 2022, 3, 181–191. [Google Scholar] [CrossRef]
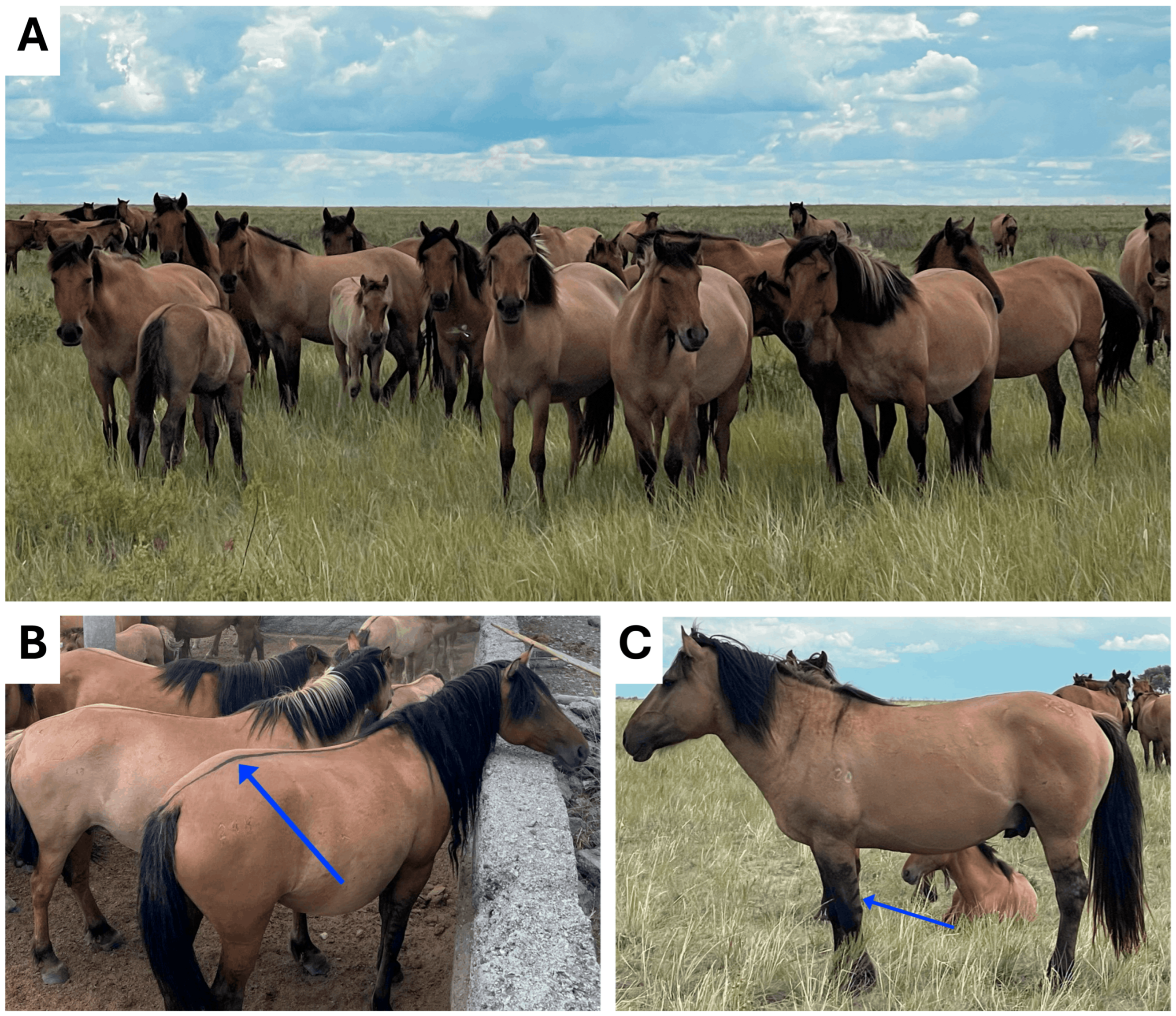
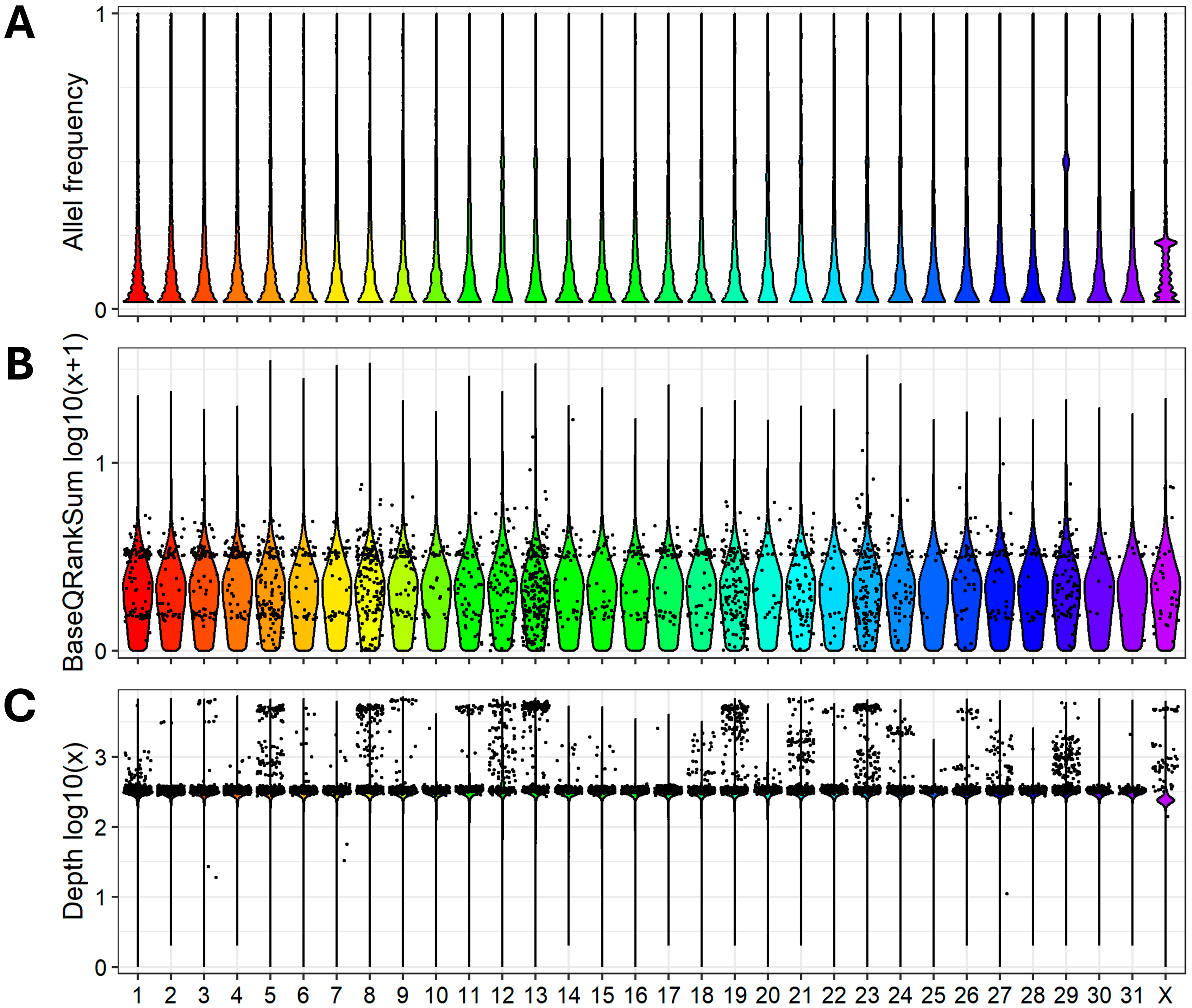
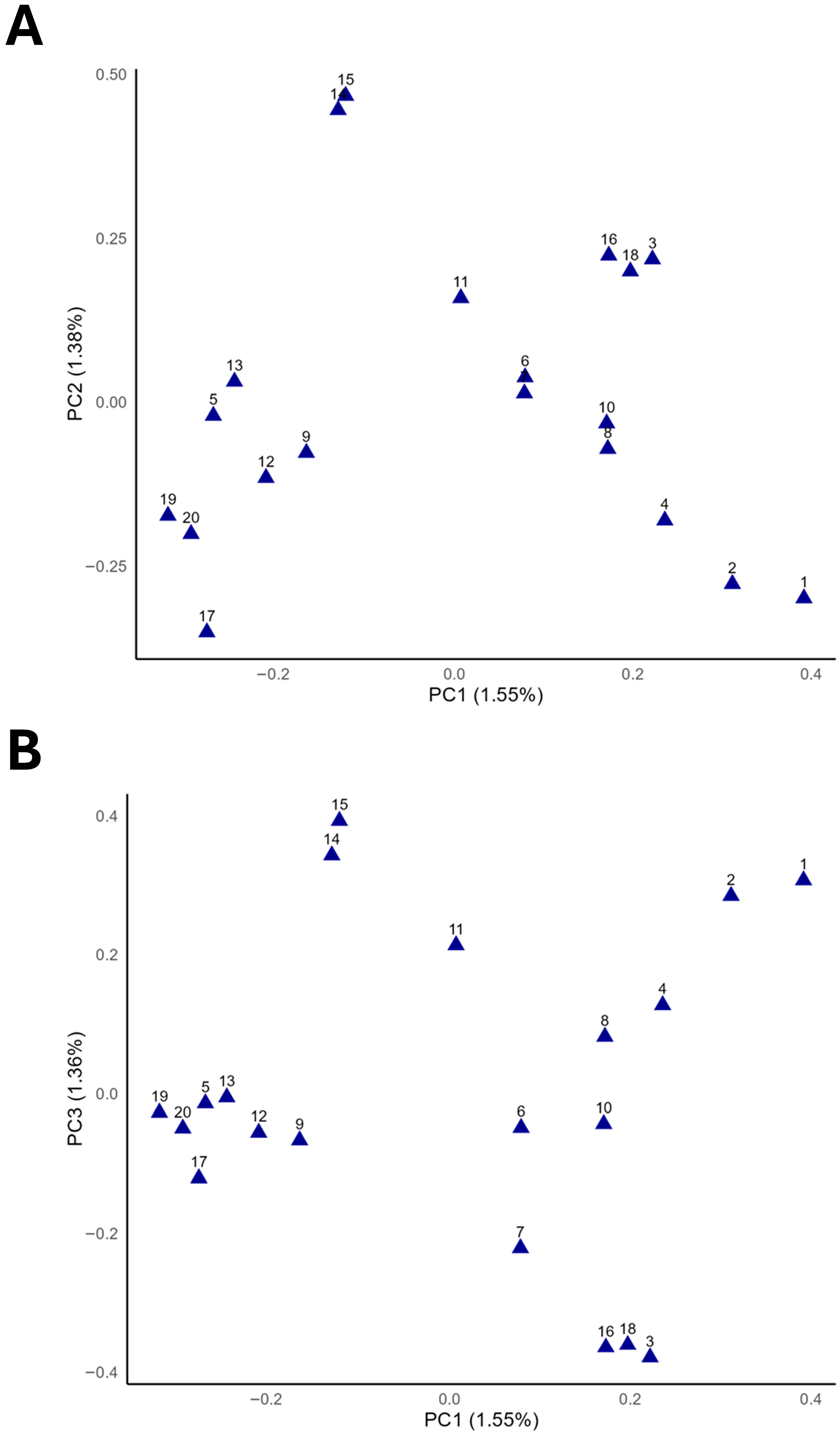
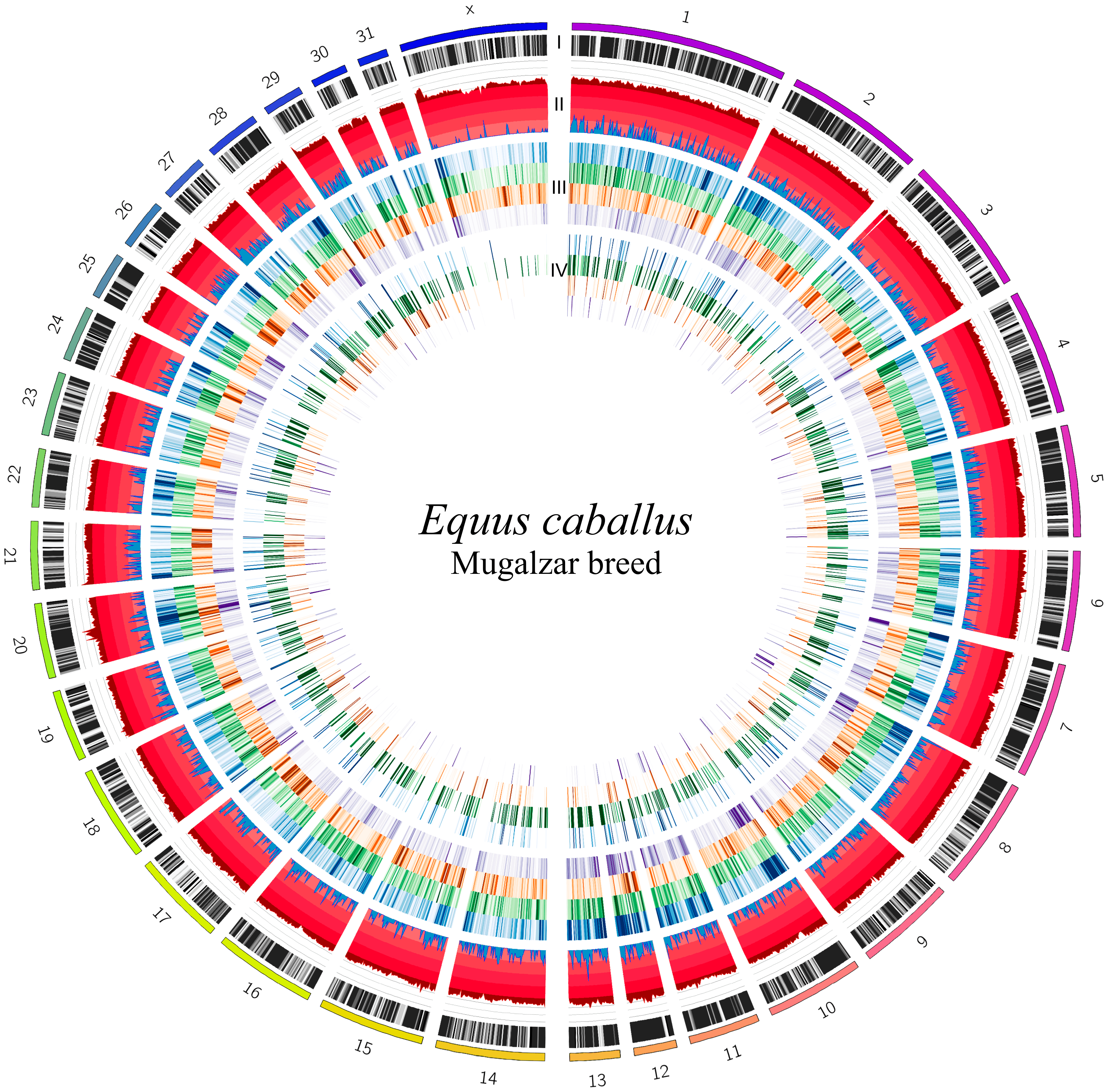
| Horse ID | Read Count | Read (Raw/Trim) | QC > 20, % | QC > 35, % | Alignment, % |
|---|---|---|---|---|---|
| 1 | 421,952,515 | 150/140 | 98.63 | 90.94 | 99.69 |
| 2 | 420,140,194 | 150/140 | 98.67 | 90.78 | 99.77 |
| 3 | 389,171,234 | 150/140 | 99.01 | 93.05 | 99.81 |
| 4 | 562,962,552 | 150/140 | 98.82 | 92.10 | 99.74 |
| 5 | 266,970,766 | 150/140 | 99.11 | 93.09 | 99.80 |
| 6 | 466,017,248 | 150/140 | 99.00 | 92.65 | 99.79 |
| 7 | 232,870,671 | 150/140 | 99.25 | 94.84 | 99.86 |
| 8 | 502,411,868 | 150/140 | 98.89 | 92.63 | 99.80 |
| 9 | 362,727,174 | 150/140 | 99.26 | 94.58 | 99.83 |
| 10 | 429,967,714 | 150/140 | 98.95 | 93.05 | 99.78 |
| 11 | 363,588,160 | 150/140 | 98.64 | 90.78 | 99.78 |
| 12 | 400,995,480 | 150/140 | 98.96 | 92.99 | 99.82 |
| 13 | 337,511,396 | 150/140 | 98.57 | 90.67 | 99.78 |
| 14 | 335,204,785 | 150/140 | 98.90 | 92.65 | 99.80 |
| 15 | 347,165,492 | 150/140 | 98.95 | 92.70 | 99.79 |
| 16 | 525,540,100 | 150/140 | 98.77 | 91.61 | 99.75 |
| 17 | 216,143,288 | 150/140 | 98.87 | 92.82 | 99.75 |
| 18 | 417,983,452 | 150/140 | 99.13 | 93.70 | 99.82 |
| 19 | 339,261,443 | 150/140 | 98.87 | 92.59 | 99.72 |
| 20 | 271,018,576 | 150/140 | 99.13 | 93.60 | 99.86 |
| ECC 1 | All Variants | Bi-Allelic Variants | ||||
|---|---|---|---|---|---|---|
| Indels | SNPs | Total | Indels | SNPs | Total | |
| 1 (NC_009144.3 2) | 147,722 | 1,357,453 | 1,505,175 | 126,369 | 1,349,565 | 1,475,934 |
| 2 (NC_009145.3) | 97,645 | 899,301 | 996,946 | 83,781 | 893,743 | 977,524 |
| 3 (NC_009146.3) | 93,450 | 868,813 | 962,263 | 80,354 | 863,936 | 944,290 |
| 4 (NC_009147.3) | 91,653 | 852,027 | 943,680 | 78,769 | 846,792 | 925,561 |
| 5 (NC_009148.3) | 76,667 | 690,517 | 767,184 | 65,553 | 686,424 | 751,977 |
| 6 (NC_009149.3) | 74,571 | 674,296 | 748,867 | 63,869 | 670,070 | 733,939 |
| 7 (NC_009150.3) | 79,382 | 727,068 | 806,450 | 68,070 | 722,632 | 790,702 |
| 8 (NC_009151.3) | 80,305 | 776,161 | 856,466 | 69,441 | 771,166 | 840,607 |
| 9 (NC_009152.3) | 64,599 | 603,074 | 667,673 | 55,591 | 599,656 | 655,247 |
| 10 (NC_009153.3) | 72,890 | 649,547 | 722,437 | 62,265 | 645,511 | 707,776 |
| 11 (NC_009154.3) | 48,745 | 419,897 | 468,642 | 41,227 | 417,041 | 458,268 |
| 12 (NC_009155.3) | 41,910 | 427,686 | 469,596 | 37,294 | 423,529 | 460,823 |
| 13 (NC_009156.3) | 38,426 | 363,283 | 401,709 | 32,834 | 360,942 | 393,776 |
| 14 (NC_009157.3) | 73,042 | 668,303 | 741,345 | 62,609 | 664,494 | 727,103 |
| 15 (NC_009158.3) | 72,635 | 679,471 | 752,106 | 62,273 | 675,408 | 737,681 |
| 16 (NC_009159.3) | 68,916 | 635,961 | 704,877 | 58,733 | 632,351 | 691,084 |
| 17 (NC_009160.3) | 69,046 | 642,029 | 711,075 | 59,593 | 638,266 | 697,859 |
| 18 (NC_009161.3) | 71,768 | 664,346 | 736,114 | 61,759 | 660,204 | 721,963 |
| 19 (NC_009162.3) | 54,275 | 508,176 | 562,451 | 46,645 | 505,024 | 551,669 |
| 20 (NC_009163.3) | 74,882 | 693,542 | 768,424 | 66,477 | 683,845 | 750,322 |
| 21 (NC_009164.3) | 50,108 | 484,203 | 534,311 | 43,275 | 481,038 | 524,313 |
| 22 (NC_009165.3) | 39,885 | 385,028 | 424,913 | 34,402 | 382,654 | 417,056 |
| 23 (NC_009166.3) | 43,983 | 398,492 | 442,475 | 37,631 | 396,114 | 433,745 |
| 24 (NC_009167.3) | 40,132 | 376,887 | 417,019 | 34,381 | 374,407 | 408,788 |
| 25 (NC_009168.3) | 30,733 | 291,433 | 322,166 | 26,275 | 289,800 | 316,075 |
| 26 (NC_009169.3) | 38,294 | 381,680 | 419,974 | 33,366 | 379,107 | 412,473 |
| 27 (NC_009170.3) | 36,748 | 348,404 | 385,152 | 31,760 | 346,037 | 377,797 |
| 28 (NC_009171.3) | 36,445 | 347,184 | 383,629 | 31,480 | 345,095 | 376,575 |
| 29 (NC_009172.3) | 32,311 | 313,570 | 345,881 | 28,037 | 311,406 | 339,443 |
| 30 (NC_009173.3) | 28,951 | 269,626 | 298,577 | 25,025 | 267,744 | 292,769 |
| 31 (NC_009174.3) | 22,581 | 212,724 | 235,305 | 19,245 | 211,489 | 230,734 |
| X (NC_009175.3) | 97,952 | 754,114 | 852,066 | 84,521 | 749,848 | 834,369 |
| Total | 1,990,652 | 18,364,296 | 20,354,948 | 1,712,904 | 18,245,338 | 19,958,242 |
| Functional Ontology Class | SNPs | Indels | Total |
|---|---|---|---|
| intergenic variant | 13,745,973 | 1,243,963 | 14,989,936 |
| intron variant | 3,359,772 | 351,967 | 3,711,739 |
| upstream gene variant | 557,181 | 60,416 | 617,597 |
| downstream gene variant | 229,356 | 24,047 | 253,403 |
| 5′ UTR variant | 132,691 | 14,854 | 147,545 |
| 3′ UTR variant | 71,784 | 8073 | 79,857 |
| missense variant | 62,111 | 0 | 62,111 |
| synonymous variant | 42,319 | 0 | 42,319 |
| non coding transcript exon variant | 23,580 | 1289 | 24,869 |
| splice region variant | 13,331 | 2009 | 15,340 |
| frameshift variant | 0 | 4444 | 4444 |
| splice donor variant | 2610 | 323 | 2933 |
| stop gained | 2336 | 64 | 2400 |
| start lost | 988 | 49 | 1037 |
| splice acceptor variant | 669 | 156 | 825 |
| inframe deletion | 0 | 801 | 801 |
| stop lost | 509 | 12 | 521 |
| inframe insertion | 0 | 403 | 403 |
| stop retained variant | 126 | 7 | 133 |
| protein altering variant | 0 | 22 | 22 |
| coding sequence variant | 2 | 5 | 7 |
| Total | 18,245,338 | 1,712,904 | 19,958,242 |
| Inbreeding Estimation Method 1 | Minimum | Maximum | Average |
|---|---|---|---|
| FGRM | −0.120 | 0.062 | −0.038 |
| FHOM | −0.149 | 0.188 | −0.033 |
| FUNI | −0.057 | 0.058 | −0.033 |
| Gene Symbol | Ensembl Gene ID | No. of Variants | ECA 1 | Start | End | No. of Orthologues | No. of Paralogues |
|---|---|---|---|---|---|---|---|
| SCAPER | ENSECAG00000017272 | 9 | 1 | 117,976,410 | 118,465,952 | 207 | 1 |
| FHAD1 | ENSECAG00000025126 | 8 | 2 | 37,672,050 | 37,824,768 | 142 | 1 |
| MMP15 | ENSECAG00000000196 | 6 | 3 | 10,831,201 | 10,851,185 | 273 | 22 |
| ADGRE1 | ENSECAG00000017237 | 5 | 7 | 4,879,448 | 4,948,792 | 103 | 50 |
| CMKLR1 | ENSECAG00000049382 | 10 | 8 | 14,730,554 | 14,789,710 | 344 | 7 |
| MRPL15 | ENSECAG00000012110 | 15 | 9 | 30,176,710 | 30,221,285 | 225 | – |
| ZNF667 | ENSECAG00000010995 | 6 | 10 | 25,714,426 | 25,740,647 | 175 | 7 |
| CCDC66 | ENSECAG00000018662 | 8 | 16 | 33,029,040 | 33,134,439 | 179 | – |
| LOC100055310 | ENSECAG00000035870 | 6 | 23 | 6,312,930 | 6,557,200 | 30 | 25 |
| Variants 1 | A1 2 | A2 2 | MAF 3 | No. Heterozygotes | Type of Variant | Gene | Phenotype |
|---|---|---|---|---|---|---|---|
| ECA3:g.36979560C > T | T | C | 0.100 | 4 | missense | MC1R | coat color, chestnut |
| ECA3:g.79538738C > T | T | C | 0.025 | 1 | missense | KIT | white spotting |
| ECA3:g.79548220T > C | T | C | 0.025 | 1 | missense | KIT | coat color, dominant white |
| ECA3:g.79566881T > C | C | T | 0.025 | 1 | missense | KIT | increased white spotting |
| ECA16:g.21555811delinsAAAT | A | C | 0.025 | 1 | deletion | MITF | splashed white |
| ECA16:g.21608936C > T | C | A | 0.075 | 3 | regulatory | MITF | white splashing |
| ECA23:g.22391254C > A | A | C | 0.025 | 1 | stop-gain | DMRT3 | gaitedness |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kassymbekova, S.N.; Bimenova, Z.Z.; Iskhan, K.Z.; Sobiech, P.; Jastrzebski, J.P.; Brym, P.; Babis, W.; Kalykova, A.S.; Otebayev, Z.M.; Kabylbekova, D.I.; et al. Uncovering Genetic Diversity and Adaptive Candidate Genes in the Mugalzhar Horse Breed Using Whole-Genome Sequencing Data. Animals 2025, 15, 2667. https://doi.org/10.3390/ani15182667
Kassymbekova SN, Bimenova ZZ, Iskhan KZ, Sobiech P, Jastrzebski JP, Brym P, Babis W, Kalykova AS, Otebayev ZM, Kabylbekova DI, et al. Uncovering Genetic Diversity and Adaptive Candidate Genes in the Mugalzhar Horse Breed Using Whole-Genome Sequencing Data. Animals. 2025; 15(18):2667. https://doi.org/10.3390/ani15182667
Chicago/Turabian StyleKassymbekova, Shinara N., Zhanat Z. Bimenova, Kairat Z. Iskhan, Przemyslaw Sobiech, Jan P. Jastrzebski, Pawel Brym, Wiktor Babis, Assem S. Kalykova, Zhassulan M. Otebayev, Dinara I. Kabylbekova, and et al. 2025. "Uncovering Genetic Diversity and Adaptive Candidate Genes in the Mugalzhar Horse Breed Using Whole-Genome Sequencing Data" Animals 15, no. 18: 2667. https://doi.org/10.3390/ani15182667
APA StyleKassymbekova, S. N., Bimenova, Z. Z., Iskhan, K. Z., Sobiech, P., Jastrzebski, J. P., Brym, P., Babis, W., Kalykova, A. S., Otebayev, Z. M., Kabylbekova, D. I., Baneh, H., & Romanov, M. N. (2025). Uncovering Genetic Diversity and Adaptive Candidate Genes in the Mugalzhar Horse Breed Using Whole-Genome Sequencing Data. Animals, 15(18), 2667. https://doi.org/10.3390/ani15182667










