Genetic Diversity and Population Structure of Five Pig Breeds from Chongqing, China
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. DNA Extraction
2.3. SNP Genotyping and Quality Control
2.4. Genetic Diversity Analysis
2.5. Analysis of Linkage Disequilibrium (LD)
2.6. Genetic Differentiation Index
2.7. Principal Component Analysis (PCA)
2.8. Phylogenetic Tree Analysis
2.9. Population Structure Analysis
2.10. TreeMix Analysis
2.11. Genetic Relationships Analysis
3. Results
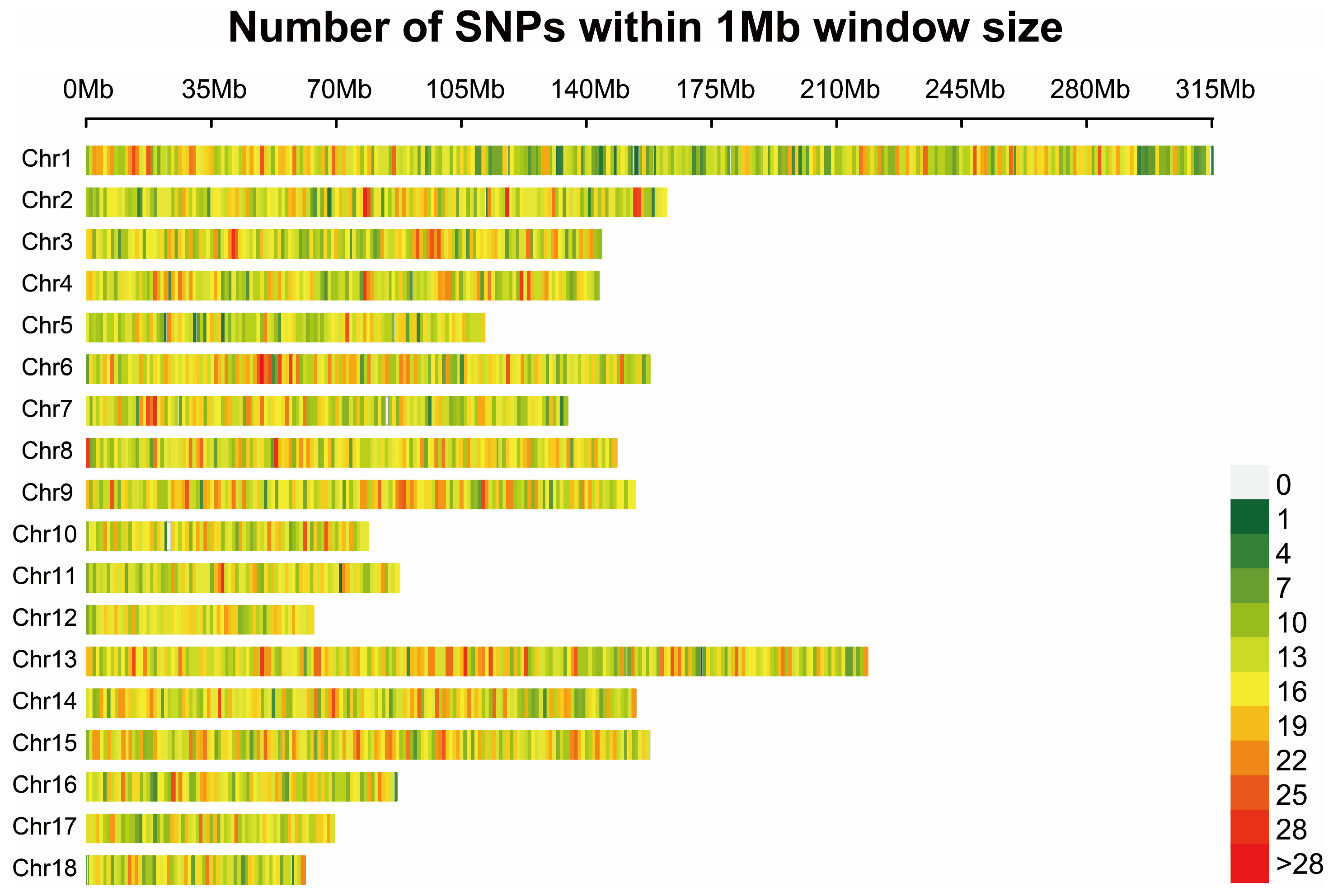
3.1. Quality Control of SNPs
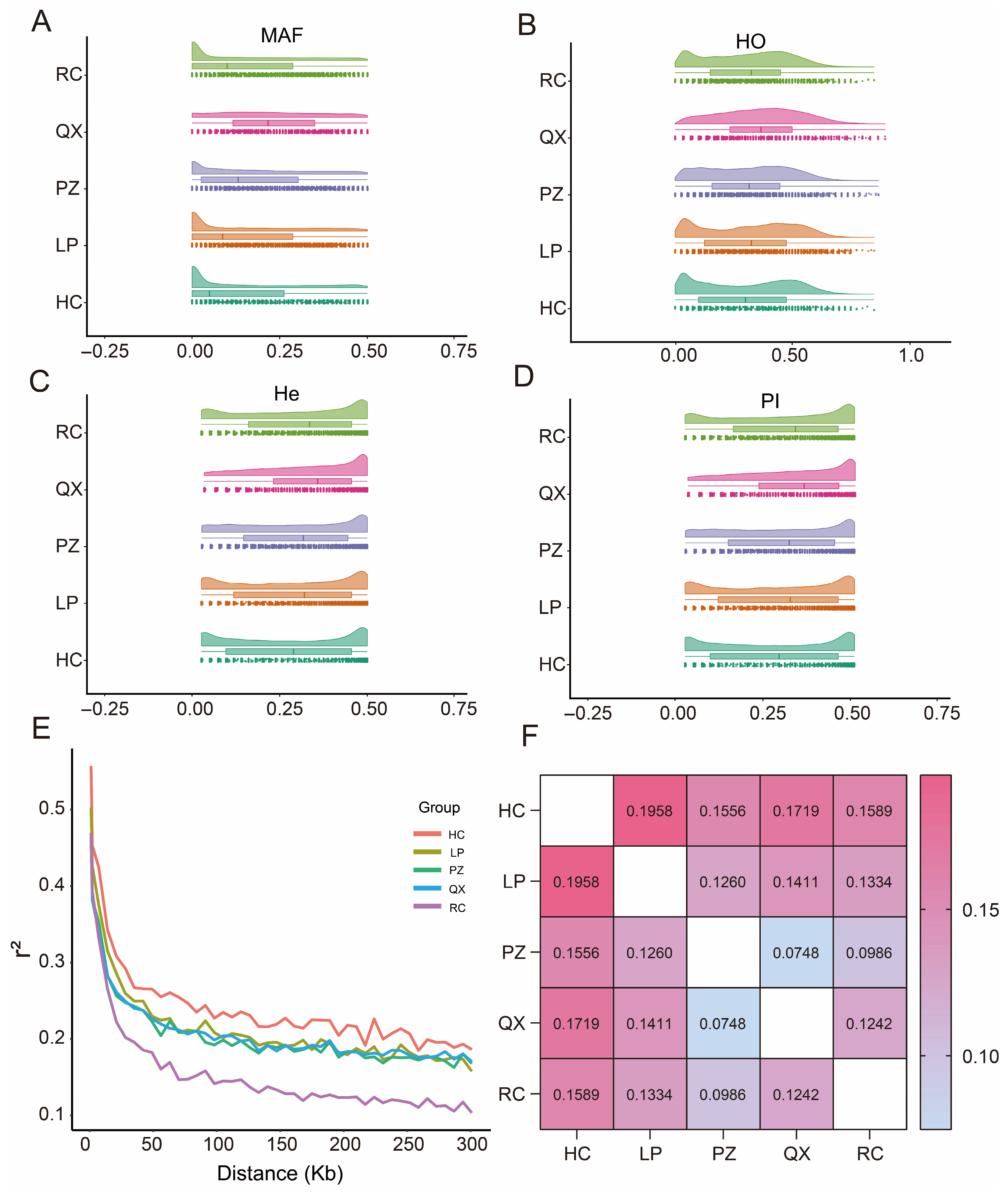
3.2. Analysis of Genetic Diversity
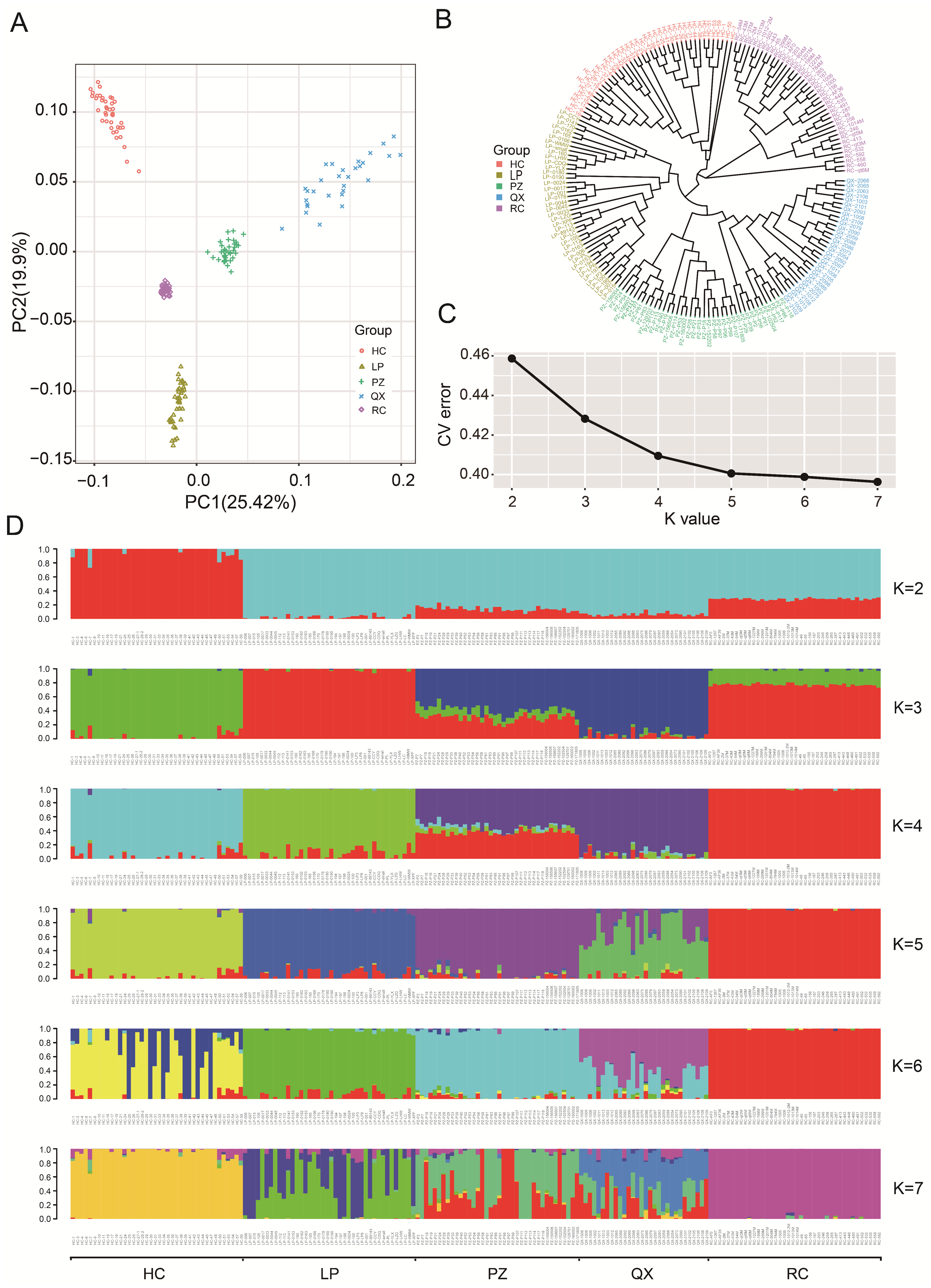
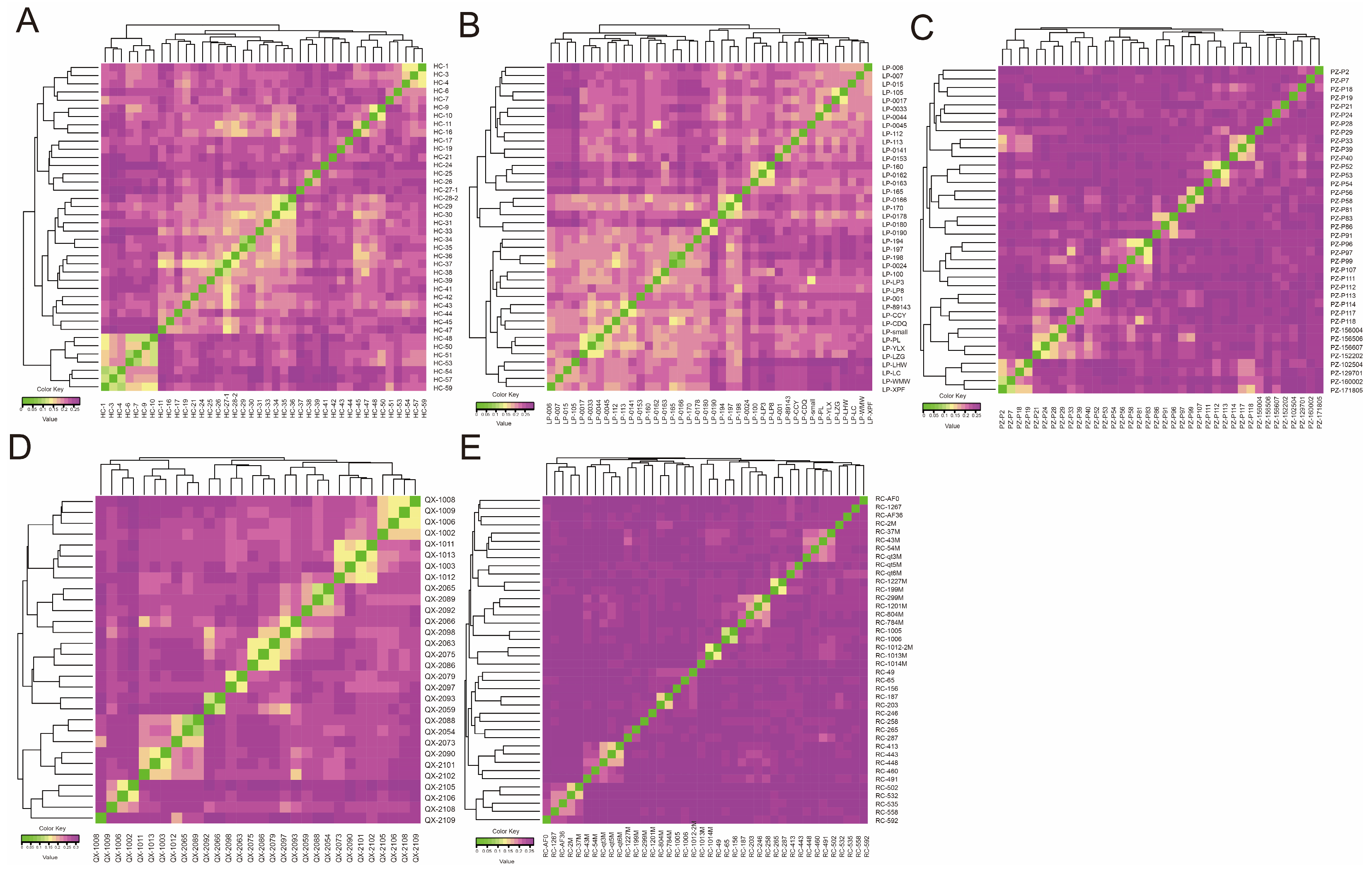
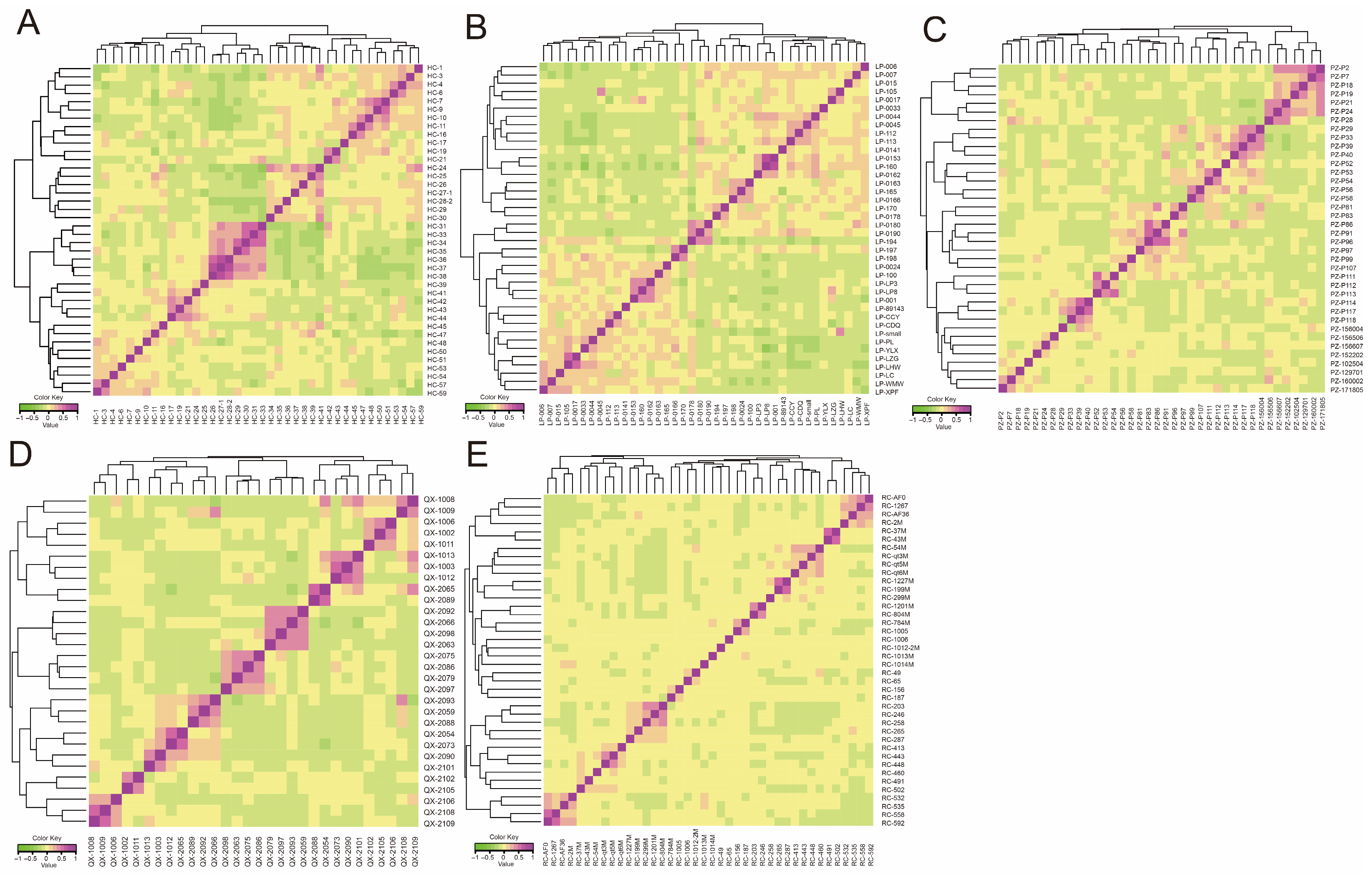
3.3. Analysis of the Population Genetic Structure and Phylogenetic Relationships
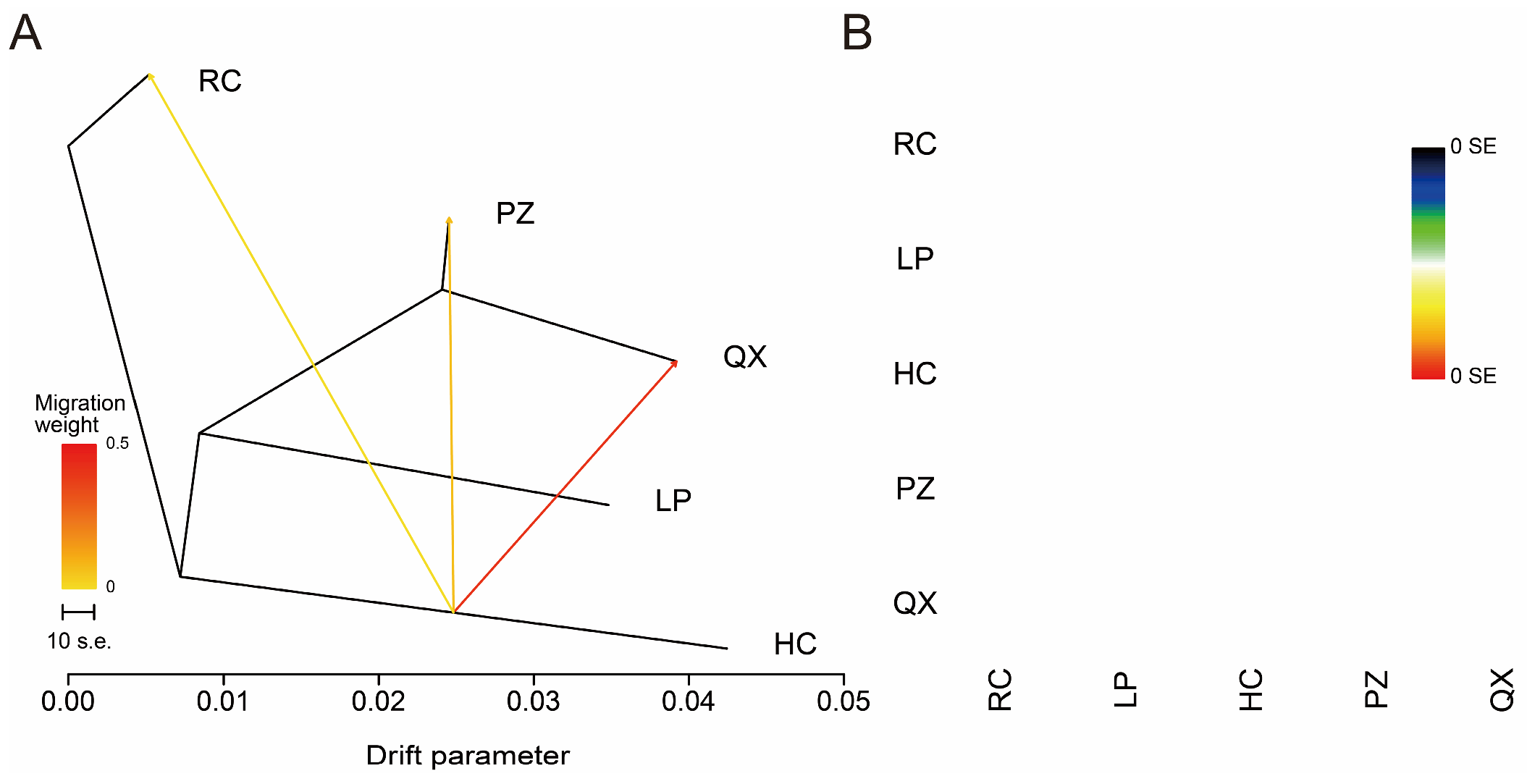
3.4. Analysis of Genetic Relationships
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cucchi, T.; Hulme-Beaman, A.; Yuan, J.; Dobney, K. Early Neolithic pig domestication at Jiahu, Henan Province, China: Clues from molar shape analyses using geometric morphometric approaches. J. Archaeol. Sci. 2011, 38, 11–22. [Google Scholar] [CrossRef]
- Wang, J.; Tang, Y.; Zheng, Y.; Jiang, L.; Ma, X.; Hou, Y.; Sun, G. Early evidence for pig domestication (8,000 cal. BP) in the Lower Yangtze, South China. Proc. Natl. Acad. Sci. USA 2025, 122, e2507123122. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Zhang, Z.; Cai, J.; Wang, C.; Liu, D.; Liu, Z.; Xu, C. Coevolution of specific gut microbiota of Min pig with host cold adaptation through enhanced vitamin B1 synthesis. Front. Microbiol. 2024, 15, 1448090. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Pan, D.; Xie, X.; Zhong, Z.; Wang, F.; Xiao, Q. Genome-wide detection of runs of homozygosity in Ding’an pigs revealed candidate genes relating to meat quality traits. BMC Genom. 2025, 26, 316. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Zha, Z.; He, Y.; Li, J.; Zhong, Z.; Xiao, Q.; Tan, Z. Genome-Wide Re-Sequencing Data Reveals the Population Structure and Selection Signatures of Tunchang Pigs in China. Animals 2023, 13, 1835. [Google Scholar] [CrossRef]
- Wu, D.D.; Yang, C.P.; Wang, M.S.; Dong, K.Z.; Yan, D.W.; Hao, Z.Q.; Fan, S.Q.; Chu, S.Z.; Shen, Q.S.; Jiang, L.P.; et al. Convergent genomic signatures of high-altitude adaptation among domestic mammals. Natl. Sci. Rev. 2020, 7, 952–963. [Google Scholar] [CrossRef]
- Ai, H.; Yang, B.; Li, J.; Xie, X.; Chen, H.; Ren, J. Population history and genomic signatures for high-altitude adaptation in Tibetan pigs. BMC Genom. 2014, 15, 834. [Google Scholar] [CrossRef]
- Zhao, P.; Yu, Y.; Feng, W.; Du, H.; Yu, J.; Kang, H.; Zheng, X.; Wang, Z.; Liu, G.E.; Ernst, C.W.; et al. Evidence of evolutionary history and selective sweeps in the genome of Meishan pig reveals its genetic and phenotypic characterization. GigaScience 2018, 7, giy058. [Google Scholar] [CrossRef]
- Zhang, Q.; Du, M.; Wei, S.; Zhu, L.; Yan, R.; Jin, M.; Wang, Y. Variation of Meat Quality and Relationship to Gut Microbiota Among Different Pig Breeds. Microb. Biotechnol. 2025, 18, e70139. [Google Scholar] [CrossRef]
- Pu, G.; Hou, L.; Zhao, Q.; Liu, G.; Wang, Z.; Zhou, W.; Niu, P.; Wu, C.; Li, P.; Huang, R. Interactions between gut microbes and host promote degradation of various fiber components in Meishan pigs. mSystems 2025, 10, e0150024. [Google Scholar] [CrossRef]
- Liu, H.; He, J.; Yuan, Z.; Xie, K.; He, Z.; Zhou, X.; Wang, M.; He, J. Metabolomics Analysis Provides Novel Insights into the Difference in Meat Quality between Different Pig Breeds. Foods 2023, 12, 3476. [Google Scholar] [CrossRef]
- Xu, B.; Qin, W.; Chen, Y.; Tang, Y.; Zhou, S.; Huang, J.; Ma, L.; Yan, X. Multi-omics analysis reveals gut microbiota-ovary axis contributed to the follicular development difference between Meishan and Landrace × Yorkshire sows. J. Anim. Sci. Biotechnol. 2023, 14, 68. [Google Scholar] [CrossRef]
- Bosse, M.; Megens, H.J.; Frantz, L.A.; Madsen, O.; Larson, G.; Paudel, Y.; Duijvesteijn, N.; Harlizius, B.; Hagemeijer, Y.; Crooijmans, R.P.; et al. Genomic analysis reveals selection for Asian genes in European pigs following human-mediated introgression. Nat. Commun. 2014, 5, 4392. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, S.; Lu, Z.H.; Megens, H.J.; Archibald, A.L.; Haley, C.; Jackson, I.J.; Groenen, M.A.; Crooijmans, R.P.; Ogden, R.; Wiener, P. Signatures of diversifying selection in European pig breeds. PLoS Genet. 2013, 9, e1003453. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Liu, Z.; Lu, S.Y.; Jiang, L.; Zhou, L.; Liu, J.F. Genomic evidence for human-mediated introgressive hybridization and selection in the developed breed. BMC Genom. 2024, 25, 331. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Wang, L.; Ma, Z.; Mao, Y.; Wang, G.; Zheng, R.; Zuo, B.; Wang, Y. Analysis of the Genetic Diversity and Genetic Structure of Jiangshan Black Pigs Using Single Nucleotide Polymorphism (SNP) Chips. Animals 2024, 14, 2660. [Google Scholar] [CrossRef]
- Hu, Z.; Su, Y.; Zong, W.; Niu, N.; Zhao, R.; Liang, R.; Wang, L.; Zhang, Y.; Zhang, L. Unveiling the Genetic Secrets of Chinese Indigenous Pigs from Guizhou Province: Diversity, Evolution and Candidate Genes Affecting Pig Coat Color. Animals 2024, 14, 699. [Google Scholar] [CrossRef]
- Yan, Z.; Song, K.; Wang, P.; Gun, S.; Long, X. Evaluation of the Genetic Diversity and Population Structure of Four Native Pig Populations in Gansu Province. Int. J. Mol. Sci. 2023, 24, 17154. [Google Scholar] [CrossRef]
- Ma, X.; Chen, Z.; Zhang, Z.; Liu, S.; Wang, M.; Zhang, X.; Shi, J.; Gao, H.; Gu, J.; Han, H.; et al. Comprehensive genomic analysis and selection signature detection in endangered Beigang pigs using whole-genome sequencing data. Anim. Genet. 2025, 56, e13502. [Google Scholar] [CrossRef]
- National Committee of Animal Genetic Resources Organization in China. Animal Genetic Resources in China: Pigs; China Agriculture Press: Beijing, China, 2011. [Google Scholar]
- Xing, L.; Lu, X.; Zhang, W.; Wang, Q.; Zhang, W. Genetic Structure and Genome-Wide Association Analysis of Growth and Reproductive Traits in Fengjing Pigs. Animals 2024, 14, 2449. [Google Scholar] [CrossRef]
- Yuan, J.; Zhou, X.; Xu, G.; Xu, S.; Liu, B. Genetic diversity and population structure of Tongcheng pigs in China using whole-genome SNP chip. Front. Genet. 2022, 13, 910521. [Google Scholar] [CrossRef]
- Meng, F.; Cai, J.; Wang, C.; Fu, D.; Di, S.; Wang, X.; Chang, Y.; Xu, C. Single nucleotide polymorphism-based analysis of the genetic structure of the Min pig conserved population. Anim. Biosci. 2022, 35, 1839–1849. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, H.; Huang, M.; Tang, J.; Yang, L.; Yu, Z.; Li, D.; Li, G.; Jiang, Y.; Sun, Y.; et al. Whole-genome SNP markers reveal conservation status, signatures of selection, and introgression in Chinese Laiwu pigs. Evol. Appl. 2021, 14, 383–398. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Jing, W.; Samuels, D.C.; Sheng, Q.; Shyr, Y.; Guo, Y. Strategies for processing and quality control of Illumina genotyping arrays. Brief. Bioinform. 2018, 19, 765–775. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Barbato, M.; Orozco-terWengel, P.; Tapio, M.; Bruford, M.W. SNeP: A tool to estimate trends in recent effective population size trajectories using genome-wide SNP data. Front. Genet. 2015, 6, 109. [Google Scholar] [CrossRef]
- Zhang, C.; Dong, S.S.; Xu, J.Y.; He, W.M.; Yang, T.L. PopLDdecay: A fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics 2019, 35, 1786–1788. [Google Scholar] [CrossRef]
- Yang, J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Alexander, D.H.; Lange, K. Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinform. 2011, 12, 246. [Google Scholar] [CrossRef] [PubMed]
- Pickrell, J.K.; Pritchard, J.K. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 2012, 8, e1002967. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.X.; Wang, Z.G.; Sun, F.Z.; Chen, W.S.; Yang, G.Y.; Guo, S.J.; Li, Y.J.; Zhao, X.L.; Zhang, Y.; Sun, J.; et al. Genetic diversity of microsatellite loci in fifty-six Chinese native pig breeds. Yi Chuan Xue Bao 2003, 30, 225–233. [Google Scholar] [PubMed]
- Wang, Y.; Dong, R.; Li, X.; Cui, C.; Yu, G. Analysis of the Genetic Diversity and Family Structure of the Licha Black Pig Population on Jiaodong Peninsula, Shandong Province, China. Animals 2022, 12, 1045. [Google Scholar] [CrossRef]
- Yang, S.L.; Wang, Z.G.; Liu, B.; Zhang, G.X.; Zhao, S.H.; Yu, M.; Fan, B.; Li, M.H.; Xiong, T.A.; Li, K. Genetic variation and relationships of eighteen Chinese indigenous pig breeds. Genet. Sel. Evol. 2003, 35, 657–671. [Google Scholar] [CrossRef]
- Liu, B.; Shen, L.; Guo, Z.; Gan, M.; Chen, Y.; Yang, R.; Niu, L.; Jiang, D.; Zhong, Z.; Li, X.; et al. Single nucleotide polymorphism-based analysis of the genetic structure of Liangshan pig population. Anim. Biosci. 2021, 34, 1105–1115. [Google Scholar] [CrossRef]
- Xu, P.; Wang, X.; Ni, L.; Zhang, W.; Lu, C.; Zhao, X.; Zhao, X.; Ren, J. Genome-wide genotyping uncovers genetic diversity, phylogeny, signatures of selection, and population structure of Chinese Jiangquhai pigs in a global perspective1. J. Anim. Sci. 2019, 97, 1491–1500. [Google Scholar] [CrossRef]
- Mariani, E.; Summer, A.; Ablondi, M.; Sabbioni, A. Genetic Variability and Management in Nero di Parma Swine Breed to Preserve Local Diversity. Animals 2020, 10, 538. [Google Scholar] [CrossRef]
- Muñoz, M.; Bozzi, R.; García-Casco, J.; Núñez, Y.; Ribani, A.; Franci, O.; García, F.; Škrlep, M.; Schiavo, G.; Bovo, S.; et al. Genomic diversity, linkage disequilibrium and selection signatures in European local pig breeds assessed with a high density SNP chip. Sci. Rep. 2019, 9, 13546. [Google Scholar] [CrossRef]
- Hedrick, P.W. Assortative Mating and Linkage Disequilibrium. G3 2017, 7, 55–62. [Google Scholar] [CrossRef]
- Ai, H.; Huang, L.; Ren, J. Genetic diversity, linkage disequilibrium and selection signatures in chinese and Western pigs revealed by genome-wide SNP markers. PLoS ONE 2013, 8, e56001. [Google Scholar] [CrossRef]
- Amaral, A.J.; Megens, H.J.; Crooijmans, R.P.; Heuven, H.C.; Groenen, M.A. Linkage disequilibrium decay and haplotype block structure in the pig. Genetics 2008, 179, 569–579. [Google Scholar] [CrossRef]
- Yuan, H.; Wei, W.; Zhang, Y.; Li, C.; Zhao, S.; Chao, Z.; Xia, C.; Quan, J.; Gao, C. Unveiling the Influence of Copy Number Variations on Genetic Diversity and Adaptive Evolution in China’s Native Pig Breeds via Whole-Genome Resequencing. Int. J. Mol. Sci. 2024, 25, 5843. [Google Scholar] [CrossRef]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef]
| Breed | Number of Individuals | Location |
|---|---|---|
| HC | 40 | Hechuan |
| LP | 40 | Tongnan |
| PZ | 38 | Fulin |
| QX | 30 | Fengdu |
| RC | 40 | Rongchang |
| Breed | MAF | He | Ho | Ne | PN | Pi |
|---|---|---|---|---|---|---|
| HC | 0.135 | 0.2751 | 0.2958 | 2.9 | 0.5090 | 0.2785 |
| LP | 0.150 | 0.2884 | 0.3144 | 4.1 | 0.5644 | 0.2921 |
| PZ | 0.173 | 0.2948 | 0.3084 | 3.5 | 0.7002 | 0.2987 |
| QX | 0.229 | 0.3302 | 0.3624 | 2.7 | 0.8946 | 0.3359 |
| RC | 0.153 | 0.3012 | 0.3044 | 4.6 | 0.5744 | 0.305 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Long, X.; Zhang, L.; Pan, Y.; Zhang, L.; Tu, Z.; Zhang, L.; Wang, Q.; Pan, H.; Guo, Z. Genetic Diversity and Population Structure of Five Pig Breeds from Chongqing, China. Animals 2025, 15, 2610. https://doi.org/10.3390/ani15172610
Long X, Zhang L, Pan Y, Zhang L, Tu Z, Zhang L, Wang Q, Pan H, Guo Z. Genetic Diversity and Population Structure of Five Pig Breeds from Chongqing, China. Animals. 2025; 15(17):2610. https://doi.org/10.3390/ani15172610
Chicago/Turabian StyleLong, Xi, Lidan Zhang, Yu Pan, Liang Zhang, Zhi Tu, Lijuan Zhang, Qing Wang, Hongmei Pan, and Zongyi Guo. 2025. "Genetic Diversity and Population Structure of Five Pig Breeds from Chongqing, China" Animals 15, no. 17: 2610. https://doi.org/10.3390/ani15172610
APA StyleLong, X., Zhang, L., Pan, Y., Zhang, L., Tu, Z., Zhang, L., Wang, Q., Pan, H., & Guo, Z. (2025). Genetic Diversity and Population Structure of Five Pig Breeds from Chongqing, China. Animals, 15(17), 2610. https://doi.org/10.3390/ani15172610





