Transcriptome and Metabolome Profiles Reveal the Underlying Mechanism of Fat Deposition Changes in Three-Way Crossbred Yak for High-Quality Beef Production
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Animals and Data Collection
2.3. RNA Extraction and Transcriptome Analysis
2.4. Non-Targeted LC-MS/MS Metabolomics Analysis
2.5. Integrated Analysis of the Transcriptome and Metabolome
2.6. Statistical Analyses
3. Results
3.1. XF Possess Enhanced Growth Characteristics and Meat Quality
3.2. Identification of Differentially Expressed Genes (DEGs)
3.3. DEGs Are Enriched in Immune, Oxidation, and Fat Metabolism
3.4. Identification of Differential Metabolites
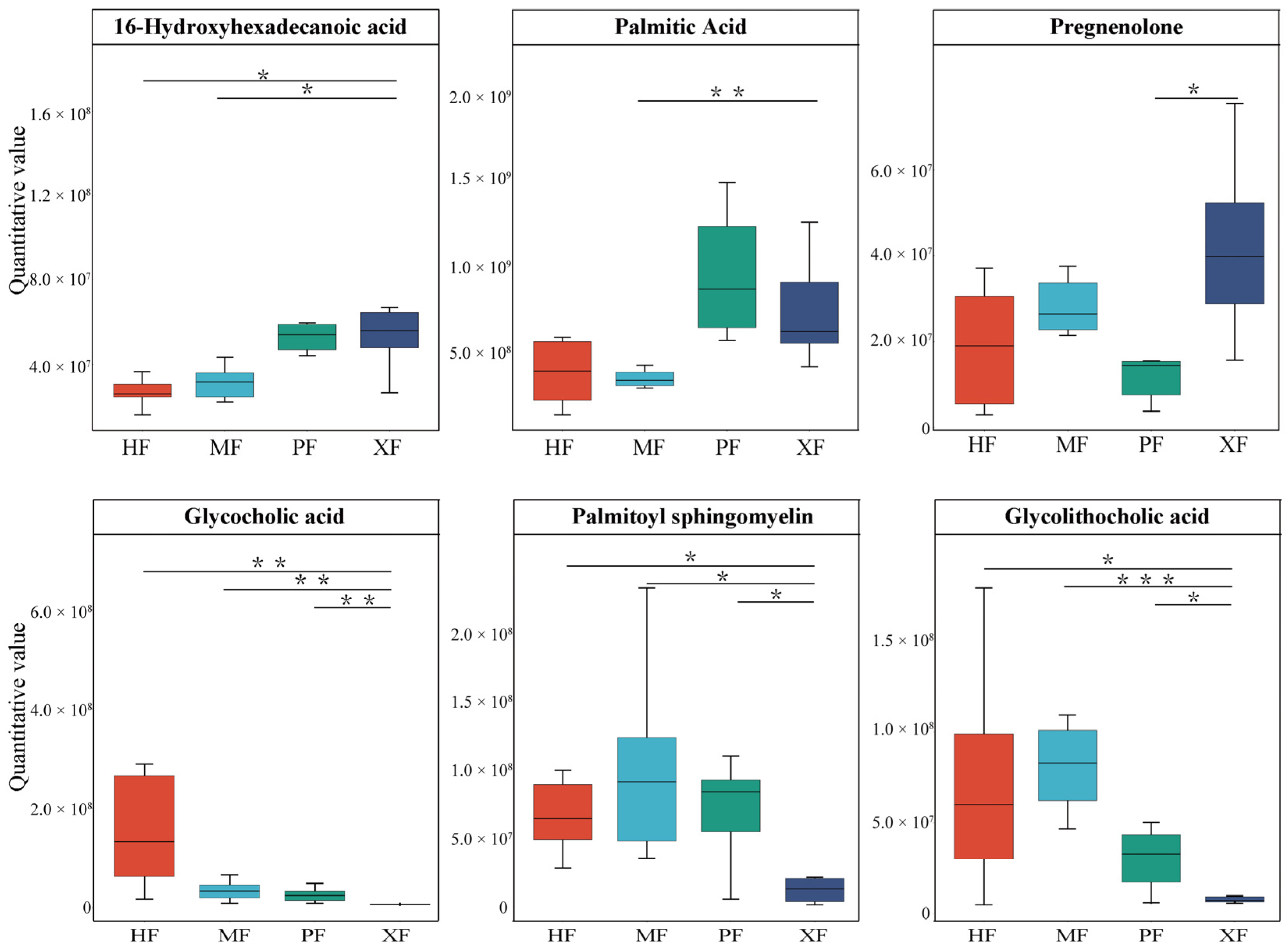
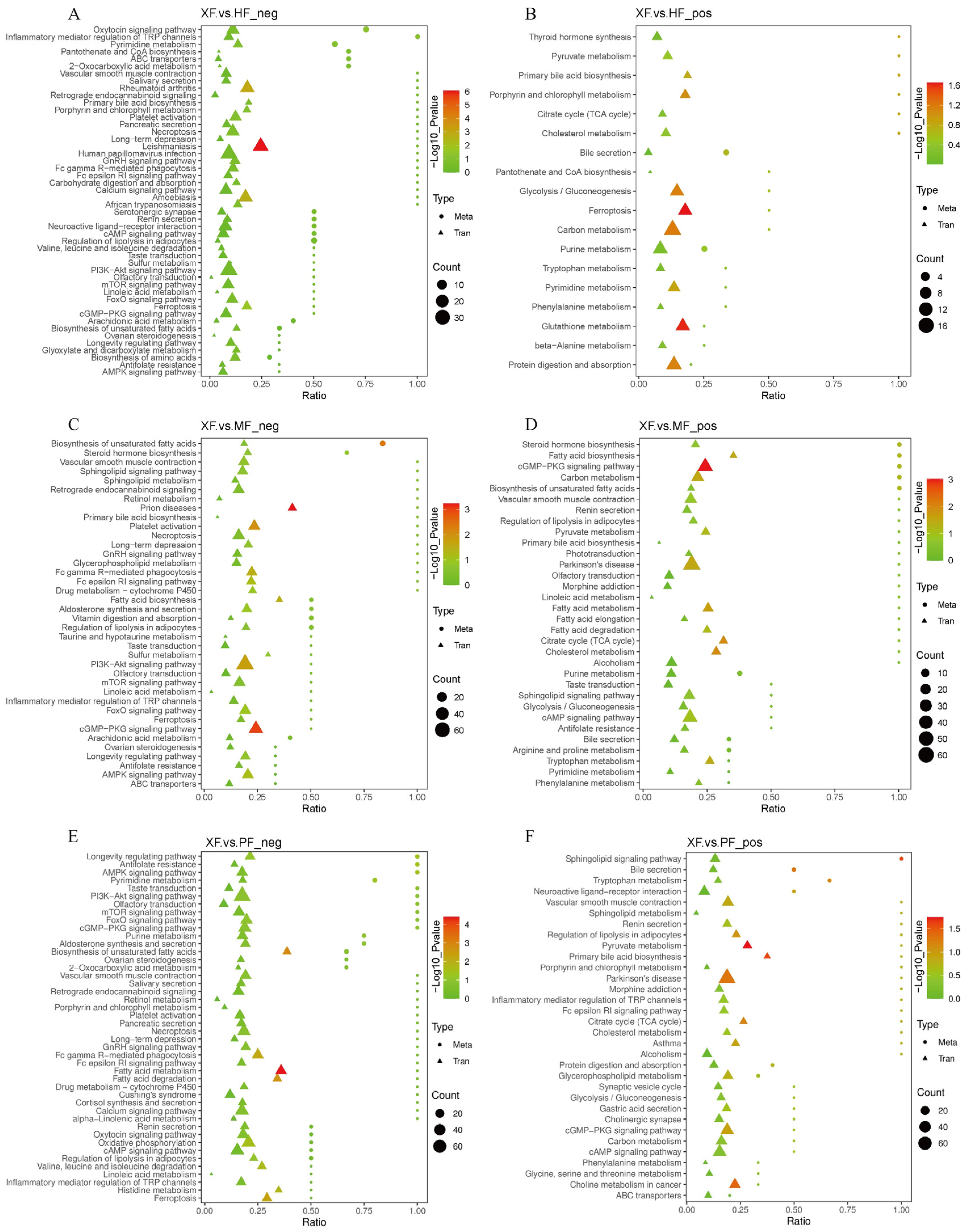
3.5. DMs Are Enriched in Fatty Acid Biosynthesis and Hormone Metabolism
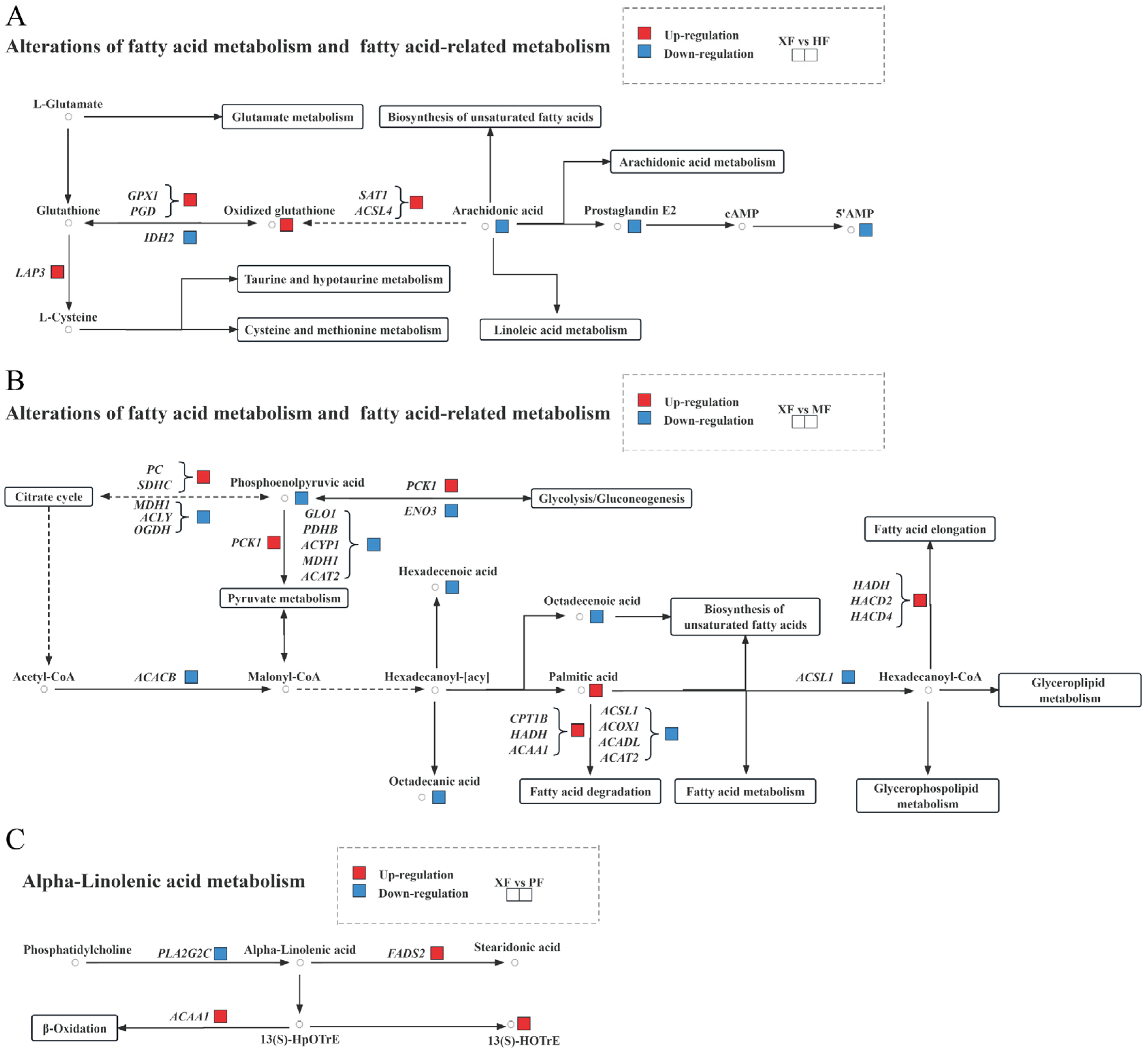
3.6. Integrated Analysis Reveals Promising Candidates Associated with Meat Quality
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| DEG | differentially expressed gene |
| DM | differential metabolite |
| FC | fold change |
| FUFA | functional unsaturated fatty acid |
| GC-MS | gas chromatography–mass spectrometer |
| GO | gene ontology |
| GSEA | gene set enrichment analysis |
| HF | Tibetan yellow cattle |
| LC-MS | liquid chromatography–mass spectrometry |
| LC-MS/MS | liquid chromatography with tandem mass spectrometry |
| MF | Tibetan yak |
| MUFA | monounsaturated fatty acid |
| PCA | principal component analysis |
| PF | cattle–yak |
| PLS-DA | partial least squares discriminant analysis |
| PUFA | polyunsaturated fatty acid |
| SFA | saturated fatty acid |
| VIP | variable importance in projection |
| XF | Yajiangxue cattle |
References
- Royden, L.H.; Burchfiel, B.C.; van der Hilst, R.D. The geological evolution of the Tibetan Plateau. Science 2008, 321, 1054–1058. [Google Scholar] [CrossRef]
- Kang, S.; Xu, Y.; You, Q.; Flügel, W.-A.; Pepin, N.; Yao, T. Review of climate and cryospheric change in the Tibetan Plateau. Environ. Res. Lett. 2010, 5, 015101. [Google Scholar] [CrossRef]
- Cao, X.-K.; Cheng, J.; Huang, Y.-Z.; Wang, X.-G.; Ma, Y.-L.; Peng, S.-J.; Chaogetu, B.; Zhuoma, Z.; Chen, H. Growth performance and meat quality evaluations in three-way cross cattle developed for the Tibetan Plateau and their molecular understanding by integrative omics analysis. J. Agric. Food Chem. 2018, 67, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Wood, J.; Enser, M.; Fisher, A.; Nute, G.; Sheard, P.; Richardson, R.; Hughes, S.; Whittington, F. Fat deposition, fatty acid composition and meat quality: A review. Meat Sci. 2008, 78, 343–358. [Google Scholar] [CrossRef] [PubMed]
- Pitchford, W.; Deland, M.; Siebert, B.; Malau-Aduliand, A.; Bottema, C. Genetic variation in fatness and fatty acid composition of crossbred cattle. J. Anim. Sci. 2002, 80, 2825–2832. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Huang, Y.; Yang, Z.; Ma, Y.; Chaogetu, B.; Zhuoma, Z.; Chen, H. RNA-Seq analysis identifies differentially expressed genes in subcutaneous adipose tissue in qaidaford cattle, cattle-yak, and angus cattle. Animals 2019, 9, 1077. [Google Scholar] [CrossRef]
- You, Z.; Jin, Z.; Mingliang, Z. Research progress on interspecific hybridization and utilization of yak. Cao Xue 2025, 1, 67–70+74. [Google Scholar]
- Motoyama, M.; Sasaki, K.; Watanabe, A. Wagyu and the factors contributing to its beef quality: A Japanese industry overview. Meat Sci. 2016, 120, 10–18. [Google Scholar] [CrossRef]
- Wood, J.; Richardson, R.; Nute, G.; Fisher, A.; Campo, M.; Kasapidou, E.; Sheard, P.; Enser, M. Effects of fatty acids on meat quality: A review. Meat Sci. 2004, 66, 21–32. [Google Scholar] [CrossRef]
- Wang, Q.; Wu, J.; Zhang, S.; Zhang, Y.; Zhang, H.; Fan, E. GC analysis of the fatty acid composition of yak kidney. Chromatographia 2009, 69, 139–143. [Google Scholar] [CrossRef]
- Gu, X.; Sun, W.; Yi, K.; Yang, L.; Chi, F.; Luo, Z.; Wang, J.; Zhang, J.; Wang, W.; Yang, T. Comparison of muscle lipidomes between cattle-yak, yak, and cattle using UPLC–MS/MS. J. Food Compos. Anal. 2021, 103, 104113. [Google Scholar] [CrossRef]
- Li, W.; Wang, J.; Zhang, C.; Wang, N.; Zhang, C.; Chen, W.; Wu, T. Using an integrated feature-based molecular network and lipidomics approach to reveal the differential lipids in yak shanks and flanks. Food Chem. 2023, 403, 134352. [Google Scholar] [CrossRef]
- Xiong, L.; Pei, J.; Wang, X.; Guo, S.; Guo, X.; Yan, P. Lipidomics and transcriptome reveal the effects of feeding systems on fatty acids in yak’s meat. Foods 2022, 11, 2582. [Google Scholar] [CrossRef]
- Xiong, L.; Pei, J.; Wang, X.; Guo, S.; Guo, X.; Yan, P. Effect of lipids in yak muscle under different feeding systems on meat quality based on untargeted lipidomics. Animals 2022, 12, 2814. [Google Scholar] [CrossRef]
- Li, J.; Zhang, D.; Yin, L.; Li, Z.; Yu, C.; Du, H.; Jiang, X.; Yang, C.; Liu, Y. Integration analysis of metabolome and transcriptome profiles revealed the age-dependent dynamic change in chicken meat. Food Res. Int. 2022, 156, 111171. [Google Scholar] [CrossRef]
- Bischof, G.; Witte, F.; Terjung, N.; Januschewski, E.; Heinz, V.; Juadjur, A.; Gibis, M. Effect of sampling position in fresh, dry-aged and wet-aged beef from M. Longissimus dorsi of Simmental cattle analyzed by 1H NMR spectroscopy. Food Res. Int. 2022, 156, 111334. [Google Scholar] [CrossRef]
- de Oliveira, E.S.; Lião, L.M.; Silva, A.K.; Prado, C.S.; Sena, M.M.; Oliveira, G.d.A.R. A t-test ranking-based discriminant analysis for classification of free-range and barn-raised broiler chickens by 1H NMR spectroscopy. Food Chem. 2023, 399, 134004. [Google Scholar] [CrossRef]
- Xiao, Z.; Ge, C.; Zhou, G.; Zhang, W.; Liao, G. 1H NMR-based metabolic characterization of Chinese Wuding chicken meat. Food Chem. 2019, 274, 574–582. [Google Scholar] [CrossRef]
- Yuan, X.; Shi, W.; Jiang, J.; Li, Z.; Fu, P.; Yang, C.; Rehman, S.U.; Pauciullo, A.; Liu, Q.; Shi, D. Comparative metabolomics analysis of milk components between Italian Mediterranean buffaloes and Chinese Holstein cows based on LC-MS/MS technology. PLoS ONE 2022, 17, e0262878. [Google Scholar] [CrossRef]
- Chen, X.; Cao, J.; Geng, A.; Zhang, X.; Wang, H.; Chu, Q.; Yan, Z.; Zhang, Y.; Liu, H.; Zhang, J. Integration of GC-MS and LC-MS for metabolite characteristics of thigh meat between fast-and slow-growing broilers at marketable age. Food Chem. 2023, 403, 134362. [Google Scholar] [CrossRef]
- Cheng, J.; Pan, Y.; Yang, S.; Wei, Y.; Lv, Q.; Xing, Q.; Zhang, R.; Sun, L.; Qin, G.; Shi, D. Integration of transcriptomics and non-targeted metabolomics reveals the underlying mechanism of follicular atresia in Chinese buffalo. J. Steroid Biochem. Mol. Biol. 2021, 212, 105944. [Google Scholar] [CrossRef]
- Zhan, H.; Xiong, Y.; Wang, Z.; Dong, W.; Zhou, Q.; Xie, S.; Li, X.; Zhao, S.; Ma, Y. Integrative analysis of transcriptomic and metabolomic profiles reveal the complex molecular regulatory network of meat quality in Enshi black pigs. Meat Sci. 2022, 183, 108642. [Google Scholar] [CrossRef]
- Eom, J.S.; Lee, S.J.; Gu, B.-H.; Lee, S.J.; Lee, S.-S.; Kim, S.-H.; Kim, B.-W.; Lee, S.S.; Kim, M. Metabolomic and transcriptomic study to understand changes in metabolic and immune responses in steers under heat stress. Anim. Nutr. 2022, 11, 87–101. [Google Scholar] [CrossRef]
- NY/T 815-2004; Feeding Standard of Beef Cattle. MAPRC: Beijing, China, 2004.
- GB/T 29392-2022; Quality Grading for Livestock and Poultry Meat-Beef. Standards Press of China: Beijing, China, 2022.
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Want, E.J.; Masson, P.; Michopoulos, F.; Wilson, I.D.; Theodoridis, G.; Plumb, R.S.; Shockcor, J.; Loftus, N.; Holmes, E.; Nicholson, J.K. Global metabolic profiling of animal and human tissues via UPLC-MS. Nat. Protoc. 2013, 8, 17–32. [Google Scholar] [CrossRef]
- Wen, B.; Mei, Z.; Zeng, C.; Liu, S. metaX: A flexible and comprehensive software for processing metabolomics data. BMC Bioinform. 2017, 18, 183. [Google Scholar] [CrossRef]
- Zhou, J.; Liu, H.; Zhong, C.; Degen, A.; Yang, G.; Zhang, Y.; Qian, J.; Wang, W.; Hao, L.; Qiu, Q. Apparent digestibility, rumen fermentation, digestive enzymes and urinary purine derivatives in yaks and Qaidam cattle offered forage-concentrate diets differing in nitrogen concentration. Livest. Sci. 2018, 208, 14–21. [Google Scholar] [CrossRef]
- Sierra, V.; Guerrero, L.; Fernández-Suárez, V.; Martínez, A.; Castro, P.; Osoro, K.; Rodríguez-Colunga, M.; Coto-Montes, A.; Oliván, M. Eating quality of beef from biotypes included in the PGI “Ternera Asturiana” showing distinct physicochemical characteristics and tenderization pattern. Meat Sci. 2010, 86, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Wang, L.; Liu, S.; Shan, T. Adipose tissue adipokines and lipokines: Functions and regulatory mechanism in skeletal muscle development and homeostasis. Metabolism 2023, 139, 155379. [Google Scholar] [CrossRef] [PubMed]
- White, U.A.; Stephens, J.M. Transcriptional factors that promote formation of white adipose tissue. Mol. Cell. Endocrinol. 2010, 318, 10–14. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Shang, P.; Li, Q.; Wang, L.; Chamba, Y.; Zhang, B.; Zhang, H.; Wu, C. iTRAQ-based proteomic analysis reveals key proteins affecting muscle growth and lipid deposition in pigs. Sci. Rep. 2017, 7, 46717. [Google Scholar] [CrossRef]
- Wang, X.; Fang, C.; He, H.; Cao, H.; Liu, L.; Jiang, L.; Ma, Y.; Liu, W. Identification of key genes in sheep fat tail evolution Based on RNA-seq. Gene 2021, 781, 145492. [Google Scholar] [CrossRef]
- Bickhart, D.M.; Hou, Y.; Schroeder, S.G.; Alkan, C.; Cardone, M.F.; Matukumalli, L.K.; Song, J.; Schnabel, R.D.; Ventura, M.; Taylor, J.F. Copy number variation of individual cattle genomes using next-generation sequencing. Genome Res. 2012, 22, 778–790. [Google Scholar] [CrossRef]
- Poleti, M.D.; Regitano, L.C.; Souza, G.H.; Cesar, A.S.; Simas, R.C.; Silva-Vignato, B.; Oliveira, G.B.; Andrade, S.C.; Cameron, L.C.; Coutinho, L.L. Longissimus dorsi muscle label-free quantitative proteomic reveals biological mechanisms associated with intramuscular fat deposition. J. Proteom. 2018, 179, 30–41. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Song, T.-J.; Li, X.; Hu, L.; He, Q.; Liu, M.; Lane, M.D.; Tang, Q.-Q. BMP signaling pathway is required for commitment of C3H10T1/2 pluripotent stem cells to the adipocyte lineage. Proc. Natl. Acad. Sci. USA 2009, 106, 12670–12675. [Google Scholar]
- Jonker, J.W.; Suh, J.M.; Atkins, A.R.; Ahmadian, M.; Li, P.; Whyte, J.; He, M.; Juguilon, H.; Yin, Y.-Q.; Phillips, C.T. A PPARγ–FGF1 axis is required for adaptive adipose remodelling and metabolic homeostasis. Nature 2012, 485, 391–394. [Google Scholar] [CrossRef]
- Han, M.S.; White, A.; Perry, R.J.; Camporez, J.-P.; Hidalgo, J.; Shulman, G.I.; Davis, R.J. Regulation of adipose tissue inflammation by interleukin 6. Proc. Natl. Acad. Sci. USA 2020, 117, 2751–2760. [Google Scholar] [CrossRef]
- Huang, R.-L.; Sun, Y.; Ho, C.-K.; Liu, K.; Tang, Q.-Q.; Xie, Y.; Li, Q. IL-6 potentiates BMP-2-induced osteogenesis and adipogenesis via two different BMPR1A-mediated pathways. Cell Death Dis. 2018, 9, 144. [Google Scholar] [CrossRef]
- Ighodaro, O.; Akinloye, O. First line defence antioxidants-superoxide dismutase (SOD), catalase (CAT) and glutathione peroxidase (GPX): Their fundamental role in the entire antioxidant defence grid. Alex. J. Med. 2018, 54, 287–293. [Google Scholar] [CrossRef]
- Di Pasquale, M.G. The essentials of essential fatty acids. J. Diet. Suppl. 2009, 6, 143–161. [Google Scholar] [CrossRef] [PubMed]
- Hammarstedt, A.; Syed, I.; Vijayakumar, A.; Eliasson, B.; Gogg, S.; Kahn, B.B.; Smith, U. Adipose tissue dysfunction is associated with low levels of the novel Palmitic Acid Hydroxystearic Acids. Sci. Rep. 2018, 8, 15757. [Google Scholar] [CrossRef]
- Domínguez, R.; Pateiro, M.; Gagaoua, M.; Barba, F.J.; Zhang, W.; Lorenzo, J.M. A comprehensive review on lipid oxidation in meat and meat products. Antioxidants 2019, 8, 429. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Santoro, A.; Peroni, O.D.; Nelson, A.T.; Saghatelian, A.; Siegel, D.; Kahn, B.B. PAHSAs enhance hepatic and systemic insulin sensitivity through direct and indirect mechanisms. J. Clin. Investig. 2019, 129, 4138–4150. [Google Scholar] [CrossRef]
- Liu, L.; Liu, X.; Cui, H.; Liu, R.; Zhao, G.; Wen, J. Transcriptional insights into key genes and pathways controlling muscle lipid metabolism in broiler chickens. BMC Genom. 2019, 20, 863. [Google Scholar] [CrossRef]
- Hocquette, J.-F.; Gondret, F.; Baéza, E.; Médale, F.; Jurie, C.; Pethick, D. Intramuscular fat content in meat-producing animals: Development, genetic and nutritional control, and identification of putative markers. Animal 2010, 4, 303–319. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-C.; Li, H.-J.; He, Z.-F.; Wang, T.; Qin, G. Study on the flavor contribution of phospholipids and triglycerides to pork. Food Sci. Biotechnol. 2010, 19, 1267–1276. [Google Scholar] [CrossRef]
- Wang, W.; Zhou, X.; Liu, Y. Characterization and evaluation of umami taste: A review. TrAC Trends Anal. Chem. 2020, 127, 115876. [Google Scholar] [CrossRef]
- Moya, V.-J.; Flores, M.; Aristoy, M.; Toldrá, F. Pork meat quality affects peptide and amino acid profiles during the ageing process. Meat Sci. 2001, 58, 197–206. [Google Scholar] [CrossRef]
- Listrat, A.; Lebret, B.; Louveau, I.; Astruc, T.; Bonnet, M.; Lefaucheur, L.; Picard, B.; Bugeon, J. How muscle structure and composition influence meat and flesh quality. Sci. World J. 2016, 2016, 3182746. [Google Scholar] [CrossRef]
- Klamt, S.; Stelling, J. Two approaches for metabolic pathway analysis? Trends Biotechnol. 2003, 21, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Williams, P. Nutritional composition of red meat. Nutr. Diet. 2007, 64, S113–S119. [Google Scholar] [CrossRef]
- Song, Q.; Guo, J.; Zhang, Y.; Chen, W. The beneficial effects of taurine in alleviating fatty liver disease. J. Funct. Foods 2021, 77, 104351. [Google Scholar] [CrossRef]
- Sordillo, L.M.; Aitken, S.L. Impact of oxidative stress on the health and immune function of dairy cattle. Vet. Immunol. Immunopathol. 2009, 128, 104–109. [Google Scholar] [CrossRef]
- Gaucher, C.; Boudier, A.; Bonetti, J.; Clarot, I.; Leroy, P.; Parent, M. Glutathione: Antioxidant properties dedicated to nanotechnologies. Antioxidants 2018, 7, 62. [Google Scholar] [CrossRef] [PubMed]
- Correa, L.B.; Netto, A.S.; da Silva, J.S.; Consolo, N.R.B.; Pugine, S.M.P.; de Melo, M.P.; de Souza Santana, R.S.; Zanetti, M.A. Changes on meat fatty acid profile, cholesterol and hepatic metabolism associated with antioxidants and canola oil supplementation for Nellore cattle. Livest. Sci. 2022, 257, 104850. [Google Scholar] [CrossRef]
- Lonergan, E.H.; Zhang, W.; Lonergan, S.M. Biochemistry of postmortem muscle—Lessons on mechanisms of meat tenderization. Meat Sci. 2010, 86, 184–195. [Google Scholar] [CrossRef] [PubMed]
- Imanari, M.; Higuchi, M.; Shiba, N.; Watanabe, A. Accurate analysis of taurine, anserine, carnosine and free amino acids in a cattle muscle biopsy sample. Anim. Sci. J. 2010, 81, 369–376. [Google Scholar] [CrossRef] [PubMed]
- Murakami, S. Role of taurine in the pathogenesis of obesity. Mol. Nutr. Food Res. 2015, 59, 1353–1363. [Google Scholar] [CrossRef] [PubMed]
- Ueda, Y.; Yonemitsu, M.; Tsubuku, T.; Sakaguchi, M.; Miyajima, R. Flavor characteristics of glutathione in raw and cooked foodstuffs. Biosci. Biotechnol. Biochem. 1997, 61, 1977–1980. [Google Scholar] [CrossRef] [PubMed]
- Egelandsdal, B.; Oostindjer, M.; Hovland, E.-M.; Okholm, B.; Saarem, K.; Bjerke, F.; Ruud, L.; Grabež, V.; Haug, A. Identifying labelling and marketing advantages of nutrients in minced beef meat: A case study. Meat Sci. 2020, 159, 107920. [Google Scholar] [CrossRef] [PubMed]
- Thakur, K.; Tomar, S.K.; Singh, A.K.; Mandal, S.; Arora, S. Riboflavin and health: A review of recent human research. Crit. Rev. Food Sci. Nutr. 2017, 57, 3650–3660. [Google Scholar] [CrossRef] [PubMed]
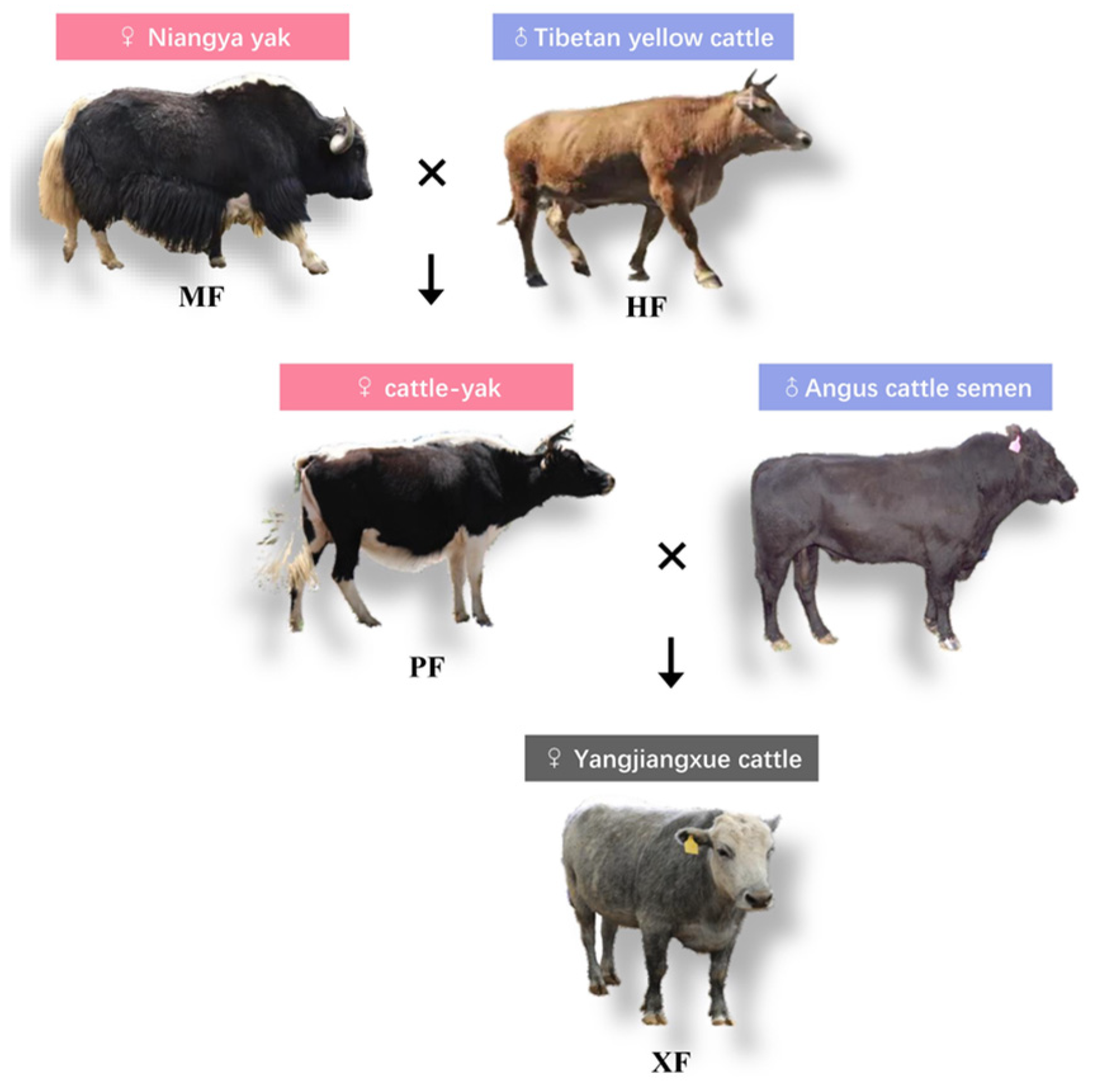



| Economic Traits | Breeds/Populations (LSM ± SE) | p | |||
|---|---|---|---|---|---|
| MF (n = 6) | HF (n = 5) | XF (n = 6) | PF (n = 6) | ||
| Head weight | 27.91 ± 1.10 Bc | 19.74 ± 1.32 Cd | 44.44 ± 1.23 Aa | 32.51 ± 2.13 Bb | 0.000 |
| Front hoof weight | 4.53 ± 0.26 Bb | 2.76 ± 0.31 Cc | 8.07 ± 0.29 Aa | 5.24 ± 0.50 Bb | 0.000 |
| Back hoof weight | 4.53 ± 0.28 Bb | 2.82 ± 0.34 Cc | 8.06 ± 0.32 Aa | 5.00 ± 0.55 BCb | 0.000 |
| Tare weight | 31.46 ± 1.85 Bc | 20.22 ± 2.22 Cd | 56.27 ± 2.06 Aa | 38.84 ± 3.58 Bb | 0.000 |
| Heart weight | 1.74 ± 0.13 Bb | 1.06 ± 0.15 Cc | 4.93 ± 0.14 Aa | 2.27 ± 0.25 Bb | 0.000 |
| Liver weight | 5.74 ± 0.50 Bb | 3.92 ± 0.60 Bc | 11.57 ± 0.56 Aa | 7.22 ± 0.97 Bb | 0.000 |
| Lung weight | 4.89 ± 0.48 Bb | 2.45 ± 0.58 Cc | 7.53 ± 0.54 Aa | 5.13 ± 0.94 ABCb | 0.000 |
| Spleen weight | 0.58 ± 0.08 Bc | 0.36 ± 0.10 Bc | 1.69 ± 0.09 Aa | 1.20 ± 0.16 Ab | 0.000 |
| Kidney weight | 0.86 ± 0.07 Bb | 0.61 ± 0.09 Bc | 1.52 ± 0.08 Aa | 1.01 ± 0.14 ABbc | 0.000 |
| Stomach weight | 15.34 ± 1.14 Bb | 12.37 ± 1.37 Bb | 32.00 ± 1.27 Aa | 17.39 ± 2.21 Bb | 0.000 |
| Visceral fat | na | na | 53.94 ± 4.27 Aa | 11.82 ± 4.57 Bb | 0.001 |
| Oxtail | 0.85 ± 0.43 Bb | 0.69 ± 0.52 Bb | 4.31 ± 0.48 Aa | 1.46 ± 0.84 ABb | 0.000 |
| Bullwhip | 0.99 ± 2.03 b | 1.82 ± 2.32 b | 10.67 ± 1.64 a | na | 0.027 |
| Small intestine | 5.68 ± 0.57 Bb | 3.59 ± 0.68 Bc | 8.27 ± 0.64 Aa | 5.39 ± 1.10 ABabc | 0.000 |
| Large intestine | 6.61 ± 1.66 Bb | 4.61 ± 1.99 Bb | 15.96 ± 1.85 Aa | 5.60 ± 3.21 ABb | 0.000 |
| Intestinal fat | 3.76 ± 1.97 Bb | 3.14 ± 2.37 Bb | 27.42 ± 2.20 Aa | 13.34 ± 3.82 ABb | 0.000 |
| Marbling score | 0.00 ± 0.00 Bb | 1.00 ± 0.00 Bb | 3.17 ± 0.60 Aa | 1.78 ± 0.31 Aa | 0.000 |
| Fat color | 7.00 ± 0.00 Aa | 6.00 ± 0.00 Bb | 2.00 ± 0.00 Cc | 6.33 ± 0.21 Bb | 0.000 |
| Metabolites | Groups | Trend | KEGG Pathway | Significantly Related Genes (p < 0.05) |
|---|---|---|---|---|
| oxidized glutathione | XF vs. HF | up | ferroptosis | CP (gene id 514194), FTL (286861), FTH1 (281173), ACSL4 (536628), CYBB (281112), LOC788801 (788801), TF (280705), novel.1303, ATG (5532686), SAT1 (508861) |
| oxidized glutathione | XF vs. HF | up | glutathione metabolism | RRM1 (505537), GPX1 (281209), LAP3 (781648), GSTA1 (777644), PGD (514939), CHAC1 (505991), IDH2 (327669), LOC100295687 (100295687), RRM2B (528960), ODC1 (281365), and GSTA3 (768055) |
| palmitic acid | XF vs. MF | up | fatty acid biosynthesis | HSD17B8 (532422), ACACB (515338), CBR4 (533020), RPP14 (515208), ACSF3 (509209), and ACSL1 (537161) |
| palmitic acid | XF vs. MF | up | fatty acid metabolism | FADS2 (521822), ACOX1 (513996), HACD4 (618814), ACAT2 (512044), HSD17B8 (532422), ACADL (614508), CBR4 (533020), HADH (532785), CPT1B (509459), RPP14 (515208), HACD2 (613886), LOC613570 (613570), ACSF3 (509209), ELOVL1 (540348), ACSL1 (537161), and ACAA1 (508324) |
| sedoheptulose 1,7-bisphosphate | XF vs. MF | up | carbon metabolism | LOC101902656 (101902656), HK3 (510616), PSPH (533630), PDHB (613610), ACOX1 (513996), ENO3 (540303), ACAT2 (512044), PRPS2 (537688), HIBCH (535883), MDH2 (281306), HK2 (788926), PSAT1 (533044), ALDH6A1 (327692), IDH3B (613338), MDH1 (535182), LOC788293 (788293), PCCA (614302), GPI (280808), GOT2 (286886), GLYCTK (507949), OGDH (534599), LOC614208 (614208), FBP1 (513483), PHGDH (505103), IDH3G (614145), SDHC (327696), LOC616200 (616200), GCSH (317723), and PC (338471) |
| 13(S)-HOTrE | XF vs. PF | up | alpha-linolenic acid metabolism | ACAA1 (508324), PLA2G2C (504978), and FADS2 (521822) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, X.; Ru, W.; Cheng, J.; Sun, L.; Zhang, N.; Zhaxi, L.; Dunzhu, R.; Sun, F.; Yang, K.; Gao, Y.; et al. Transcriptome and Metabolome Profiles Reveal the Underlying Mechanism of Fat Deposition Changes in Three-Way Crossbred Yak for High-Quality Beef Production. Animals 2025, 15, 2599. https://doi.org/10.3390/ani15172599
Cao X, Ru W, Cheng J, Sun L, Zhang N, Zhaxi L, Dunzhu R, Sun F, Yang K, Gao Y, et al. Transcriptome and Metabolome Profiles Reveal the Underlying Mechanism of Fat Deposition Changes in Three-Way Crossbred Yak for High-Quality Beef Production. Animals. 2025; 15(17):2599. https://doi.org/10.3390/ani15172599
Chicago/Turabian StyleCao, Xiukai, Wenxiu Ru, Jie Cheng, Le Sun, Nan Zhang, Lawang Zhaxi, Renzeng Dunzhu, Fengbo Sun, Kai Yang, Yue’e Gao, and et al. 2025. "Transcriptome and Metabolome Profiles Reveal the Underlying Mechanism of Fat Deposition Changes in Three-Way Crossbred Yak for High-Quality Beef Production" Animals 15, no. 17: 2599. https://doi.org/10.3390/ani15172599
APA StyleCao, X., Ru, W., Cheng, J., Sun, L., Zhang, N., Zhaxi, L., Dunzhu, R., Sun, F., Yang, K., Gao, Y., Huang, X., Huang, B., & Chen, H. (2025). Transcriptome and Metabolome Profiles Reveal the Underlying Mechanism of Fat Deposition Changes in Three-Way Crossbred Yak for High-Quality Beef Production. Animals, 15(17), 2599. https://doi.org/10.3390/ani15172599





