Bacterial Isolates Associated with Mortality Events in Brown Trout (Salmo trutta) Restocking Farms in Spain: A Descriptive Field Study
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
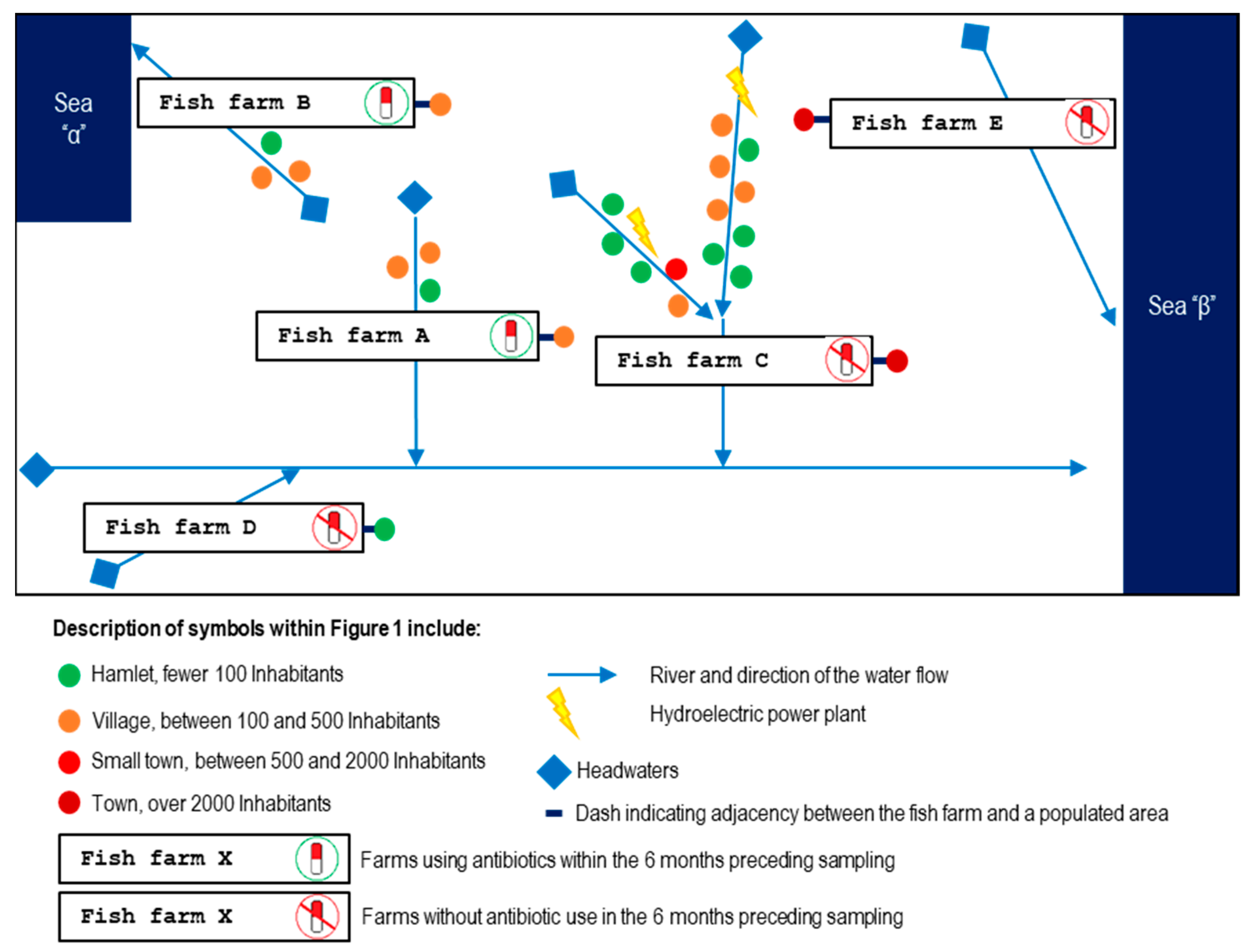
2.1. Farms Visited
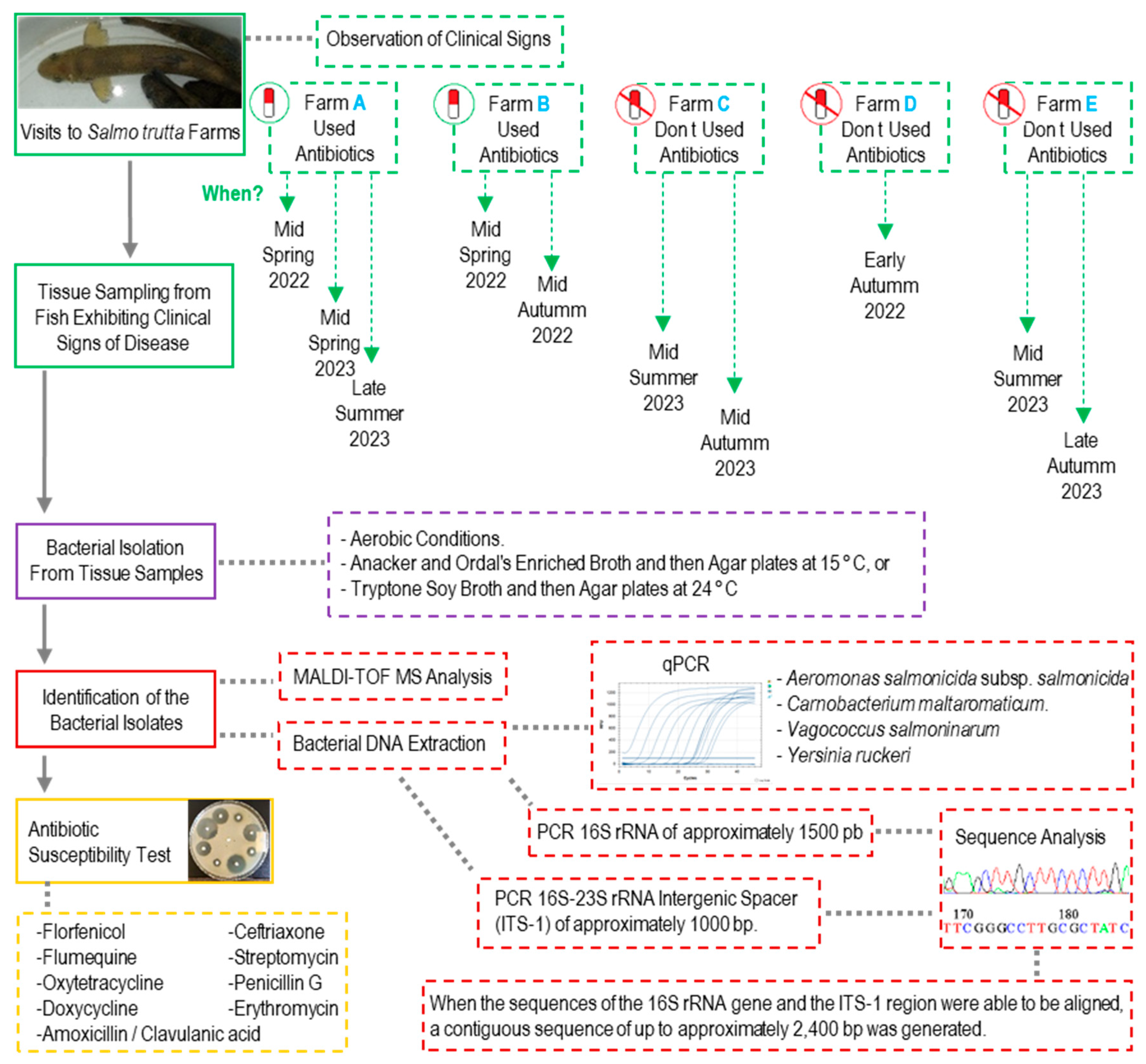
2.2. Sample Collection
2.3. Bacterial Isolation from Infected Tissue Samples
2.4. DNA Extraction and Identification of the Bacterial Isolates by qPCR
2.5. Identification of the Bacterial Isolates by 16S rRNA and ITS-1 Sequencing
| Target | Primer | Sequence 5′ to 3′ | Amplicon/Reference |
|---|---|---|---|
| Aeromonas salmonicida subsp. salmonicida | aopP F | 5′-CGGAACGTAATCTGAATTGTTCTTTTC-3′ | 340 pb [16] |
| aopP R | 5′-ATTGCTTATCGAGGCAGCCAAC-3′ | ||
| Carnobacterium maltaromaticum | 16S F | 5′-GAGGGTCATTGGAAACTGGA-3′ | 219 pb [14] |
| 16S R | 5′-CGGAAACCCTCCAACACTTA-3′ | ||
| Vagococcus salmoninarum | sal F | 5′-GACGCTTTCGGGTGTCACTA-3′ | 543 pb [15] |
| sal R | 5′-CAGACCAGAGAGTCGCCTTC-3′ | ||
| Yersinia ruckeri | glnA F | 5′-TCCAGCACCAAATACGAAGG-3′ | 113 pb [17] |
| glnA R | 5′-ACATGGCAGAACGCAGATC-3′ | ||
| glnA P | HEX-5′-AAGGCGGTTACTTCCCGGTTCC-3′-BHQ1 | ||
| 16S rRNA gene | 27F | 5′-AGAGTTTGATCCTGGCTCAG-3′ | 1465 pb [20] |
| 1492R | 5′-ACGGCTACCTTGTTACGACTT-3′ | ||
| 16S rRNA-23S rRNA intergenic spacer | R1391 | 5′-TTGTACACACCGCCCGTC-3′ | 1000 pb [13] |
| 473F | 5′-TTGTACACACCGCCCGTC-3′ |
2.6. Identification of the Bacterial Isolates by MALDI-TOF MS
2.7. Phylogenetic Analysis
2.8. Antibiotic Susceptibility Test
3. Results
3.1. Fish Farm A: Observations and Bacterial Diagnosis
3.1.1. First Visit, Mid-Spring 2022
3.1.2. Second Visit, Mid-Spring 2023
3.1.3. Third Visit, Late Summer 2023
3.2. Fish Farm B: Observations and Bacterial Diagnosis
3.2.1. First Visit, Mid-Spring 2022
3.2.2. Second Visit, Mid-Autumn 2022
3.3. Fish Farm C: Observations and Bacterial Diagnosis
3.3.1. First Visit, Mid-Summer 2023
3.3.2. Second Visit, Mid-Autumn 2023
3.4. Fish Farm D: Observations and Bacterial Diagnosis
Visit in Early Autumn 2022
3.5. Fish Farm E: Observations and Bacterial Diagnosis
3.5.1. First Visit, Mid-Summer 2023
3.5.2. Second Visit, Late Autumn 2023
3.6. Phylogenetic Analysis
3.7. Antibiotic Susceptibility Test
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- García-Vega, A.; Fuentes-Pérez, J.F.; Leunda Urretabizkaia, P.M.; Ardaiz Ganuza, J.; Sanz-Ronda, F.J. Upstream migration of anadromous and potamodromous brown trout: Patterns and triggers in a 25-year overview. Hydrobiologia 2022, 849, 197–213. [Google Scholar] [CrossRef]
- Vera, M.; Aparicio, E.; Heras, S.; Abras, A.; Casanova, A.; Roldán, M.I.; García-Marin, J.-L. Regional environmental and climatic concerns on preserving native gene pools of a least concern species: Brown trout lineages in Mediterranean streams. Sci. Total Environ. 2023, 862, 160739. [Google Scholar] [CrossRef] [PubMed]
- World Organisation for Animal Health (WOAH). Diseases of Fish. In Manual of Diagnostic Tests for Aquatic Animals, 11th ed.; World Organisation for Animal Health: Paris, France, 2024; Chapter 2.3; Available online: https://www.woah.org/fileadmin/Home/eng/Health_standards/aahm/current/2.3.0_General_info.pdf (accessed on 29 June 2025).
- European Parliament and Council of the European Union. Regulation (EU) 2016/429 of the European Parliament and of the Council of 9 March 2016 on Transmissible Animal Diseases and Amending and Repealing Certain Acts in the Area of Animal Health (“Animal Health Law”); EUR-Lex: Luxembourg, 2016; Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=celex%3A32016R0429 (accessed on 17 April 2025).
- Rathinam, R.B.; Iburahim, S.A.; Ramanan, S.S.; Tripathi, G. A scientometric mapping of research on Aeromonas infection in fish across the world (1998–2020). Aquac. Int. 2022, 30, 341–363. [Google Scholar] [CrossRef]
- Revina, O.; Avsejenko, J.; Cīrule, D.; Valdovska, A. Antimicrobial Resistance of Aeromonas spp. Isolated from the Sea Trout (Salmo trutta L.) in Latvia. 2017; pp. 271–275. Available online: http://llufb.llu.lv/conference/Research-for-Rural-Development/2017/LatviaResRuralDev_23rd_2017_vol1-271-275.pdf (accessed on 21 October 2024).
- Austin, B.; Austin, D.A. Bacterial Fish Pathogens; Springer International Publishing: Cham, Switzerland, 2016; Available online: http://link.springer.com/10.1007/978-3-319-32674-0 (accessed on 21 October 2024).
- Elgendy, M.Y.; Ali, S.E.; Abbas, W.T.; Algammal, A.M.; Abdelsalam, M. The role of marine pollution on the emergence of fish bacterial diseases. Chemosphere 2023, 344, 140366. [Google Scholar] [CrossRef] [PubMed]
- Uren Webster, T.M.; Consuegra, S.; Garcia De Leaniz, C. Early life stress causes persistent impacts on the microbiome of Atlantic salmon. Comp. Biochem. Physiol. Part D Genom. Proteom. 2021, 40, 100888. [Google Scholar] [CrossRef]
- Milijasevic, M.; Veskovic-Moracanin, S.; Babic Milijasevic, J.; Petrovic, J.; Nastasijevic, I. Antimicrobial Resistance in Aquaculture: Risk Mitigation within the One Health Context. Foods 2024, 13, 2448. [Google Scholar] [CrossRef]
- Aerts, M.; Baron, S.; Bortolaia, V.; Hendriksen, R.; Guerra, B.; Stoicescu, A.; Beloeil, P. Technical specifications for a EU-wide baseline survey of antimicrobial resistance in bacteria from aquaculture animals. EFSA J. 2024, 22, e8928. Available online: https://data.europa.eu/doi/10.2903/j.efsa.2024.8928 (accessed on 21 October 2024).
- Marti, E.; Huerta, B.; Rodríguez-Mozaz, S.; Barceló, D.; Marcé, R.; Balcázar, J.L. Abundance of antibiotic resistance genes and bacterial community composition in wild freshwater fish species. Chemosphere 2018, 196, 115–119. [Google Scholar] [CrossRef]
- Vargas-González, A.; Barajas, M.; Pérez-Sánchez, T. Isolation of Lactic Acid Bacteria (LAB) from salmonids for potential use as probiotics: In vitro assays and toxicity assessment of Salmo trutta embryonated eggs. Animals 2024, 14, 200. [Google Scholar] [CrossRef]
- Mohsina, K.; Kaur, M.; Bowman, J.P.; Powell, S.; Tamplin, M.L. qPCR quantification of Carnobacterium maltaromaticum, Brochothrix thermosphacta, and Serratia liquefaciens growth kinetics in mixed culture. J. Microbiol. Methods 2020, 175, 105961. [Google Scholar] [CrossRef] [PubMed]
- Torres-Corral, Y.; Fernández-Álvarez, C.; Santos, Y. High-throughput identification and quantification of Vagococcus salmoninarum by SYBR Green I-based real-time PCR combined with melting curve analysis. J. Fish Dis. 2019, 42, 1359–1368. [Google Scholar] [CrossRef]
- Balcázar, J.L.; Vendrell, D.; De Blas, I.; Ruiz-Zarzuela, I.; Gironés, O.; Múzquiz, J.L. Quantitative detection of Aeromonas salmonicida in fish tissue by real-time PCR using self-quenched, fluorogenic primers. J. Med. Microbiol. 2007, 56, 323–328. [Google Scholar] [CrossRef]
- Keeling, S.E.; Johnston, C.; Wallis, R.; Brosnahan, C.L.; Gudkovs, N.; McDonald, W.L. Development and validation of real-time PCR for the detection of Yersinia ruckeri: Yersinia ruckeri real-time PCR. J. Fish Dis. 2012, 35, 119–125. [Google Scholar] [CrossRef]
- Sabat, A.J.; van Zanten, E.; Akkerboom, V.; Wisselink, G.; van Slochteren, K.; de Boer, R.F.; Hendrix, R.; Friedrich, A.W.; Rossen, J.W.A.; Kooistra-Smid, A.M.D.M. Targeted next-generation sequencing of the 16S-23S rRNA region for culture-independent bacterial identification—Increased discrimination of closely related species. Sci. Rep. 2017, 7, 3434. [Google Scholar] [CrossRef]
- Janda, J.M.; Abbott, S.L. 16S rRNA Gene Sequencing for Bacterial Identification in the Diagnostic Laboratory: Pluses, Perils, and Pitfalls. J. Clin. Microbiol. 2007, 45, 2761–2764. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef]
- Çağatay, İ.T. Use of proteomic-based MALDI-TOF mass spectra for identification of bacterial pathogens in aquaculture: A review. Aquac. Int. 2024, 32, 7835–7871. [Google Scholar] [CrossRef]
- MacAulay, S.; Ellison, A.R.; Kille, P.; Cable, J. Moving towards improved surveillance and earlier diagnosis of aquatic pathogens: From traditional methods to emerging technologies. Rev. Aquac. 2022, 14, 1813–1829. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: Oxford, UK; New York, NY, USA, 2000; 333p. [Google Scholar]
- Lewis, J.S., II; Weinstein, M.P.; Bobenchik, A.M.; Campeau, S.; Cullen, S.K.; Dingle, T.; Galas, M.F.; Humphries, R.M.; Krin, T.J., Jr.; Limbago, B. M100 Performance Standards for Antimicrobial Susceptibility Testing, 33rd ed.; Clinical and Laboratory Standards Institute: Malvern, PA, USA, 2023. [Google Scholar]
- Hindler, J.A.; Humphries, R.M.; Richter, S.S.; Jorgensen, J.H.; Bernard, K.; Killian, S.B.; Bodeis-Jones, S.; Kohner, P.; Castanheira, M.; Matuschek, E. M45 Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria, 3rd ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2016; 98p. [Google Scholar]
- Miller, R.A.; Gieseker, C.M.; Avendaño-Herrera, R.; Baron, S.; Buller, N.; Burbick, C.R.; Chuanchuen, R.; Dalsgaard, I.; Declercq, A.M. VET04 Performance Standards for Antimicrobial Susceptibility Testing of Bacteria Isolated from Aquatic Animals, 3rd ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020. [Google Scholar]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Noguera, P.A. A Colour Atlas of Salmonid Diseases, 2nd ed.; Springer: Dordrecht, The Netherlands, 2013. [Google Scholar]
- Smith, S.A.; Newman, S.J.; Harrison, C.E.; Loch, T.P. First isolation of Carnobacterium maltaromaticum from farmed rainbow trout in Virginia. J. Aquat. Anim. Health 2023, 35, 3–10. [Google Scholar] [CrossRef]
- Pastorino, P.; Colussi, S.; Pizzul, E.; Varello, K.; Menconi, V.; Mugetti, D.; Tomasoni, M.; Esposito, G.; Bertoli, M.; Bozzetta, E.; et al. The unusual isolation of carnobacteria in eyes of healthy salmonids in high-mountain lakes. Sci. Rep. 2021, 11, 2314. [Google Scholar] [CrossRef] [PubMed]
- Orozova, P.; Sirakov, I.; Chikova, V.; Popova, R.; Al-Harbi, A.H.; Crumlish, M.; Austin, B. Recovery of Hafnia alvei from diseased brown trout, Salmo trutta L., and healthy noble crayfish, Astacus astacus (L.), in Bulgaria. J. Fish Dis. 2014, 37, 891–898. [Google Scholar] [CrossRef] [PubMed]
- Duman, M.; Altun, S.; Saticioglu, I.B.; Romalde, J.L. A review of bacterial disease outbreaks in rainbow trout (Oncorhynchus mykiss) reported from 2010 to 2022. J. Fish Dis. 2025, 48, e13886. [Google Scholar] [CrossRef]
- Clinton, M.; Kintner, A.H.; Delannoy, C.M.J.; Brierley, A.S.; Ferrier, D.E.K. Molecular identification of potential aquaculture pathogens adherent to cnidarian zooplankton. Aquaculture 2020, 518, 734801. [Google Scholar] [CrossRef]
- Farkas, A.; Butiuc-Keul, A.; Carpa, R.; Szekeres, E.; Teban-Man, A.; Coman, C. Overlooked Enterobacterales as hosts of antimicrobial resistance in aquatic environments. Sci. Rep. 2025, 15, 26026. [Google Scholar] [CrossRef]
- Abdel-Raheem, S.M.; Khodier, S.M.; Almathen, F.; Hanafy, A.S.T.; Abbas, S.M.; Al-Shami, S.A.; Al-Sultan, S.I.; Alfifi, A.; El-Tarabili, R.M. Dissemination, virulence characteristic, antibiotic resistance determinants of emerging linezolid and vancomycin-resistant Enterococcus spp. in fish and crustacean. Int. J. Food Microbiol. 2024, 418, 110711. [Google Scholar] [CrossRef] [PubMed]



| Species/Strain | Accession No. | Nucleotide Position |
|---|---|---|
| Aeromonas allosaccharophila strain CECT 4199 | NR_025945.2 | 79–1435 |
| Aeromonas bestiarum strain CIP 74.30 | NR_026089.2 | 79–1435 |
| Aeromonas bivalvium strain 868E | NR_043885.1 | 80–1436 |
| Aeromonas encheleia strain CECT 4342 | NR_118042.1 | 79–1435 |
| Aeromonas eucrenophila strain NCIMB 74 | NR_118946.1 | 79–1435 |
| Aeromonas hydrophila strain ATCC 7966 | NR_074841.1 | 80–1436 |
| Aeromonas molluscorum strain LMG 22214 | NR_115351.1 | 64–1420 |
| Aeromonas rivuli strain DSM 22539 | NR_116880.1 | 79–1435 |
| Aeromonas salmonicida strain CECT 894 | NR_043324.1 | 64–1420 |
| Aeromonas salmonicida subsp. achromogenes strain 6263/4/5 | NR_037011.1 | 79–1434 |
| Aeromonas salmonicida subsp. masoucida strain NBRC 13784 | NR_113635.1 | 60–1416 |
| Aeromonas sobria strain ATCC 43979 | NR_119044.1 | 85–1441 |
| Aeromonas veronii bv. veronii strain ATCC 35624 | NR_118947.1 | 79–1435 |
| Carnobacterium divergens strain NBRC 15683 | NR_113798.1 | 60–1426 |
| Carnobacterium gallinarum strain DSM 4847 | NR_042093.1 | 82–1444 |
| Carnobacterium inhibens subsp. inhibens DSM 13024 strain K1 | NR_036895.1 | 82–1450 |
| Carnobacterium maltaromaticum strain DSM 20342 | NR_044710.2 | 87–1452 |
| Enterococcus durans strain 98D | NR_036922.1 | 74–1442 |
| Enterococcus faecium strain DSM 20477 | NR_114742.1 | 71–1439 |
| Enterococcus hirae ATCC 9790 | NR_075022.1 | 85–1453 |
| Enterococcus lactis strain BT159 | NR_117562.1 | 40–1408 |
| Enterococcus pseudoavium strain LMG 11426 | NR_114785.2 | 81–1445 |
| Hafnia alvei strain JCM 1666 | NR_112985.1 | 67–1422 |
| Hafnia paralvei strain ATCC 29927 | NR_116898.1 | 59–1414 |
| Huaxiibacter chinensis strain 155047 | NR_184601.1 | 82–1435 |
| Klebsiella aerogenes strain KCTC 2190 | NR_102493.2 | 85–1438 |
| Kluyvera ascorbata strain ATCC 33433 | NR_028677.1 | 35–1388 |
| Kluyvera intermedia strain NBRC 102594 | NR_114153.1 | 58–1411 |
| Lactococcus carnosus strain TMW 2.1612 | NR_174340.1 | 75–1443 |
| Lactococcus paracarnosus strain TMW 2.1615 | NR_174341.1 | 75–1443 |
| Lactococcus piscium strain CCUG 32732 | NR_043739.1 | 1–1369 |
| Lactococcus raffinolactis strain NBRC 100932 | NR_113959.1 | 62–1427 |
| Piscirickettsia salmonis strain LF-89 | NR_025980.1 | 78–1433 |
| Plesiomonas shigelloides strain NCIMB 9242 | NR_044827.1 | 78–1431 |
| Pseudomonas ceruminis strain BML-PP028 | NR_181196.1 | 79–1431 |
| Pseudomonas entomophila strain L48 | NR_102854.1 | 76–1428 |
| Pseudomonas fulva strain IAM1529 | NR_115610.1 | 74–1425 |
| Pseudomonas juntendi strain BML3 | NR_180497.1 | 79–1431 |
| Pseudomonas monteilii strain CIP 104883 | NR_024910.1 | 67–1419 |
| Pseudomonas parafulva NBRC 16636 = DSM 17004 | NR_040859.1 | 68–1420 |
| Pseudomonas sp. URMO17WK12 I11 | LN865164.1 | 1,267,100–1,268,453 |
| Serratia inhibens strain S40 | NR_180863.1 | 83–1437 |
| Serratia liquefaciens strain ATCC 27592 | NR_122057.1 | 87–1442 |
| Isolate | qPCR Results | MALDI-TOF MS (Score) | 16S rRNA (Id. BLAST) | ITS-1 (Id. BLAST) | Isolate Identity | Genbank Accession No | Size |
|---|---|---|---|---|---|---|---|
| St-Pi3-L-87 | Negative | ND (<1.70) | P. shigelloides (97.30%) | P. shigelloides (92.48%) | Plesiomonas sp. | PQ671489.1 | 2102 nt 16SrRNA + ITS-1 |
| St-Pi3-VL-99 | Not tested | Aeromonas sobria (1.99) | A. salmonicida (100%) | A. salmonicida (99.83%) | Aeromonas sp. | PQ671490.1 | 1423 nt 16SrRNA |
| St-Pi3-OP-104 | Not tested | Hafnia alvei (2.03) | Hafnia alvei (100%) | Hafnia alvei (99.43%) | Hafnia alvei | PQ671491.1 | 1392 nt 16SrRNA |
| St-Pi5-Sp-107 | Not tested | Aeromonas encheleia (1.99) | Aeromonas encheleia (99.93%) | Aeromonas encheleia (100%) | Aeromonas encheleia | PQ671492.1 | 1538 nt 16SrRNA |
| St-Pi5-L-92 | (+) Carnobacterium maltaromaticum | C. maltaromaticum (2.41) | C. maltaromaticum (100%) | C. maltaromaticum (99.41%) | Carnobacterium maltaromaticum | PQ671493.1 | 2384 nt 16SrRNA + ITS-1 |
| St-Pi5-Sk-98 | Negative | Pseudomonas fulva (2.22) | Pseudomonas fulva (100%) | Pseudomonas fulva (99.62%) | Pseudomonas fulva | PQ671494.1 | 2162 nt 16SrRNA + ITS-1 |
| St-Pi5-Sk-89 | (+) A. salmonicida subsp. salmonicida | Aeromonas bestiarum (1.72) | Aeromonas salmonicida (100%) | Aeromonas bestiarum (97.36%) | A. salmonicida subsp. salmonicida | PQ671495.1 | 2437 nt 16SrRNA + ITS-1 |
| St-Pi6-OP-110 | Negative | Aeromonas bestiarum (1.76) | Aeromonas salmonicida (100%) | Aeromonas bestiarum (99.83%) | Aeromonas sp. | PQ671496.1 | 1376 nt 16SrRNA |
| St-Pi6-OP-111 | Negative | Aeromonas hydrophila (2.01) | Aeromonas hydrophila (100%) | Aeromonas hydrophila (99.83%) | Aeromonas hydrophila | PQ671497.1 | 1528 nt 16SrRNA |
| St-Pi6-OP-113 | Negative | Kluyvera intermedia (2.06) | Kluyvera intermedia (100%) | Kluyvera intermedia (98.31%) | Kluyvera intermedia | PQ671498.1 | 2274 nt 16SrRNA + ITS-1 |
| St-Pi6-OP-105 | Not tested | Kluyvera intermedia (1.74) | Kluyvera intermedia (99.93%) | Kluyvera intermedia (99.08%) | Kluyvera intermedia | PQ671499.1 | 1397 nt 16SrRNA |
| St-Pi2-Sp-101 | Not tested | Aeromonas eucrenophila (2.05) | A. salmonicida (99.86%) | Aeromonas bestiarum (98.03%) | Aeromonas sp. | PQ671500.1 | 1419 nt 16SrRNA |
| St-Pi7-OP-96 | Negative | Enterococcus faecium (2.12) | Enterococcus faecium (100%) | Enterococcus faecium (100%) | Enterococcus faecium | PQ671501.1 | 2337 nt 16SrRNA + ITS-1 |
| St-Pi7-OP-97 | Negative | Aeromonas bestiarum (1.92) | Aeromonas sobria (100%) | A. salmonicida (96.79%) | Aeromonas sp. | PQ671502.1 | 2378 nt 16SrRNA + ITS-1 |
| St-Pi7-OP-114 | Negative | Lactococcus raffinolactis (1.71) | Lactococcus raffinolactis (100%) | Lactococcus raffinolactis (99.18%) | Lactococcus raffinolactis | PQ671503.1 | 2312 nt 16SrRNA + ITS-1 |
| St-Pi7-OP-115 | Negative | Hafnia alvei (2.05) | Hafnia alvei (100%) | Hafnia alvei (98.59%) | Hafnia alvei | PQ671504.1 | 2375 nt 16SrRNA + ITS-1 |
| St-Pi7-OP-116 | Negative | Hafnia alvei (2.15) | Hafnia alvei (100%) | Hafnia alvei (98.55%) | Hafnia alvei | PQ671505.1 | 2358 nt 16SrRNA + ITS-1 |
| St-Pi7-OP-118 | (+) Carnobacterium maltaromaticum | Not tested | C. maltaromaticum (99.93%) | C. maltaromaticum (99.18%) | Carnobacterium maltaromaticum | PQ671506.1 | 2343 nt 16SrRNA + ITS-1 |
| St-Pi7-OP-119 | Negative | Aeromonas bestiarum (1.83) | Aeromonas salmonicida (100%) | Aeromonas bestiarum (96.58%) | Aeromonas sp. | PQ671507.1 | 2408 nt 16SrRNA + ITS-1 |
| Isolate | Amphenicols | Quinolones | Tetracyclines | Macrolides | Cephalosporins | Aminoglycosides | Penicillin | Combination Agents | |
|---|---|---|---|---|---|---|---|---|---|
| Florfenicol (30 µg) | Flumequine (30 µg) | Oxytetracycline (30 µg) | Doxycycline (30 µg) | Erythromycin (15 µg) | Ceftriaxone (30 µg) | Streptomycin (10 µg) | Penicillin G (10 Units) | Amoxicillin/Clavulanic Acid (30 µg) | |
| St-Pi3-L-87 Plesiomonas sp. | 36.7 ± 1.2 | 39.0 ± 0.0 | 33.0 ± 0.0 | 32.0 ± 0.8 | 19.3 ± 0.9 | 37.3 ± 0.5 | 10.7 ± 0.5 | 8.3 ± 0.5 | 29.0 ± 0.8 |
| a S | a S | a S | b S | b Ri | b S | b R | b Ri | b S | |
| St-Pi3-VL-99 Aeromonas sp. | 33.7 ± 0.5 | 39.0 ± 1.6 | 29.0 ± 0.0 | 26.3 ± 0.5 | 18.7 ± 0.5 | 36.7 ± 0.5 | 8.7 ± 0.5 | 0.0 ± 0.0 | 15.0 ± 0.8 |
| a S c (WT) | a S | c S c (WT) | a S | a ND c (WT) | d S | a ND | d Ri | d Ri | |
| St-Pi3-OP-104 Hafnia alvei | 16.7 ± 1.2 | 35.3 ± 0.5 | 21.0 ± 0.8 | 14.3 ± 0.5 | 0.0 ± 0.0 | 25.3 ± 0.9 | 13.7 ± 0.5 | 0.0 ± 0.0 | 8.0 ± 0.0 |
| a ND | a S | a ND | b S | b Ri | b Sx | b I | b Ri | b Ri | |
| St-Pi5-Sp-107 A. encheleia | 28.7 ± 0.9 | 37.7 ± 0.5 | 29.0 ± 0.8 | 25.0 ± 0.8 | 12.3 ± 0.5 | 36.7 ± 0.5 | 14.3 ± 0.5 | 0.0 ± 0.0 | 14.7 ± 0.5 |
| a S | a S | a S | a S | a ND | d S | a ND | d Ri | d Ri | |
| St-Pi5-L-92 C. maltaromaticum | 34.0 ± 0.8 | 14.0 ± 0.8 | 34.7 ± 0.9 | 35.3 ± 0.5 | 33.3 ± 0.5 | 0.0 ± 0.0 | 0.0 ± 0.0 | 22.0 ± 0.8 | 30.0 ± 1.6 |
| a S | a ND | a S | e S | e S | e Ri | e Ri | e S | a S | |
| St-Pi5-Sk-98 Pseudomonas fulva | 34.0 ± 0.8 | 39.7 ± 0.5 | 32.0 ± 0.8 | 28.0 ± 0.8 | 16.0 ± 0.8 | 36.0 ± 0.8 | 14.7 ± 0.5 | 0.0 ± 0.0 | 12.0 ± 0.8 |
| a S | a S | a S | a S | a ND | a S | a ND | a R | a ND | |
| St-Pi5-Sk-89 A. salmonicida subsp. salmonicida | 34.0 ± 0.8 | 39.3 ± 0.5 | 31.7 ± 0.5 | 28.0 ± 0.0 | 16.3 ± 0.5 | 35.3 ± 0.5 | 14.7 ± 0.5 | 0.0 ± 0.0 | 12.7 ± 0.5 |
| a S c (WT) | a S | c S c (WT) | a S | a ND c (WT) | d S | a ND | d Ri | d Ri | |
| St-Pi6-OP-110 Aeromonas sp. | 33.7 ± 0.5 | 44.3 ± 0.5 | 30.3 ± 0.5 | 28.0 ± 0.8 | 18.0 ± 0.0 | 34.0 ± 0.0 | 14.3 ± 0.5 | 0.0 ± 0.0 | 15.0 ± 0.8 |
| a S c (WT) | a S | c S c (WT) | a S | a ND c (WT) | d S | a ND | d Ri | d Ri | |
| St-Pi6-OP-111 A. hydrophila | 30.3 ± 0.9 | 13.7 ± 0.5 | 9.7 ± 0.5 | 15.7 ± 0.5 | 0.0 ± 0.0 | 34.3 ± 0.5 | 17.0 ± 0.0 | 0.0 ± 0.0 | 13.7 ± 0.5 |
| a S f (WT) | a ND | a R f (NWT) | a ND | a R | d S | a ND | d Ri | d Ri | |
| St-Pi6-OP-113 K. intermedia | 18.7 ± 0.5 | 29.0 ± 0.8 | 24.3 ± 0.5 | 20.7 ± 0.5 | 0.0 ± 0.0 | 33.7 ± 0.5 | 14.7 ± 0.9 | 0.0 ± 0.0 | 13.3 ± 1.2 |
| a ND | a S | a ND | b S | b Ri | b S | b I | b Ri | b R | |
| St-Pi6-OP-105 K. intermedia | 18.0 ± 0.0 | 22.3 ± 0.9 | 19.0 ± 0.8 | 16.3 ± 1.2 | 0.0 ± 0.0 | 27.3 ± 1.7 | 12.7 ± 0.5 | 0.0 ± 0.0 | 13.7 ± 0.5 |
| a ND | a ND | a ND | b S | b Ri | b S | b I | b Ri | b R | |
| St-Pi2-Sp-101 Aeromonas sp. | 31.7 ± 0.5 | 40.3 ± 0.5 | 29.7 ± 0.5 | 27.0 ± 0.8 | 15.0 ± 0.8 | 36.0 ± 0.0 | 15.7 ± 0.9 | 0.0 ± 0.0 | 15.3 ± 0.9 |
| a S | a S | a S | a S | a ND | d S | a ND | d Ri | d Ri | |
| St-Pi7-OP-96 E. faecium | 32.7 ± 0.9 | 11.3 ± 0.5 | 0.0 ± 0.0 | 12.0 ± 0.8 | 23.3 ± 0.5 | 12.3 ± 0.5 | 0.0 ± 0.0 | 20.7 ± 0.9 | 27.7 ± 0.5 |
| a S | a ND | a R | g R | g S | g Ri | g Ri | g S | a S | |
| St-Pi7-OP-97 Aeromonas sp. | 0.0 ± 0.0 | 21.0 ± 0.8 | 21.3 ± 0.5 | 21.7 ± 0.5 | 0.0 ± 0.0 | 20.3 ± 0.5 | 10.3 ± 0.5 | 0.0 ± 0.0 | 11.0 ± 0.0 |
| a R | a ND | a ND | a ND | a R | d I | a ND | d Ri | d Ri | |
| St-Pi7-OP-114 L. raffinolactis | 35.3 ± 1.2 | 0.0 ± 0.0 | 36.0 ± 0.8 | 36.3 ± 0.9 | 37.3 ± 0.5 | 34.0 ± 0.8 | 16.3 ± 0.5 | 35.3 ± 1.2 | 39.7 ± 1.7 |
| a S | a R | a S | i S | h S | i S | a ND | h S | a S | |
| St-Pi7-OP-115 Hafnia alvei | 16.7 ± 0.5 | 36.3 ± 0.5 | 20.0 ± 0.0 | 15.0 ± 0.8 | 0.0 ± 0.0 | 28.0 ± 0.8 | 14.7 ± 0.5 | 0.0 ± 0.0 | 8.7 ± 0.5 |
| a ND | a S | a ND | b S | b Ri | b Sx | b I | b Ri | b Ri | |
| St-Pi7-OP-116 Hafnia alvei | 12.0 ± 0.8 | 33.3 ± 0.5 | 18.3 ± 0.9 | 16.7 ± 0.5 | 8.0 ± 0.0 | 28.7 ± 0.9 | 16.7 ± 0.5 | 9.3 ± 0.9 | 18.7 ± 0.5 |
| a ND | a S | a ND | b S | b Ri | b Sx | b S | b Ri | b Ri | |
| St-Pi7-OP-118 C. maltaromaticum | 30.3 ± 2.1 | 10.3 ± 0.5 | 32.3 ± 2.1 | 33.7 ± 0.9 | 32.0 ± 0.8 | 0.0 ± 0.0 | 0.0 ± 0.0 | 23.0 ± 0.8 | 31.3 ± 0.9 |
| a S | a ND | a S | e S | e S | e Ri | e Ri | e S | a S | |
| St-Pi7-OP-119 Aeromonas sp. | 32.0 ± 0.8 | 36.0 ± 0.0 | 29.3 ± 0.5 | 26.0 ± 0.8 | 15.0 ± 0.8 | 31.7 ± 0.5 | 16.0 ± 0.0 | 0.0 ± 0.0 | 14.0 ± 0.8 |
| a S | a S | a S | a S | a ND | d S | a ND | d Ri | d Ri | |
| Farm | Adjacent and Upstream Population | Visit | Date | Weather Events | Symptoms | Bacterial Isolates Identified | AMR |
|---|---|---|---|---|---|---|---|
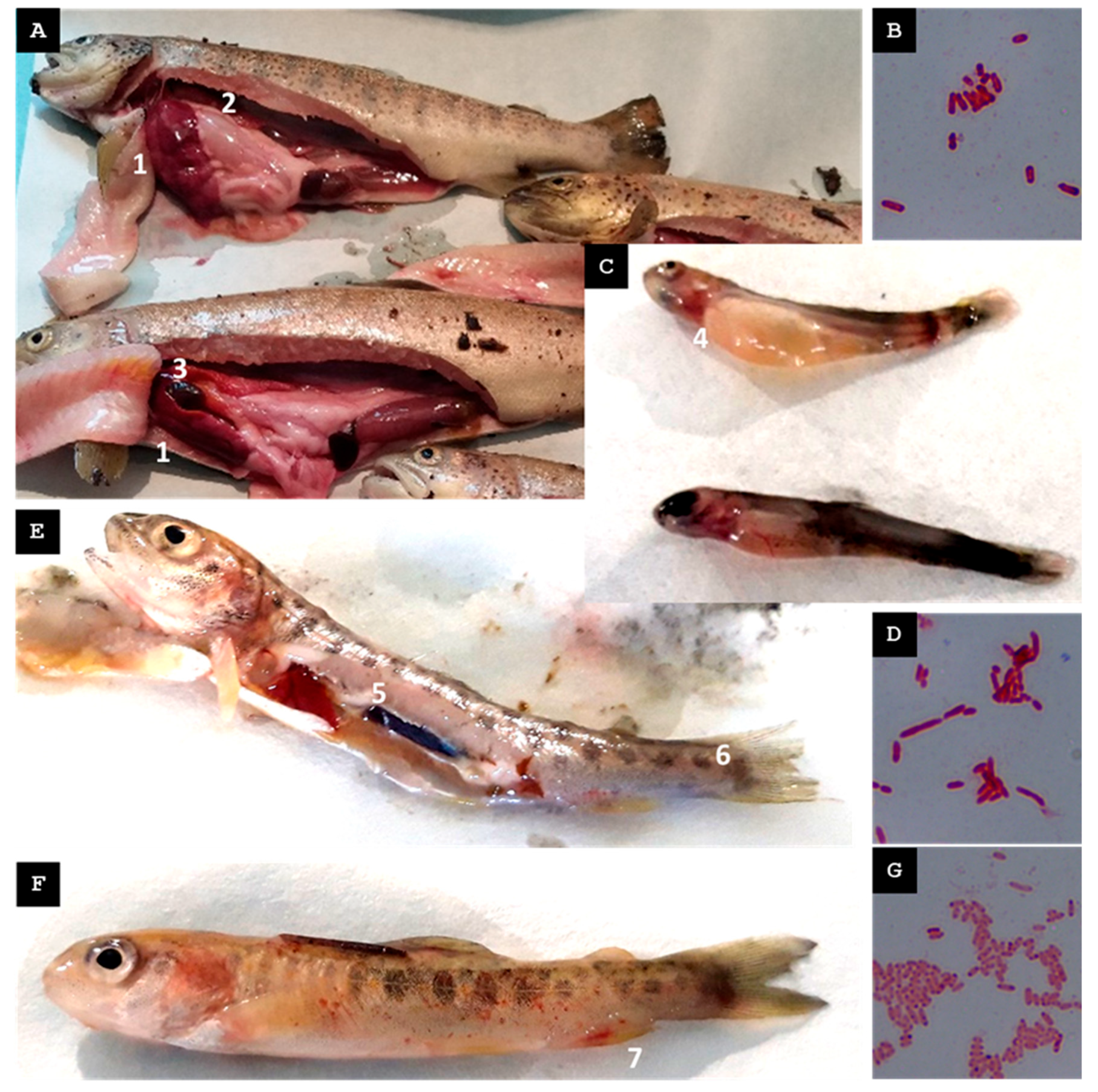
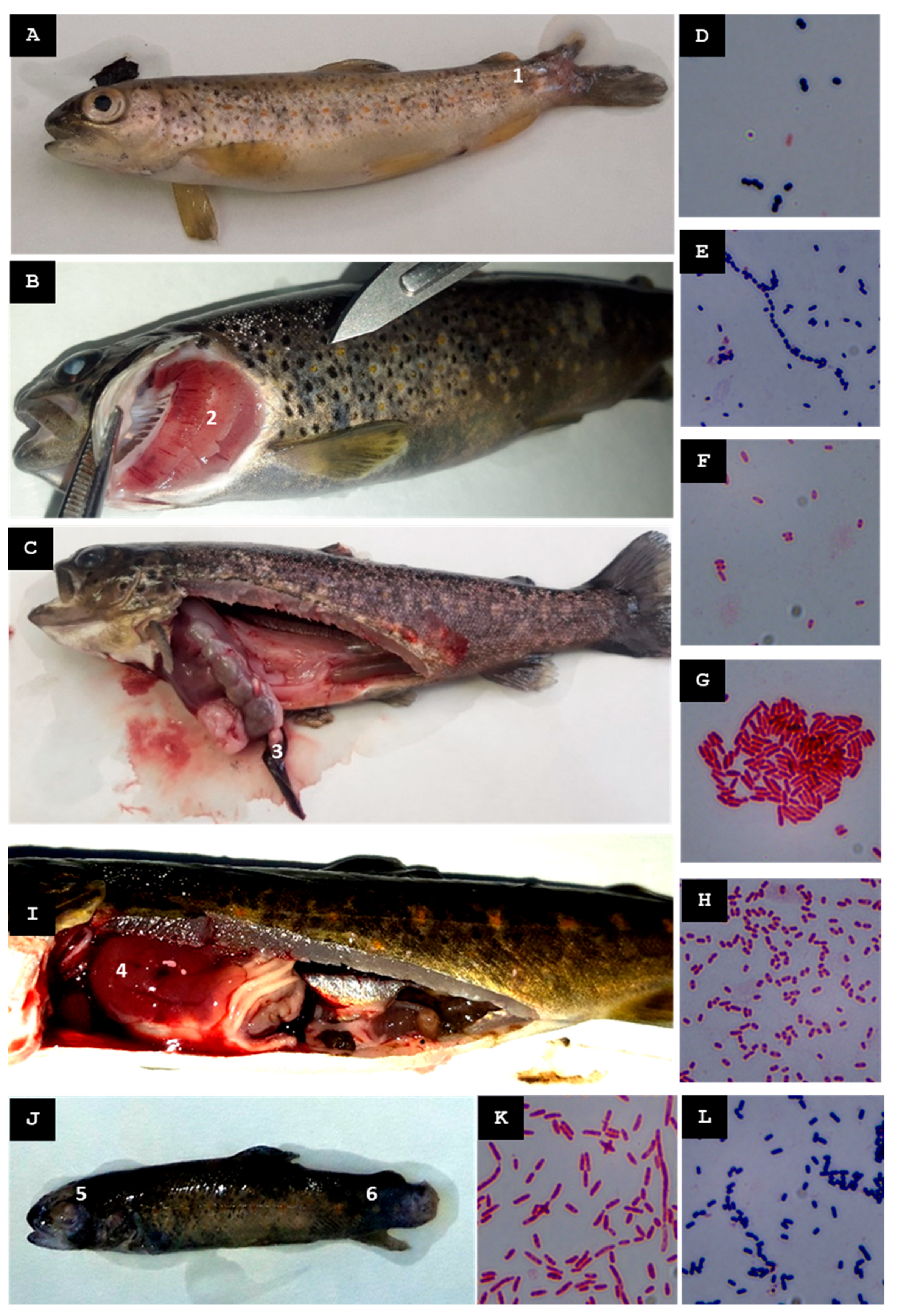
| A | c. 1600 inhabitants | 1st | Mid-Spring 2022 | Water temp. 4 °C above the usual levels | Broodstock exhibiting erratic swimming, ascitic fluid in the peritoneal cavity, hepatomegaly, hepatic hemorrhages, stomach dilation, and gallbladder distension | St-Pi3-L-87 Plesiomonas sp. (hepatic tissue) | Streptomycin |
| 2nd | Mid-Spring 2023 | Increase in water temperature | Yolk-sac fry showing melanosis and hemorrhagic lesions around the head and gill regions | St-Pi3-VL-99 Aeromonas sp. (sac fry) | None | ||
| 3rd | Late Summer 2023 | Increase in water temperature | Fry exhibiting erratic swimming, petechial hemorrhages along the lateral line, gill redness, liver hemorrhages, and enlarged posterior kidney | St-Pi3-OP-104 Hafnia alvei (organs pool; Pi3.L1) | None | ||
| B | c. 1600 inhabitants | 1st | Mid-Spring 2022 | Increase in the turbidity of the river water | Fry exhibiting erratic swimming, ocular hemorrhages, loss of the eyeballs, gill pallor, hemorrhagic skin lesions, hepatomegaly, and gastric dilation with mucous content | St-Pi5-Sp-107 Aeromonas encheleia (pooled spleen tissue) | None |
| 2nd | Mid-Autumn 2022 | None | Juveniles (batch Pi5.L1) exhibiting furuncles, petechiae in the liver and intestines, and splenomegaly | St-Pi5-Sk-98 Pseudomonas fulva (skin boils and abscesses) | Penicillin G | ||
| St-Pi5-Sk-89 A. salmonicida subsp. salmonicida (skin boils and abscesses) | None | ||||||
| Adult (batch Pi5.L2) displaying melanosis, petechial hemorrhages in liver and intestines | St-Pi5-L-92 Carnobacterium maltaromaticum (hepatic tissue) | None | |||||
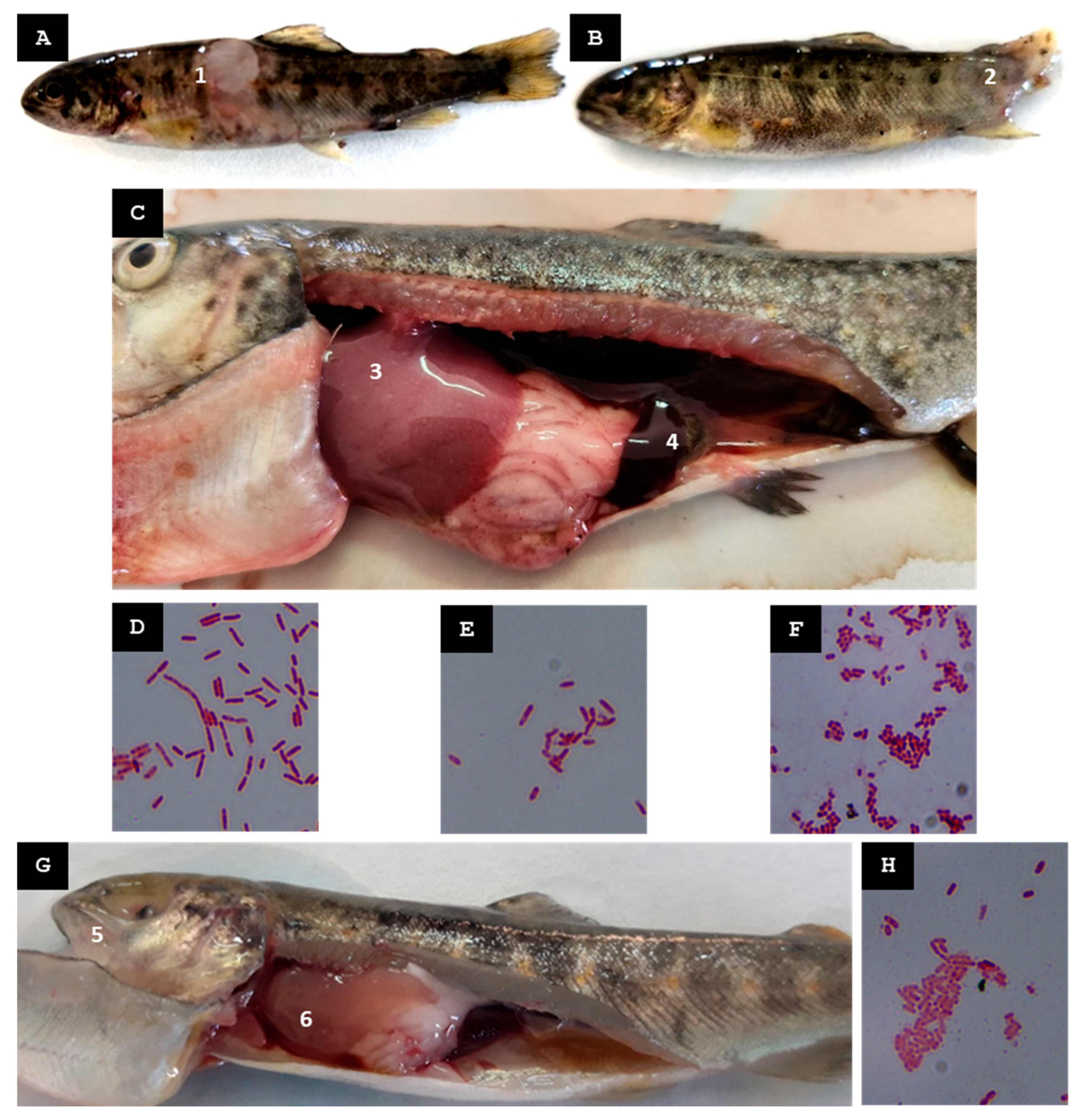
| C | c. 7300 inhabitants | 1st | Mid-Summer 2023 | None | Fry (batch Pi6.L1) exhibiting deep cutaneous ulcers, loss of muscle tissue, erosion and disappearance of the caudal fin | St-Pi6-OP-110 Aeromonas sp. (pooled tissue samples of liver and cutaneous lesions) | None |
| St-Pi6-OP-111 Aeromonas hydrophila (pooled tissue samples of liver and cutaneous lesions) | Oxytetracycline Erythromycin | ||||||
| Juveniles (batch Pi6.L2) exhibiting hepatomegaly and splenomegaly | St-Pi6-OP-113 Kluyvera intermedia (pooled tissue samples of liver, kidney, spleen, and ocular lesions) | Amoxicillin/clav. acid | |||||
| 2nd | Mid-Autumn 2023 | Heavy rainfall event increased the turbidity of the incoming water | Juveniles exhibited rupture of one or both eyeballs, hepatomegaly, and liver discoloration | St-Pi6-OP-105 Kluyvera intermedia (pooled tissue samples of liver, kidney, spleen, and muscle) | Amoxicillin/clav. acid | ||
| D | c. 100 inhabitants | 1st | Early Autumn 2022 | None | Juveniles exhibiting melanosis; no internal lesions were detected | St-Pi2-Sp-101 Aeromonas sp. (pooled tissue from liver and spleen) | None |
| E | c. 2100 inhabitants | 1st | Mid-Summer 2023 | None | Juveniles (batch Pi7.L1; size 9–10 cm) exhibiting cutaneous wounds with loss of muscle or fin tissue but no internal organ lesions | St-Pi7-OP-96 Enterococcus faecium (pooled tissue samples of liver, spleen, kidney, and wounded skin) | Oxytetracycline Doxycycline |
| St-Pi7-OP-115 Hafnia alvei (pooled tissue samples of liver, spleen, kidney, and wounded skin) | None | ||||||
| St-Pi7-OP-114 Lactococcus raffinolactis (pooled tissue samples of liver, spleen, kidney, and wounded skin) | Flumequine | ||||||
| Juveniles (batch Pi7.L2; size 12–16 cm) exhibiting dorsal fin hemorrhages and gill petechiae, distended gallbladder, and abundant mucous gastric contents | St-Pi7-OP-97 Aeromonas sp. (pooled tissue samples of wounded skin, kidney, liver, and spleen) | Florfenicol Erythromycin | |||||
| Juveniles (batch Pi7.L3; size 17–18 cm) exhibiting fin hemorrhages and splenomegaly | St-Pi7-OP-116 Hafnia alvei (pooled tissue samples of liver, kidney, and spleen) | None | |||||
| 2nd | Late Autumn 2023 | None | Juveniles exhibiting ruptured eyeballs, erosion of the caudal fin, and hepatomegaly with hepatic hemorrhages | St-Pi7-OP-118 Carnobacterium malta-romaticum (pooled tissue samples of liver and kidney) | None | ||
| St-Pi7-OP-119 Aeromonas sp. (pooled tissue samples of liver and kidney) | None |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vargas-González, A.; Barajas, M.; Pérez-Sánchez, T. Bacterial Isolates Associated with Mortality Events in Brown Trout (Salmo trutta) Restocking Farms in Spain: A Descriptive Field Study. Animals 2025, 15, 2532. https://doi.org/10.3390/ani15172532
Vargas-González A, Barajas M, Pérez-Sánchez T. Bacterial Isolates Associated with Mortality Events in Brown Trout (Salmo trutta) Restocking Farms in Spain: A Descriptive Field Study. Animals. 2025; 15(17):2532. https://doi.org/10.3390/ani15172532
Chicago/Turabian StyleVargas-González, Augusto, Miguel Barajas, and Tania Pérez-Sánchez. 2025. "Bacterial Isolates Associated with Mortality Events in Brown Trout (Salmo trutta) Restocking Farms in Spain: A Descriptive Field Study" Animals 15, no. 17: 2532. https://doi.org/10.3390/ani15172532
APA StyleVargas-González, A., Barajas, M., & Pérez-Sánchez, T. (2025). Bacterial Isolates Associated with Mortality Events in Brown Trout (Salmo trutta) Restocking Farms in Spain: A Descriptive Field Study. Animals, 15(17), 2532. https://doi.org/10.3390/ani15172532






