Population Genetic Structure, Historical Effective Population Size, and Dairy Trait Selection Signatures in Chinese Red Steppe and Holstein Cattle
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples and Sequencing Data
2.2. Data Quality Control
2.3. Population Genetic Structure Analysis
2.4. Identification of Selective Sweeps
2.5. Candidate Region Annotation and Functional Enrichment Analysis
3. Results and Analysis
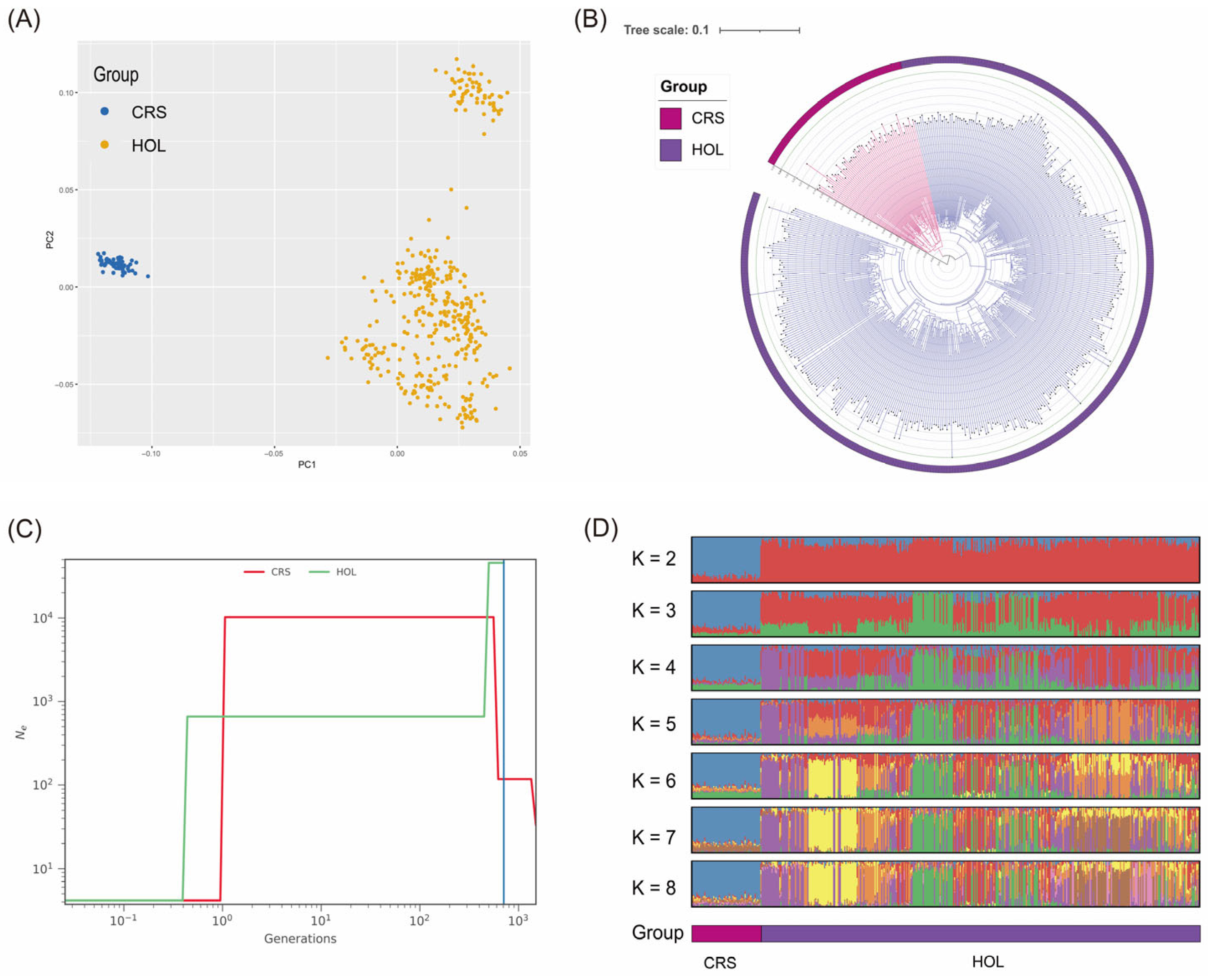
3.1. Population Structure
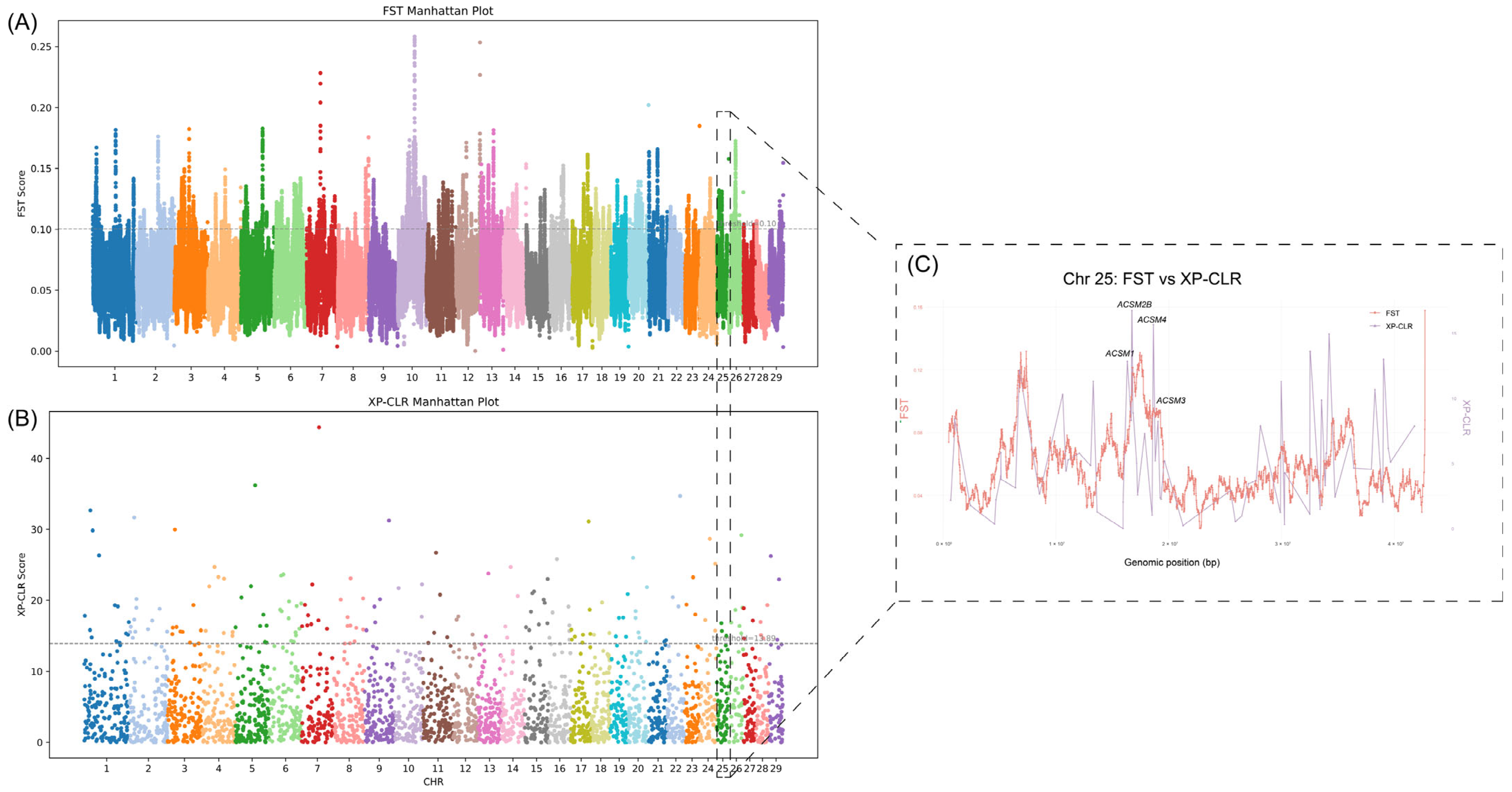
3.2. Selection Signal Detection
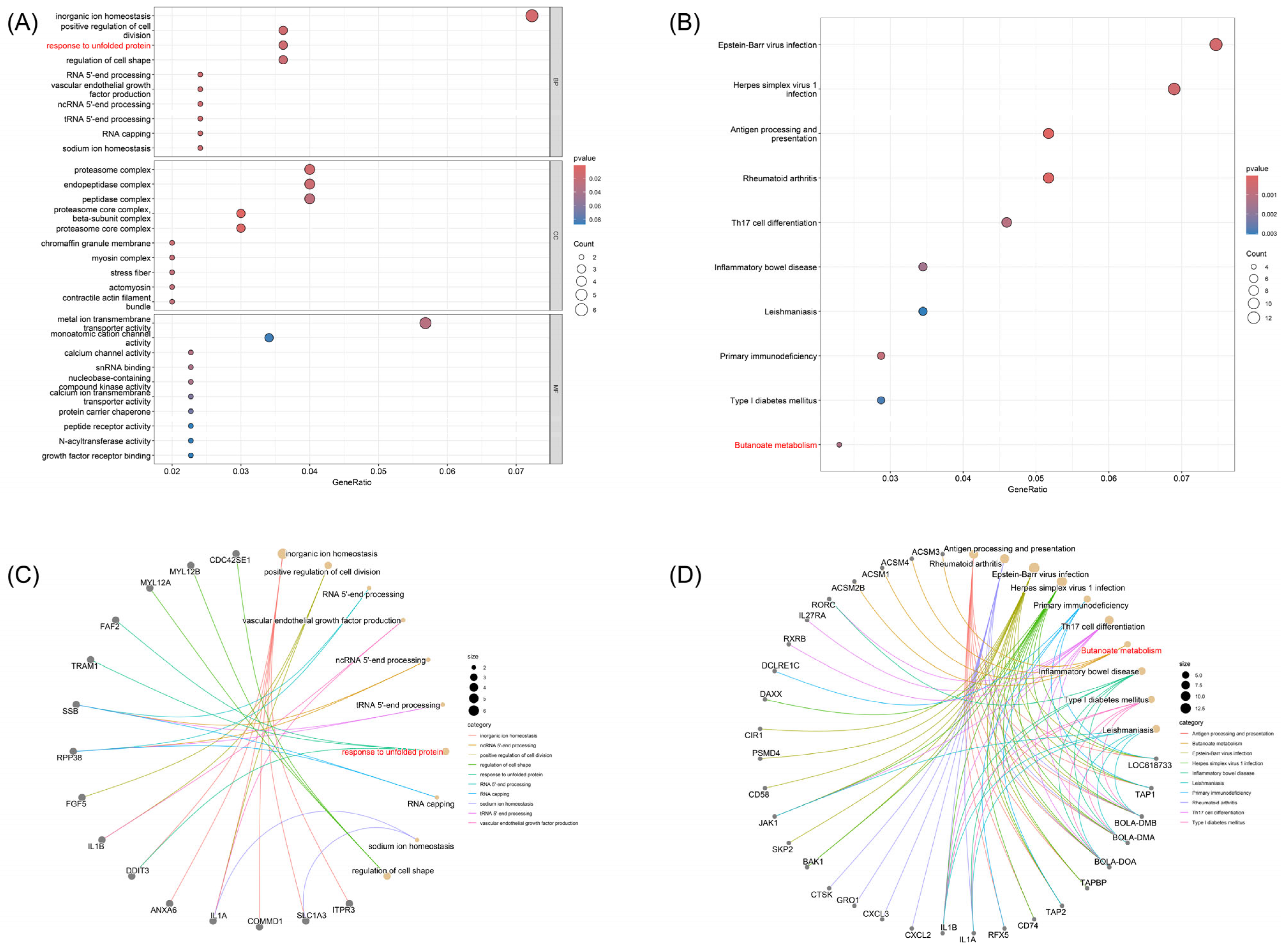
3.3. Enrichment Analysis of Milk Production-Related Pathways and Regulatory Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Nguyen, B.T.; Briggs, K.R.; Nydam, D.V. Dairy production sustainability through a one-health lens. J. Am. Vet. Med. Assoc. 2022, 261, 12–16. [Google Scholar] [CrossRef]
- Bhuvanendran, R.K.; Bhuvaneshwari, S. Hybrid electrocoagulation reactor for dairy wastewater treatment and methodology for sludge reusability for the development of vermicompost. Environ. Sci. Pollut. Res. Int. 2023, 30, 90960–90979. [Google Scholar] [CrossRef]
- Lopez-Gatius, F. Advances in Dairy Cattle Reproduction-A Foreword. Animals 2024, 14, 2650. [Google Scholar] [CrossRef]
- Braga, L.G.; Schenkel, F.S.; Chud, T.C.S.; Rodrigues, J.L.; Saada, B.; Machado, M.A.; Panetto, J.C.C.; Silva, M.; Munari, D.P. Selection signatures in Gir and Holstein cattle. J. Dairy Sci. 2025, 108, 9876–9900. [Google Scholar] [CrossRef]
- Tenhunen, S.; Thomasen, J.R.; Sorensen, L.P.; Berg, P.; Kargo, M. Genomic analysis of inbreeding and coancestry in Nordic Jersey and Holstein dairy cattle populations. J. Dairy Sci. 2024, 107, 5897–5912. [Google Scholar] [CrossRef] [PubMed]
- Lozada-Soto, E.A.; Tiezzi, F.; Jiang, J.; Cole, J.B.; VanRaden, P.M.; Maltecca, C. Genomic characterization of autozygosity and recent inbreeding trends in all major breeds of US dairy cattle. J. Dairy Sci. 2022, 105, 8956–8971. [Google Scholar] [CrossRef]
- Hu, M.; Jiang, H.; Lai, W.; Shi, L.; Yi, W.; Sun, H.; Chen, C.; Yuan, B.; Yan, S.; Zhang, J. Assessing Genomic Diversity and Signatures of Selection in Chinese Red Steppe Cattle Using High-Density SNP Array. Animals 2023, 13, 1717. [Google Scholar] [CrossRef]
- Fang, X.; Zhao, Z.; Yu, H.; Li, G.; Jiang, P.; Yang, Y.; Yang, R.; Yu, X. Comparative genome-wide methylation analysis of longissimus dorsi muscles between Japanese black (Wagyu) and Chinese Red Steppes cattle. PLoS ONE 2017, 12, e0182492. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Cao, Y.; Gao, Y.; Yun, J.; Yu, Y.; Zhang, L.; Hu, Z.; Liu, L.; Xue, J.; Zhang, G. Effect of ACSL3 Expression Levels on Preadipocyte Differentiation in Chinese Red Steppe Cattle. DNA Cell Biol. 2019, 38, 945–954. [Google Scholar] [CrossRef]
- Erdogan, M.; Cinkaya, S.; Brenig, B.; Celikeloglu, K.; Demirtas, M.; Sariibrahimoglu, S.; Tekerli, M. Genome-wide association studies for milk production traits and persistency of first calving Holstein cattle in Turkiye. Front. Vet. Sci. 2024, 11, 1461075. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Zhao, S.; Jia, Y.; Xu, L. Estimation of Linkage Disequilibrium, Effective Population Size, and Genetic Parameters of Phenotypic Traits in Dabieshan Cattle. Genes 2022, 14, 107. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Guan, X.; Xia, X.; Lyu, Y.; Liu, X.; Mazei, Y.; Xie, P.; Chang, F.; Zhang, X.; Chen, J.; et al. Evolution and legacy of East Asian aurochs. Sci. Bull. 2024, 69, 3425–3433. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Xia, X.; Hanif, Q.; Zhang, F.; Dang, R.; Huang, B.; Lyu, Y.; Luo, X.; Zhang, H.; Yan, H.; et al. Global genetic diversity, introgression, and evolutionary adaptation of indicine cattle revealed by whole genome sequencing. Nat. Commun. 2023, 14, 7803. [Google Scholar] [CrossRef]
- López, V.I.G.; Martínez-Rocha, R.; Domínguez, R.N.; Valverde, R.R.; Viveros, J.D.; Ceron, A.R.; Hidalgo, J. Genome-wide scan for selection signatures in Mexican Sardo Negro Zebu cattle. PLoS ONE 2024, 19, e0312453. [Google Scholar] [CrossRef]
- Hatlen, A.; Marco, A. Pervasive Selection against MicroRNA Target Sites in Human Populations. Mol. Biol. Evol. 2020, 37, 3399–3408. [Google Scholar] [CrossRef]
- Leigh, D.M.; Lischer, H.E.L.; Guillaume, F.; Grossen, C.; Gunther, T. Disentangling adaptation from drift in bottlenecked and reintroduced populations of Alpine ibex. Mol. Ecol. Resour. 2021, 21, 2350–2363. [Google Scholar] [CrossRef]
- Hejase, H.A.; Salman-Minkov, A.; Campagna, L.; Hubisz, M.J.; Lovette, I.J.; Gronau, I.; Siepel, A. Genomic islands of differentiation in a rapid avian radiation have been driven by recent selective sweeps. Proc. Natl. Acad. Sci. USA 2020, 117, 30554–30565. [Google Scholar] [CrossRef]
- Cheng, J.Y.; Stern, A.J.; Racimo, F.; Nielsen, R. Detecting Selection in Multiple Populations by Modeling Ancestral Admixture Components. Mol. Biol. Evol. 2022, 39, msab294. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Terhorst, J.; Kamm, J.A.; Song, Y.S. Robust and scalable inference of population history from hundreds of unphased whole genomes. Nat. Genet 2017, 49, 303–309. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Chen, H.; Patterson, N.; Reich, D. Population differentiation as a test for selective sweeps. Genome Res. 2010, 20, 393–402. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Cartuche-Macas, L.F.; Gutierrez-Reinoso, M.A.; Chacon, E.; Larrea-Izurieta, C.O.; Garcia-Flores, J.M.; Garcia-Herreros, M. Ecuadorian Holstein-Friesian cattle paternal lineages: Demographic structure, inbreeding evolution, and genetic diversity. PLoS ONE 2025, 20, e0318730. [Google Scholar] [CrossRef] [PubMed]
- Unlusoy, I. Determination of declined genetic diversity of Holstein stud bulls based on microsatellite markers. Anim. Biotechnol. 2023, 34, 4627–4633. [Google Scholar] [CrossRef] [PubMed]
- Shormanova, M.; Makhmutov, A.; Shormanova, A.; Muslimova, Z.; Ussenbekov, Y. Development of alternative diagnosis of HH1, HH3, HH5 and HCD fertility haplotypes and subfertility syndrome in cattle. Reprod. Domest. Anim. 2024, 59, e14533. [Google Scholar] [CrossRef]
- Ladeira, G.C.; Pinedo, P.J.; Santos, J.E.P.; Thatcher, W.W.; Rezende, F.M. Detecting and characterizing copy number variation in a large commercial U.S. Holstein cattle population. BMC Genom. 2025, 26, 381. [Google Scholar] [CrossRef]
- Sanchez-Molano, E.; Mukiibi, R.; Riggio, V.; Ogwang, J.; Kawule, L.; Benda, K.; Beine, P.; de Clare Bronsvoort, B.M.; Prendergast, J.; Doeschl-Wilson, A.B.; et al. Genomic and health characteristics of crossbred dairy cattle in central Uganda. Front. Genet. 2025, 16, 1567910. [Google Scholar] [CrossRef]
- Jaafar, M.A.; Heins, B.J.; Dechow, C.; Huson, H.J. The impact of using different ancestral reference populations in assessing crossbred population admixture and influence on performance. Front. Genet. 2022, 13, 910998. [Google Scholar] [CrossRef]
- Quenon, J.; Ingrand, S.; Magne, M.A. Assessing and explaining trends in dairy cattle herd performance variables while using three-breed rotational crossbreeding: Empirical evidence from commercial farms. Animal 2023, 17, 100983. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Zhao, Z.; Ge, F.; Yu, H.; Lyu, C.; Liu, Y.; Li, J.; Chen, Y. Deciphering the Population Characteristics of Leiqiong Cattle Using Whole-Genome Sequencing Data. Animals 2025, 15, 342. [Google Scholar] [CrossRef]
- Long, G.S.; Hider, J.; Duggan, A.T.; Klunk, J.; Eaton, K.; Karpinski, E.; Giuffra, V.; Ventura, L.; Prowse, T.L.; Fornaciari, A.; et al. A 14th century CE Brucella melitensis genome and the recent expansion of the Western Mediterranean clade. PLoS Pathog. 2023, 19, e1011538. [Google Scholar] [CrossRef]
- Huang, S.; Ma, L.; Li, B.; Dou, J.; Xu, Q.; Wang, Y. Genomic analysis reveals population structure and selection signatures in plateau dairy cattle. BMC Genom. 2025, 26, 240. [Google Scholar] [CrossRef]
- Mugambe, J.; Ahmed, R.H.; Thaller, G.; Schmidtmann, C. Impact of inbreeding on production, fertility, and health traits in German Holstein dairy cattle utilizing various inbreeding estimators. J. Dairy Sci. 2024, 107, 4714–4725. [Google Scholar] [CrossRef]
- Shrestha, R.K.; Nassar, Z.D.; Hanson, A.R.; Iggo, R.; Townley, S.L.; Dehairs, J.; Mah, C.Y.; Helm, M.; Alizadeh-Ghodsi, M.; Pickering, M.; et al. ACSM1 and ACSM3 Regulate Fatty Acid Metabolism to Support Prostate Cancer Growth and Constrain Ferroptosis. Cancer Res. 2024, 84, 2313–2332. [Google Scholar] [CrossRef]
- Kang, X.; Li, C.; Liu, S.; Baldwin, R.L.t.; Liu, G.E.; Li, C.J. Genome-Wide Acetylation Modification of H3K27ac in Bovine Rumen Cell Following Butyrate Exposure. Biomolecules 2023, 13, 1137. [Google Scholar] [CrossRef]
- Luo, C.; Li, N.; Wang, Q.; Li, C. Sodium acetate promotes fat synthesis by suppressing TATA element modulatory factor 1 in bovine mammary epithelial cells. Anim. Nutr. 2023, 13, 126–136. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Yang, B.; Qiu, L.; He, R.; Wu, Z.; Ye, M.; Zan, L.; Yang, W. Bta-miR-200a Regulates Milk Fat Biosynthesis by Targeting IRS2 to Inhibit the PI3K/Akt Signal Pathway in Bovine Mammary Epithelial Cells. J. Agric. Food Chem. 2024, 72, 16449–16460. [Google Scholar] [CrossRef] [PubMed]
- Strillacci, M.G.; Bernini, F.; Vevey, M.; Blanket, V.; Bagnato, A. The genomic comparison between autochthonous and cosmopolitan cows reveals structural variants involved in environmental adaptation. Sci. Rep. 2025, 15, 22280. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Niu, P.; Li, X.; Wang, X.; Qu, H.; Chen, H.; Huang, F.; Hu, K.; Fang, D.; Gao, Q. Population Genetic Structure, Historical Effective Population Size, and Dairy Trait Selection Signatures in Chinese Red Steppe and Holstein Cattle. Animals 2025, 15, 2516. https://doi.org/10.3390/ani15172516
Niu P, Li X, Wang X, Qu H, Chen H, Huang F, Hu K, Fang D, Gao Q. Population Genetic Structure, Historical Effective Population Size, and Dairy Trait Selection Signatures in Chinese Red Steppe and Holstein Cattle. Animals. 2025; 15(17):2516. https://doi.org/10.3390/ani15172516
Chicago/Turabian StyleNiu, Peng, Xiaopeng Li, Xueyan Wang, Huimin Qu, Hong Chen, Fei Huang, Kai Hu, Di Fang, and Qinghua Gao. 2025. "Population Genetic Structure, Historical Effective Population Size, and Dairy Trait Selection Signatures in Chinese Red Steppe and Holstein Cattle" Animals 15, no. 17: 2516. https://doi.org/10.3390/ani15172516
APA StyleNiu, P., Li, X., Wang, X., Qu, H., Chen, H., Huang, F., Hu, K., Fang, D., & Gao, Q. (2025). Population Genetic Structure, Historical Effective Population Size, and Dairy Trait Selection Signatures in Chinese Red Steppe and Holstein Cattle. Animals, 15(17), 2516. https://doi.org/10.3390/ani15172516








