The Effect of rs80860411 Polymorphism on Fattening, Slaughter, and Pork Quality Traits in Polish Large White and Pulawska Breeds
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Nutrition
2.2. Fattening Performance Test
2.3. Carcass Traits
2.4. Meat Quality Traits
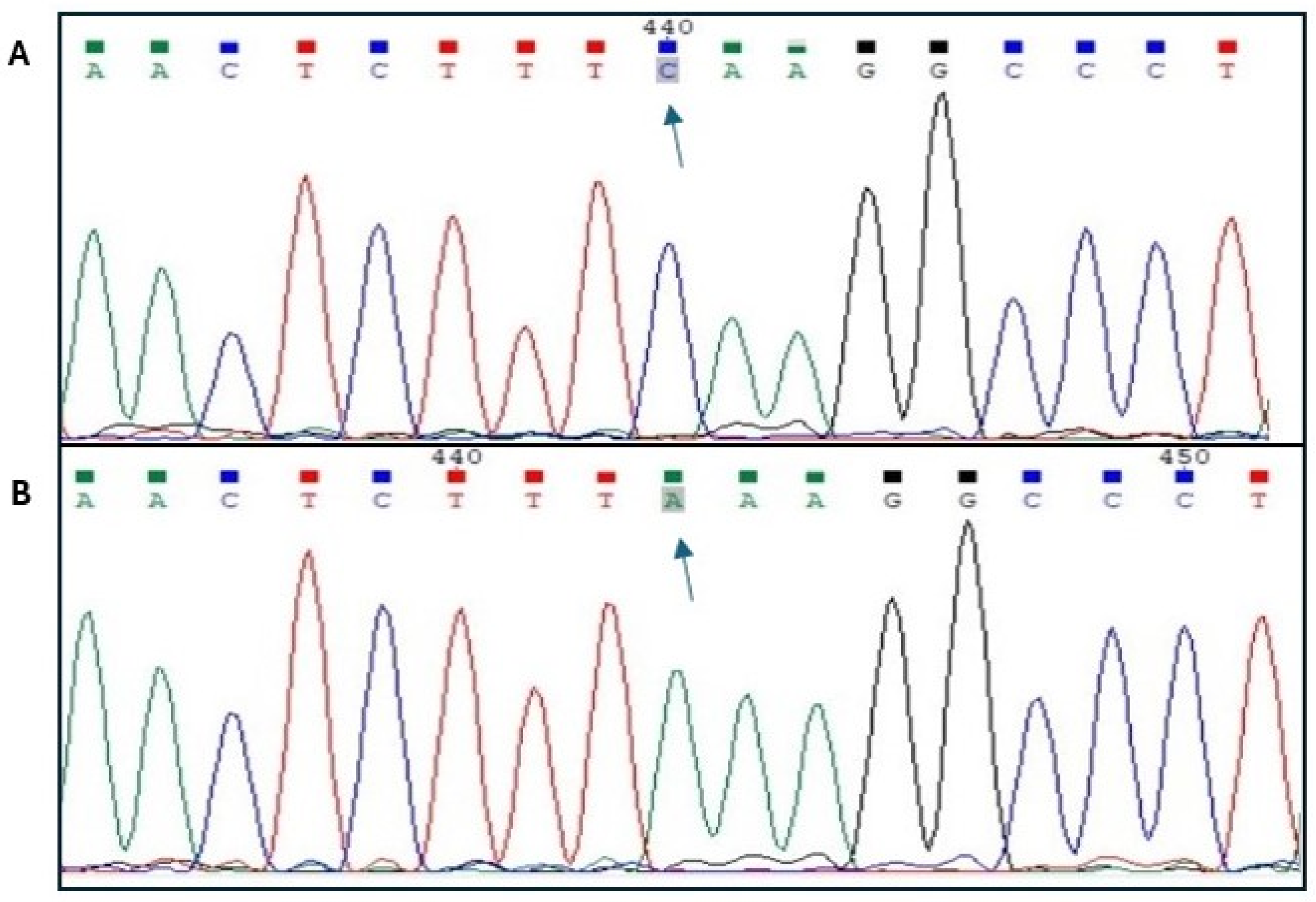
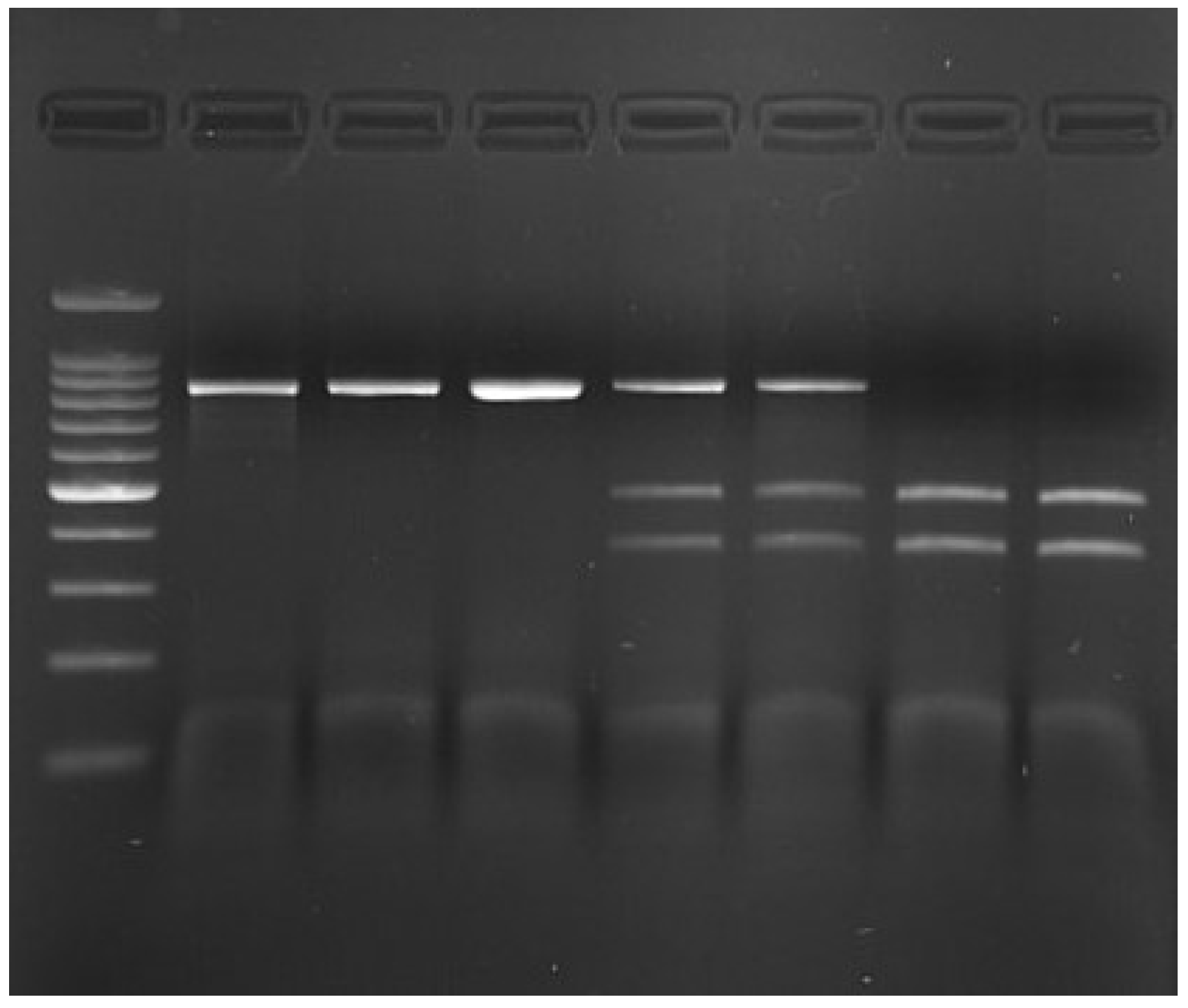
2.5. Genotyping
2.6. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Statistics Poland, G.U.S. Available online: https://stat.gov.pl/en/topics/agriculture-forestry/animal-production-farm-animals/pig-population-as-of-june-2024,3,1.html (accessed on 11 July 2025).
- Mucha, A.; Eckert, R.; Szyndler-Nędza, M.; Żak, G.; Bereta, A.; Różycki, M.; Tyra, M. Report in Pig Breeding in Poland in 2013, IZ BIP. 2014. ISSN 0239-5096. Available online: https://trzoda.izoo.krakow.pl/media/publishing/Wyniki_oceny_uzytkowej_swin_2014.pdf (accessed on 11 July 2025).
- Mucha, A.; Szyndler-Nędza, M.; Żak, G.; Tyra, M. Report in Pig Breeding in Poland in 2023, IZ BIP. 2024. ISSN 0239-5096. Available online: https://trzoda.izoo.krakow.pl/media/publishing/Wyniki_oceny_uzytkowej_swin_2024.pdf (accessed on 11 July 2025).
- Eurostat. Statistical Requirements Compendium, 2025th ed.; Eurostat: Luxembourg, 2025. [Google Scholar] [CrossRef]
- Mohammadabadi, M.; Bordbar, F.; Jensen, J.; Du, M.; Guo, W. Key Genes Regulating Skeletal Muscle Development and Growth in Farm Animals. Animals 2021, 11, 835. [Google Scholar] [CrossRef] [PubMed]
- Yan, E.; Guo, J.; Yin, J. Nutritional regulation of skeletal muscle energy metabolism, lipid accumulation and meat quality in pigs. Anim. Nutr. 2023, 14, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Bogucka, J.; Kapelański, W. Microstructure of longissimus lumborum muscle and meat quality of native Polish pig breeds: Złotnicka Spotted and Puławska. Ann. Anim. Sci. 2016, 16, 1199–1210. [Google Scholar] [CrossRef]
- Kasprzyk, A.; Bogucka, J. Meat quality of Pulawska breed pigs and image of longissimus lumborum muscle microstructure compared to commercial DanBred and Naima hybrids. Arch. Anim. Breed. 2020, 63, 293–301. [Google Scholar] [CrossRef] [PubMed]
- Sosnicki, A.; Gonzalez, J.; Fields, B.; Knap, P. A review of porcine skeletal muscle plasticity and implications for genetic improvement of carcass and meat quality. Meat Sci. 2025, 219, 109676. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-H.; Song, K.-D.; Lee, H.-K.; Cho, K.-H.; Park, H.-C.; Park, K.-D. Genetic Parameters of Reproductive and Meat Quality Traits in Korean Berkshire Pigs. Asian-Australas. J. Anim. Sci. 2015, 28, 1388–1393. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Esfandyari, H.; Thekkoot, D.; Kemp, R.; Plastow, G.; Dekkers, J. Genetic Parameters and Purebred-Crossbred Genetic Correlations for Growth, Meat Quality, and Carcass Traits in Pigs. J. Anim. Sci. 2020, 98, skaa379. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Z.; Wu, J.; Xu, C.; Ruan, D.; Qiu, Y.; Zhou, S.; Ding, R.; Quan, J.; Yang, M.; Zheng, E.; et al. The Genetic Architecture of Meat Quality Traits in a Crossbred Commercial Pig Population. Foods 2022, 11, 3143. [Google Scholar] [CrossRef] [PubMed]
- Rothschild, M.F.; Hu, Z.L.; Jiang, Z. Advances in QTL mapping in pigs. Int. J. Biol. Sci. 2007, 3, 192–197. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Q.; Du, Z.Q. Advances in the discovery of genetic elements underlying longissimus dorsi muscle growth and development in the pig. Anim. Genet. 2023, 54, 709–720. [Google Scholar] [CrossRef] [PubMed]
- Rohrer, G.A.; Nonneman, D.J.; Miller, R.K.; Zerby, H.; Moeller, S.J. Association of single nucleotide polymorphism (SNP) markers in candidate genes and QTL regions with pork quality traits in commercial pigs. Meat Sci. 2012, 92, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Fujii, J.; Otsu, K.; Zorzato, F.; De Leon, S.; Khanna, V.K.; Weiler, J.E.; O’Brien, P.J.; MacLennan, D.H. Identification of a mutation in porcine ryanodine receptor associated with malignant hyperthermia. Science 1991, 253, 448–451. [Google Scholar] [CrossRef] [PubMed]
- Nezer, C.; Moreau, L.; Brouwers, B.; Coppieters, W.; Detilleux, J.; Hanset, R.; Karim, L.; Kvasz, A.; Leroy, P.; Georges, M. An imprinted QTL with major effect on muscle mass and fat deposition maps to the IGF2 locus in pigs. Nat. Genet. 1999, 21, 155–156. [Google Scholar] [CrossRef] [PubMed]
- Van Laere, A.S.; Nguyen, M.; Braunschweig, M.; Nezer, C.; Collette, C.; Moreau, L.; Archibald, A.L.; Haley, C.S.; Buys, N.; Tally, M.; et al. A regulatory mutation in IGF2 causes a major QTL effect on muscle growth in the pig. Nature 2003, 425, 832–836. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Chai, J.; Luo, Z.; He, H.; Chen, L.; Liu, X.; Zhou, Q. Meat and nutritional quality comparison of purebred and crossbred pigs. Anim Sci. J. 2018, 89, 202–210. [Google Scholar] [CrossRef] [PubMed]
- Xiang, G.; Ren, J.; Hai, T.; Fu, R.; Yu, D.; Wang, J.; Li, W.; Wang, H.; Zhou, Q. Editing porcine IGF2 regulatory element improved meat production in Chinese Bama pigs. Cell. Mol. Life Sci. 2018, 75, 4619–4628. [Google Scholar] [CrossRef] [PubMed]
- López-Buesa, P.; Burgos, C.; Galve, A.; Varona, L. Joint analysis of additive, dominant and first-order epistatic effects of four genes (IGF2, MC4R, PRKAG3 and LEPR) with known effects on fat content and fat distribution in pigs. Anim. Genet. 2014, 45, 133–137. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Lei, H.; Li, J.; Sun, A.; Ahmed, Z.; Duan, H.; Chen, L.; Zhang, B.; Lei, C.; Yi, K. Analysis of genetic diversity and selection signals in Chaling cattle of southern China using whole-genome scan. Anim. Genet. 2023, 54, 284–294. [Google Scholar] [CrossRef] [PubMed]
- Schierding, W.; Antony, J.; Cutfield, W.S.; Horsfield, J.A.; O’Sullivan, J.M. Intergenic GWAS SNPs are key components of the spatial and regulatory network for human growth. Hum. Mol. Genet. 2016, 25, 3372–3382. [Google Scholar] [CrossRef] [PubMed]
- Ron, M.; Weller, J.I. From QTL to QTN identification in livestock--winning by points rather than knock-out: A review. Anim. Genet. 2007, 38, 429–439. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Wang, Q.; Pan, Y. From QTL to QTN: Candidate gene set approach and a case study in porcine IGF1-FoxO pathway. PLoS ONE 2013, 8, e53452. [Google Scholar] [CrossRef] [PubMed]
- Salas, R.C.; Mingala, C.N. Genetic Factors Affecting Pork Quality: Halothane and Rendement Napole Genes. Anim. Biotechnol. 2017, 28, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Ropka-Molik, K.; Pawlina-Tyszko, K.; Żukowski, K.; Tyra, M.; Derebecka, N.; Wesoły, J.; Szmatoła, T.; Piórkowska, K. Identification of Molecular Mechanisms Related to Pig Fatness at the Transcriptome and miRNAome Levels. Genes 2020, 11, 600. [Google Scholar] [CrossRef] [PubMed]
- Pocrnic, I.; Lourenco, D.; Misztal, I. SNP profile for quantitative trait nucleotide in populations with small effective size and its impact on mapping and genomic predictions. Genetics 2024, 227, iyae103. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, J.; Xiong, X.; Chen, C.; Xing, Y.; Duan, Y.; Xiao, S.; Yang, B.; Ma, J. An Integrative Analysis of Transcriptome and GWAS Data to Identify Potential Candidate Genes Influencing Meat Quality Traits in Pigs. Front. Genet. 2021, 12, 748070. [Google Scholar] [CrossRef] [PubMed]
- Blicharski, T.; Polok, P.; Snopkiewicz, M. Results of the Pig Assessment in 2017 [Wyniki Oceny Trzody Chlewnej w 2017 Roku]; Polish Association of Pig Breeders and Producers POLSUS [Polski Związek Hodowców i Producentów Trzody Chlewnej POLSUS]: Warsaw, Poland, 2018; pp. 5–35. [Google Scholar]
- Różycki, M.; Tyra, M. Results of fattening and slaughter performance assessment of pigs at control stations [Wyniki oceny użytkowości tucznej i rzeźnej świń w stacjach kontroli]. In Stan Hodowli i Wyniki Oceny Świń w roku 2012; Zespół Wydawnictw i Poligrafii IZ PIB: Kraków, Poland, 2013; pp. 49–72. (In Polish) [Google Scholar]
- Tyra, M.; Żak, G. Analysis of relationships between fattening and slaughter performance of pigs and the level of intramuscular fat (IMF) in longissimus dorsi muscle. Ann. Anim. Sci. 2012, 12, 169–178. [Google Scholar] [CrossRef]
- Oczkowicz, M.; Tyra, M.; Ropka-Molik, K.; Mucha, A.; Żukowski, K. Effect of IGF2 intron3-g.3072G>A on intramuscular fat (IMF) content in pigs raised in Poland. Livest. Sci. 2012, 149, 301–304. [Google Scholar] [CrossRef]
- Grau, R.; Hamm, R. Eine einfache Methode zur Bestimmung der Wasserbindung im Muskel. Naturwissenschaften 1953, 40, 29–30. [Google Scholar] [CrossRef]
- Ropka-Molik, K.; Bereta, A.; Tyra, M.; Różycki, M.; Piórkowska, K.; Szyndler-Nedza, M.; Szmatoła, T. Association of calpastatin gene polymorphism and meat quality traits in pig. Meat Sci. 2014, 97, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, Z.; Xiang, W.; Wang, F.; Nawaz, M.; Kuthu, Z.H.; Lei, C.; Xu, D. Whole-genome resequencing deciphers patterns of genetic diversity, phylogeny, and evolutionary dynamics in Kashmir cattle. Anim Genet. 2024, 55, 511–526. [Google Scholar] [CrossRef] [PubMed]
- Mishra, D.C.; Sikka, P.; Yadav, S.; Bhati, J.; Paul, S.S.; Jerome, A.; Singh, I.; Nath, A.; Budhlakoti, N.; Rao, A.R.; et al. Identification and characterization of trait-specific SNPs using ddRAD sequencing in water buffalo. Genomics 2020, 112, 3571–3578. [Google Scholar] [CrossRef] [PubMed]
- Mohammadpanah, M.; Ayatollahi Mehrgardi, A.; Gilbert, H.; Larzul, C.; Mercat, M.J.; Esmailizadeh, A.; Momen, M.; Tusell, L. Genic and non-genic SNP contributions to additive and dominance genetic effects in purebred and crossbred pig traits. Sci. Rep. 2022, 12, 3795. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Chen, W.; Chen, G.; Flannick, J.; Fikse, E.; Smerin, G.; Degner, K.; Yang, Y.; Xu, C.; Consortium AMP-T2D-GENES; et al. Ancestry-specific high-risk gene variant profiling unmasks diabetes-associated genes. Hum. Mol. Genet. 2024, 33, 655–666. [Google Scholar] [CrossRef] [PubMed]
- Babicz, M.; Hałabis, M.; Skałecki, P.; Domaradzki, P.; Litwińczuk, A.; Kropiwiec-Domańska, K.; Łukasik, M. Breeding and performance potential of Puławska pigs. A review. Ann. Anim. Sci. 2020, 20, 343–354. [Google Scholar] [CrossRef]
- Szyndler-Nędza, M.; Tyra, M. Effect of fattening and slaughter value of Puławska gilts on their lifetime piglet production. Animal Sci. Genet. 2023, 19, 55–67. [Google Scholar] [CrossRef]
- Babicz, M.; Szyndler-Nędza, M.; Skrzypczak, E.; Kasprzyk, A. Reproductive Performance of Native Pulawska and High Productivity Polish Landrace Sows in the Context of Stress During the Period of Early Pregnancy. Reprod. Domest. Anim. 2016, 51, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Tyra, M.; Żak, G. Analysis of the Possibility of Improving the Indicators of Pork Quality Through Selection with Particular Consideration of Intramuscular Fat (MF) Content. Ann. Anim. Sci. 2013, 13, 33–44. [Google Scholar] [CrossRef][Green Version]
- Gjerlaug-Enger, E.; Aass, L.; Odegård, J.; Vangen, O. Genetic parameters of meat quality traits in two pig breeds measured by rapid methods. Animal 2010, 4, 1832–1843. [Google Scholar] [CrossRef] [PubMed]
- Terada, K.; Ohtani, T.; Ogawa, S.; Hirooka, H. Genetic parameters for carcass and meat quality traits in Jinhua, Duroc, and their crossbred pigs. J. Anim. Breed. Genet. 2024, 141, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Qin, J.; Yao, T.; Tang, X.; Cui, D.; Chen, L.; Rao, L.; Xiao, S.; Zhang, Z.; Huang, L. Genetic dissection of 26 meat cut, meat quality and carcass traits in four pig populations. Genet. Sel. Evol. 2023, 55, 43. [Google Scholar] [CrossRef] [PubMed]
- Kasprzyk, A.; Tyra, M.; Babicz, M. Fatty acid profile of pork from a local and a commercial breed. Arch. Anim. Breed. 2015, 58, 379–385. [Google Scholar] [CrossRef]
- Channon, H.A.; Hamilton, A.J.; D’Souza, D.N.; Dunshea, F.R. Estimating the impact of various pathway parameters on tenderness, flavour and juiciness of pork using Monte Carlo simulation methods. Meat Sci. 2016, 116, 58–66, ISSN 0309-1740. [Google Scholar] [CrossRef] [PubMed]
- Yue, G.; Russo, V.; Davoli, R.; Sternstein, I.; Brunsch, C.; Schröffelova, D.; Stratil, A.; Moser, G.; Bartenschlager, H.; Reiner, G.; et al. Linkage and QTL mapping for Sus scrofa chromosome 13. J. Anim. Breed. Genet. 2003, 120, 103–110. [Google Scholar] [CrossRef]
- Mohrmann, M.; Roehe, R.; Knap, P.W.; Looft, H.; Plastow, G.S.; Kalm, E. Quantitative trait loci associated with AutoFOM grading characteristics, carcass cuts and chemical body composition during growth of Sus scrofa. Anim. Genet. 2006, 37, 435–443. [Google Scholar] [CrossRef] [PubMed]
- Long, Y.; Ruan, G.R.; Su, Y.; Xiao, S.J.; Zhang, Z.Y.; Ren, J.; Ding, N.S.; Huang, L.S. Genome-wide association study identifies QTLs for EBV of backfat thickness and average daily gain in Duroc pigs. Genetika. 2015, 51, 371–378. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Tachibana, K.; Iwasaki, H.; Kameyama, A.; Zhang, Y.; Kubota, T.; Hiruma, T.; Tachibana, K.; Kudo, T.; Guo, J.M.; et al. Characterization of a novel human UDP-GalNAc transferase, pp-GalNAc-T15. FEBS Lett. 2004, 566, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Varki, A. Biological roles of glycans. Glycobiology 2017, 27, 3–49. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.; Qing, X.; Liao, J.; Zhuo, K. Role of Protein Glycosylation in Host-Pathogen Interaction. Cells 2020, 9, 1022. [Google Scholar] [CrossRef] [PubMed]
- Dang, K.; Jiang, S.; Gao, Y.; Qian, A. The role of protein glycosylation in muscle diseases. Mol. Biol. Rep. 2022, 49, 8037–8049. [Google Scholar] [CrossRef] [PubMed]
- Cieniewski-Bernard, C.; Montel, V.; Berthoin, S.; Bastide, B. Increasing O-GlcNAcylation level on organ culture of soleus modulates the calcium activation parameters of muscle fibers. PLoS ONE 2012, 7, e48218. [Google Scholar] [CrossRef] [PubMed]
- Cieniewski-Bernard, C.; Lambert, M.; Dupont, E.; Montel, V.; Stevens, L.; Bastide, B. O-GlcNAcylation, contractile protein modifications and calcium affinity in skeletal muscle. Front. Physiol. 2014, 5, 421. [Google Scholar] [CrossRef] [PubMed]
- Jozefczuk, J.; Kashofer, K.; Ummanni, R.; Henjes, F.; Rehman, S.; Geenen, S.; Wruck, W.; Regenbrecht, C.; Daskalaki, A.; Wierling, C.; et al. A Systems Biology Approach to Deciphering the Etiology of Steatosis Employing Patient-Derived Dermal Fibroblasts and iPS Cells. Front. Physiol. 2012, 3, 339. [Google Scholar] [CrossRef] [PubMed]
- Funari, V.A.; Day, A.; Krakow, D.; Cohn, Z.A.; Chen, Z.; Nelson, S.F.; Cohn, D.H. Cartilage-selective genes identified in genome-scale analysis of non-cartilage and cartilage gene expression. BMC Genom. 2007, 8, 165. [Google Scholar] [CrossRef] [PubMed]
- Valdés-Hernández, J.; Folch, J.M.; Crespo-Piazuelo, D.; Passols, M.; Sebastià, C.; Criado-Mesas, L.; Castelló, A.; Sánchez, A.; Ramayo-Caldas, Y. Identification of candidate regulatory genes for intramuscular fatty acid composition in pigs by transcriptome analysis. Genet. Sel. Evol. 2024, 56, 12, Erratum in Genet. Sel. Evol. 2024, 56, 14. https://doi.org/10.1186/s12711-024-00885-8. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Sun, Q.; Zhang, W.; Wu, F.; Liu, G.; Wang, T.; Wang, Z.; Wang, Q.; Zhang, J. Transcriptomic analysis of the longissimus thoracis muscle in pigs has identified molecular regulatory patterns associated with meat quality. Genomics 2024, 116, 110779. [Google Scholar] [CrossRef] [PubMed]
- Jacyno, E.; Pietruszka, A.; Kawecka, M.; Biel, W.; Kołodziej-Skalska, A. Phenotypic correlations of backfat thickness with meatiness traits, intramuscular fat, Longissimus muscle cholesterol and fatty acid composition in pigs. South Afr. S. Afr. J. Anim. Sci. 2015, 45, 122–128. [Google Scholar] [CrossRef]
- Lebret, B.; Čandek-Potokar, M. Review: Pork quality attributes from farm to fork. Part I. Carcass and fresh meat. Animal 2022, 16 (Suppl. S1), 100402. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, J.; Ma, C.; Wang, W.; Wang, H.; Jiang, Y. Comparative Transcriptomic Analysis of mRNAs, miRNAs and lncRNAs in the Longissimus dorsi Muscles between Fat-Type and Lean-Type Pigs. Biomolecules 2022, 12, 1294. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, A.; Koike, R.; Watanabe, S.; Kuribayashi, K.; Wabitsch, M.; Miyamoto, M.; Komuro, A.; Seki, M.; Nashimoto, M.; Shimizu-Ibuka, A.; et al. Polypeptide N-acetylgalactosaminyltransferase-15 regulates adipogenesis in human SGBS cells. Sci. Rep. 2024, 14, 20049. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Zhang, W.; Lin, Q.; Gao, Y.; Teng, J.; Xu, Z.; Cai, X.; Zhong, Z.; Wu, J.; Liu, Y.; et al. PigBiobank: A valuable resource for understanding genetic and biological mechanisms of diverse complex traits in pigs. Nucleic Acids Res. 2024, 52, D980–D989, Erratum in Nucleic Acids Res. 2024, 52, 2758. https://doi.org/10.1093/nar/gkae074. [Google Scholar] [CrossRef] [PubMed]
- Bartonicek, N.; Clark, M.B.; Quek, X.C.; Torpy, J.R.; Pritchard, A.L.; Maag, J.L.V.; Gloss, B.S.; Crawford, J.; Taft, R.J.; Hayward, N.K.; et al. Intergenic disease-associated regions are abundant in novel transcripts. Genome Biol. 2017, 18, 241. [Google Scholar] [CrossRef] [PubMed]
| Forward Reverse | 5′-CCACCCCAGACCTCTTGAAT-3′ 5′-GACTCTAGACTGAAGGCCCC-3′ |
|---|---|
| Amplicon length (bp) | 842 |
| Restriction fragments (bp) | AA—474 and 368 AC—842, 474 and 368 CC—842 (Not digested) |
| Breed | N | Genotype | Allele | HWE | ||||
|---|---|---|---|---|---|---|---|---|
| AA | AC | CC | A | C | ꭓ2 | P | ||
| Polish Large White | 187 | 0.04 (n = 7) | 0.42 (n = 79) | 0.54 (n = 101) | 0.25 | 0.75 | 3.1896 | 0.0741 |
| Pulawska | 48 | 0.12 (n = 6) | 0.48 (n = 23) | 0.40 (n = 19) | 0.36 | 0.64 | 0.0561 | 0.8127 |
| Trait | Genotype | Polish Large White | Pulawska | Whole Population |
|---|---|---|---|---|
| Number of days on test [days] | AA | 79.354 ± 5.39 | 100.333 ± 3.40 A | 84.772 ± 1.75 |
| AC | 83.738 ± 3.65 | 85.217 ± 1.74 B | 83.695 ± 1.41 | |
| CC | 82.628 ± 3.72 | 88.105 ± 1.91 B | 83.678 ± 2.35 | |
| Daily feed intake [kg] | AA | 2.396 ± 0.15 | 2.138 ± 0.06 | 2.405 ± 0.04 |
| AC | 2.319 ± 0.10 | 2.277 ± 0.03 | 2370 ± 0.03 | |
| CC | 2.365 ± 0.10 | 2.244 ± 0.03 | 2373 ± 0.06 | |
| Lifetime daily gain [g/day] | AA | 592.653 ± 36.73 | 498.833 ± 23.20 a | 564.981 ± 9.95 |
| AC | 566.227 ± 24.86 | 569.130 ± 11.85 b | 568.687 ± 7.99 | |
| CC | 574.518 ± 25.33 | 535.684 ± 13.04 b | 549.649 ± 14.49 | |
| Test daily gain [g/day] | AA | 883.968 ± 58.63 | 721.167 ± 27.90 Aa | 853.293 ± 18.8 |
| AC | 858.058 ± 39.69 | 829.30 ± 14.25 B | 857.247 ± 14.62 | |
| CC | 866.307 ± 40.44 | 805.158 ± 15.64 b | 854.655 ± 26.48 | |
| Feed conversion rate [kg/kg gain] | AA | 2.708 ± 0.11 | 2.988 ± 0.07 a | 2.842 ± 0.03 |
| AC | 2.710 ± 0.08 | 2.750 ± 0.04 b | 2.780 ± 0.03 | |
| CC | 2.737 ± 0.08 | 2.798 ± 0.04 a | 2.792 ± 0.05 | |
| Age at slaughter [days] | AA | 169.980 ± 11.07 | 204.833 ± 7.98 a | 180.146 ± 3.43 |
| AC | 180.444 ± 7.49 | 178.304 ± 4.08 b | 178.925 ± 2.52 | |
| CC | 176.930 ± 7.64 | 189.105 ± 4.48 b | 184.490 ± 4.56 |
| Trait | Genotype | Polish Large White | Pulawska | Whole Population |
|---|---|---|---|---|
| Carcass yield [%] | AA | 75.973 ± 0.40 | 76.230 ± 0.30 | 76.798 ± 0.14 |
| AC | 76.031 ± 0.27 | 76.420 ± 0.15 | 76.799 ± 0.11 | |
| CC | 76.097 ± 0.28 | 76.393 ± 0.17 | 76.814 ± 0.21 | |
| Middle length of carcass [cm] | AA | 77.603 ± 1.04 | 78.625 ± 0.85 | 79.181 ± 0.77 |
| AC | 78.764 ± 0.70 | 78.452 ± 0.44 | 79.563 ± 0.62 | |
| CC | 78.491 ± 0.71 | 78.203 ± 0.48 | 79.169 ± 1.12 | |
| Loin weight [kg] | AA | 7.439 ± 0.27 | 7.272 ± 0.23 | 7.772 ± 0.11 |
| AC | 7.327 ± 0.18 | 7.700 ± 0.12 | 7.721 ± 0.09 | |
| CC | 7.355 ± 0.19 | 7.545 ± 0.13 | 7.660 ± 0.16 | |
| Loin weight without skin and backfat [kg] | AA | 5.882 ± 0.26 | 5.736 ± 0.20 | 6.093 ± 0.09 |
| AC | 5.865 ± 0.17 | 6.079 ± 0.10 | 6.065 ± 0.08 | |
| CC | 5.855 ± 0.18 | 5.840 ± 0.11 | 5.899 ± 0.14 | |
| Ham weight without skin and backfat [kg] | AA | 9.538 ± 0.26 | 9.184 ± 0.22 | 9.213 ± 0.10 b |
| AC | 9.294 ± 0.17 | 9.214 ± 0.11 | 9.234 ± 0.08 b | |
| CC | 9.310 ± 0.18 | 8.823 ± 0.12 | 8.859 ± 0.15 a | |
| Loin eye area [cm2] | AA | 53.631 ± 2.35 | 52.217 ± 2.36 a | 51.379 ± 0.86 B |
| AC | 51.196 ± 1.58 | 53.972 ± 1.20 a | 53.057 ± 0.69 B | |
| CC | 51.518 ± 1.61 | 49.587 ± 1.32 b | 48.675 ± 1.25 A | |
| Width of loin eye [cm] | AA | 10.188 ± 0.35 | 10.431 ± 0.32 A | 10.373 ± 0.13 B |
| AC | 10.195 ± 0.23 | 10.554 ± 0.16 A | 10.581 ± 0.10 B | |
| CC | 10.295 ± 0.24 | 9.778 ± 0.18 B | 9.790 ± 0.18 A | |
| Height of loin eye [cm] | AA | 6.741 ± 0.28 | 6.977 ± 0.25 | 6.919 ± 1.10 |
| AC | 6.562 ± 0.18 | 6.924 ± 0.13 | 6.875 ± 0.03 | |
| CC | 6.608 ± 0.19 | 6.772 ± 0.14 | 6.718 ± 0.15 | |
| Average backfat thickness of five measurements [cm] | AA | 1.123 ± 0.14 | 1.295 ± 0.13 | 1.395 ± 0.05 |
| AC | 1.122 ± 0.09 | 1.371 ± 0.07 | 1.361 ± 0.04 | |
| CC | 1.177 ± 0.10 | 1.527 ± 0.07 | 1.538 ± 0.08 | |
| Carcass meat content [%] | AA | 63.155 ± 1.35 | 61.339 ± 1.18 A | 61.765 ± 0.51 B |
| AC | 61.939 ± 0.90 | 62.276 ± 0.60 A | 61.986 ± 0.41 B | |
| CC | 62.117 ± 0.92 | 59.254 ± 0.66 B | 59.131 ± 0.74 A | |
| Weight of primary cuts [kg] | AA | 24.261 ± 0.53 | 23.621 ± 0.45 A | 23.952 ± 0.20 b |
| AC | 23.797 ± 0.35 | 23.993 ± 0.23 A | 24.044 ± 0.16 b | |
| CC | 23.859 ± 0.36 | 22.838 ± 0.25 B | 22.954 ± 0.29 A |
| Trait | Genotype | Polish Large White | Pulawska | Whole Population |
|---|---|---|---|---|
| Intramuscular fat content [%] | AA | 1.542 ± 0.09 | 1.184 ± 0.08 | 1.149 ± 0.03 |
| AC | 1.180 ± 0.05 | 1.210 ± 0.04 | 1.200 ± 0.03 | |
| CC | 1.166 ± 0.05 | 1.190 ± 0.04 | 1.176 ± 0.05 | |
| Meat color— luminosity [L*] | AA | 51.345 ± 1.25 | 53.463 ± 0.72 | 52.780 ± 0.36 |
| AC | 51.692 ± 0.85 | 53.943 ± 0.37 | 52.567 ± 0.29 | |
| CC | 51.935 ± 0.86 | 54.600 ± 0.40 | 53.371 ± 0.52 | |
| Meat color— redness [a*] | AA | 17.653 ± 0.97 | 17.612 ± 0.40 a | 18.306 ± 0.23 |
| AC | 17.917 ± 0.66 | 16.450 ± 0.20 b | 18.213 ± 0.19 | |
| CC | 17.699 ± 0.67 | 16.545 ± 0.22 b | 18.086 ± 0.34 | |
| Meat color— yellowness [b*] | AA | 4.135 ± 0.82 | 2.208 ± 0.25 | 5.149 ± 0.18 |
| AC | 4.325 ± 0.58 | 2.297 ± 0.13 | 5.189 ± 0.14 | |
| CC | 4.381 ± 0.60 | 2.372 ± 0.14 | 5.273 ± 0.26 | |
| pH45 M. longissimus lumborum | AA | 6.055 ± 0.10 | 6.338 ± 0.07 | 6.199 ± 0.04 |
| AC | 6.090 ± 0.07 | 6.272 ± 0.03 | 6.215 ± 0.02 | |
| CC | 6.104 ± 0.07 | 6.319 ± 0.04 | 6.245 ± 0.05 | |
| pH24 M. longissimus lumborum | AA | 5.557 ± 0.04 | 5.665 ± 0.03 | 5.601 ± 0.02 |
| AC | 5.557 ± 0.05 | 5.637 ± 0.02 | 5.591 ± 0.01 | |
| CC | 5.569 ± 0.04 | 5.640 ± 0.02 | 5.590 ± 0.02 | |
| pH45 M. semimembranosus | AA | 6.288 ± 0.09 | 6.243 ± 0.06 | 6.356 ± 0.03 |
| AC | 6.358 ± 0.06 | 6.266 ± 0.03 | 6.345 ± 0.03 | |
| CC | 6.360 ± 0.06 | 6.283 ± 0.03 | 6.369 ± 0.48 | |
| pH24 M. semimembranosus | AA | 5.685 ± 0.05 | 5.623 ± 0.06 | 5.676 ± 0.02 |
| AC | 5.681 ± 0.03 | 5.632 ± 0.04 | 5.648 ± 0.02 | |
| CC | 5.692 ± 0.03 | 5.679 ± 0.03 | 5.702 ± 0.03 | |
| Cooking loss—loin (%) | AA | 28.241 ± 1.46 | 27.080 ± 1.58 | 27.879 ± 0.72 |
| AC | 28.163 ± 0.49 | 27.328 ± 0.84 | 27.868 ± 0.50 | |
| CC | 28.271 ± 0.42 | 27.877 ± 0.91 | 28.475 ± 1.15 | |
| Cooking loss—ham (%) | AA | 32.720 ± 3.29 | 32.620 ± 1.60 | 33.441 ± 0.55 |
| AC | 34.397 ± 0.99 | 32.927 ± 0.82 | 32.748 ± 0.37 | |
| CC | 33.301 ± 0.78 | 32.264 ± 0.93 | 32.292 ± 0.88 | |
| Water holding capacity | AA | 24.532 ± 3.08 | 32.273 ± 2.83 | 25.577 ± 1.14 |
| AC | 26.420 ± 2.09 | 33.995 ± 1.47 | 26.377 ± 0.92 | |
| CC | 26.394 ± 2.12 | 36.115 ± 1.60 | 28.687 ± 1.67 |
| Trait | Genotype | Polish Large White | Pulawska | Whole Population |
|---|---|---|---|---|
| Firmness (r) | AA | 18.534 ± 4.27 b | 16.030 ± 4.10 | 24.025 ± 1.59 |
| AC | 21.113 ± 1.42 b | 19.210 ± 2.19 | 22.625 ± 1.11 | |
| CC | 25.893 ± 1.23 a | 23.628 ± 2.37 | 28.061 ± 2.57 | |
| Toughness (r) | AA | 56.779 ± 12.73 | 53.800 ± 12.47 | 64.987 ± 4.71 |
| AC | 59.880 ± 4.24 | 50.448 ± 6.67 | 62.226 ± 3.27 | |
| CC | 69.097 ± 3.65 | 70.088 ± 7.20 | 81.735 ± 7.57 | |
| Firmness | AA | 63.713 ± 8.38 | 63.498 ± 7.18 | 76.140 ± 3.32 |
| AC | 79.516 ± 2.79 | 78.843 ± 3.84 | 79.691 ± 2.31 | |
| CC | 78.498 ± 2.40 | 72.262 ± 4.15 | 75.730 ± 5.34 | |
| Toughness | AA | 161.377 ± 21.77 | 146.483 ± 18.11 | 179.135 ± 9.09 |
| AC | 187.019 ± 7.26 | 192.796 ± 9.68 | 193.171 ± 6.32 | |
| CC | 185.649 ± 6.25 | 166.577 ± 10.46 | 174.124 ± 14.63 | |
| Hardness | AA | 5.665 ± 1.64 | 6.796 ± 1.42 | 7.243 ± 0.66 |
| AC | 7.046 ± 0.55 | 7.168 ± 0.76 | 7.943 ± 0.46 | |
| CC | 7.430 ± 0.47 | 7.369 ± 0.82 | 8.072 ± 1.06 | |
| Springiness | AA | 0.714 ± 0.03 | 0.686 ± 0.02 | 0.696 ± 0.01 |
| AC | 0.684 ± 0.01 | 0.686 ± 0.01 | 0.694 ± 0.01 | |
| CC | 0.686 ± 0.01 | 0.671 ± 0.01 | 0.680 ± 0.02 | |
| Cohesiveness | AA | 0.655 ± 0.02 | 0.637 ± 0.02 | 0.634 ± 0.01 |
| AC | 0.623 ± 0.01 | 0.631 ± 0.01 | 0.638 ± 0.01 | |
| CC | 0.623 ± 0.01 | 0.612 ± 0.01 | 0.616 ± 0.02 | |
| Chewiness | AA | 2.614 ± 0.82 | 3.081 ± 0.71 | 3.334 ± 0.34 |
| AC | 3.380 ± 0.27 | 3.196 ± 0.38 | 3.652 ± 0.24 | |
| CC | 3.393 ± 0.24 | 3.237 ± 0.41 | 3.648 ± 0.55 | |
| Resilience | AA | 0.278 ± 0.01 | 0.274 ± 0.01 | 0.272 ± 0.01 |
| AC | 0.266 ± 0.01 | 0.271 ± 0.01 | 0.275 ± 0.01 | |
| CC | 0.267 ± 0.01 | 0.259 ± 0.01 | 0.262 ± 0.01 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Antonyk, A.; Terman, A.; Tyra, M.; Żak, G.; Polasik, D.; Szyndler-Nędza, M.; Kulig, H.; Dybus, A. The Effect of rs80860411 Polymorphism on Fattening, Slaughter, and Pork Quality Traits in Polish Large White and Pulawska Breeds. Animals 2025, 15, 2090. https://doi.org/10.3390/ani15142090
Antonyk A, Terman A, Tyra M, Żak G, Polasik D, Szyndler-Nędza M, Kulig H, Dybus A. The Effect of rs80860411 Polymorphism on Fattening, Slaughter, and Pork Quality Traits in Polish Large White and Pulawska Breeds. Animals. 2025; 15(14):2090. https://doi.org/10.3390/ani15142090
Chicago/Turabian StyleAntonyk, Anna, Arkadiusz Terman, Mirosław Tyra, Grzegorz Żak, Daniel Polasik, Magdalena Szyndler-Nędza, Hanna Kulig, and Andrzej Dybus. 2025. "The Effect of rs80860411 Polymorphism on Fattening, Slaughter, and Pork Quality Traits in Polish Large White and Pulawska Breeds" Animals 15, no. 14: 2090. https://doi.org/10.3390/ani15142090
APA StyleAntonyk, A., Terman, A., Tyra, M., Żak, G., Polasik, D., Szyndler-Nędza, M., Kulig, H., & Dybus, A. (2025). The Effect of rs80860411 Polymorphism on Fattening, Slaughter, and Pork Quality Traits in Polish Large White and Pulawska Breeds. Animals, 15(14), 2090. https://doi.org/10.3390/ani15142090






