Integrated Transcriptomic and Proteomic Analysis Reveals Differential Gene and Protein Expression and Signaling Pathways During a 20 Km Endurance Exercise and Recovery in Mongolian Horses
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Muscle Sample Collection
2.2. RNA Extraction from Muscle Tissue and Reverse Transcription
2.3. Real-Time Quantitative PCR
2.4. RNA Transcriptome Sequencing (RNA-Seq)
2.5. Protein Extraction from Muscle Tissue
2.6. Proteomics Sequencing
2.7. Bioinformatics Analysis
2.7.1. Statistics of Raw Sequencing Data
2.7.2. Quality Control of Raw Sequencing Data
2.7.3. Expression Level Analysis
2.7.4. Differential Expression Analysis
2.7.5. GO Annotation and KEGG Enrichment Analysis of Differential Genes
2.7.6. Protein–Protein Interaction Network Analysis
3. Results
3.1. Transcriptome Changes in Muscles of Mongolian Horses Pre- and Post-Exercise
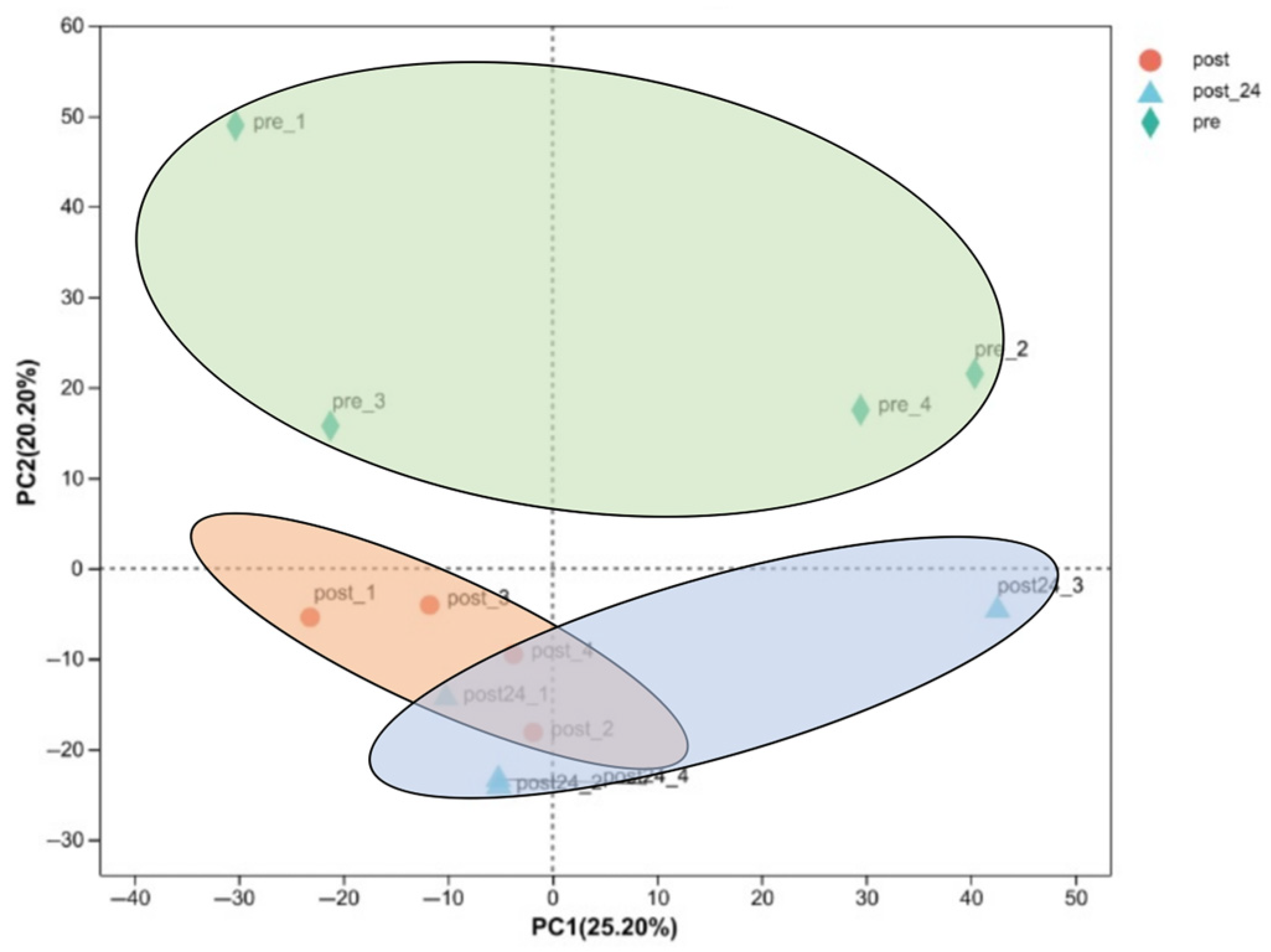
3.1.1. Comparative PCA of the Transcriptome Pre- and Post-Exercise
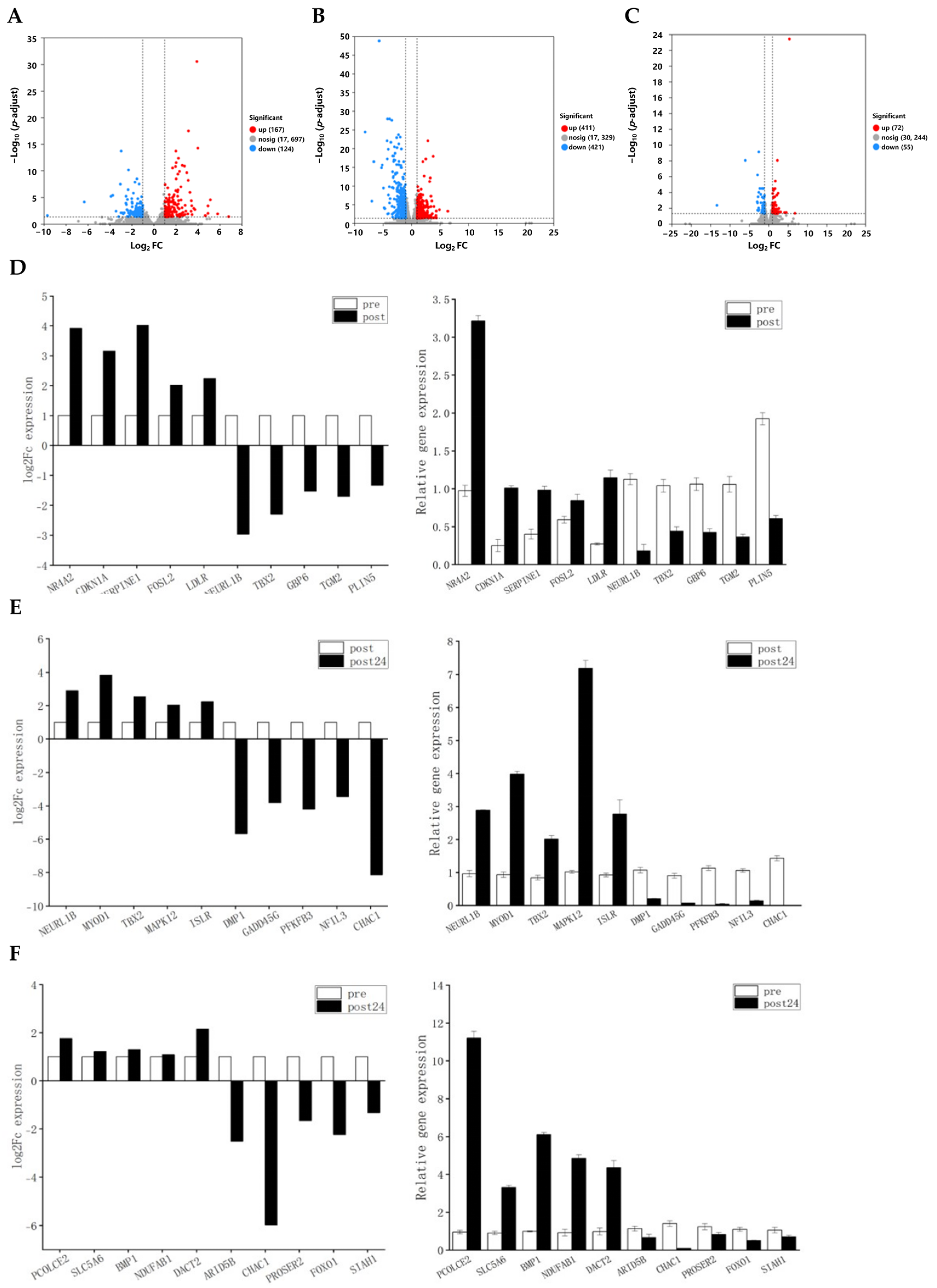
3.1.2. Differential Gene Expression (DEG) Analysis Pre- and Post-Exercise with Validation by Real-Time Fluorescent Quantitative PCR (RT-qPCR)
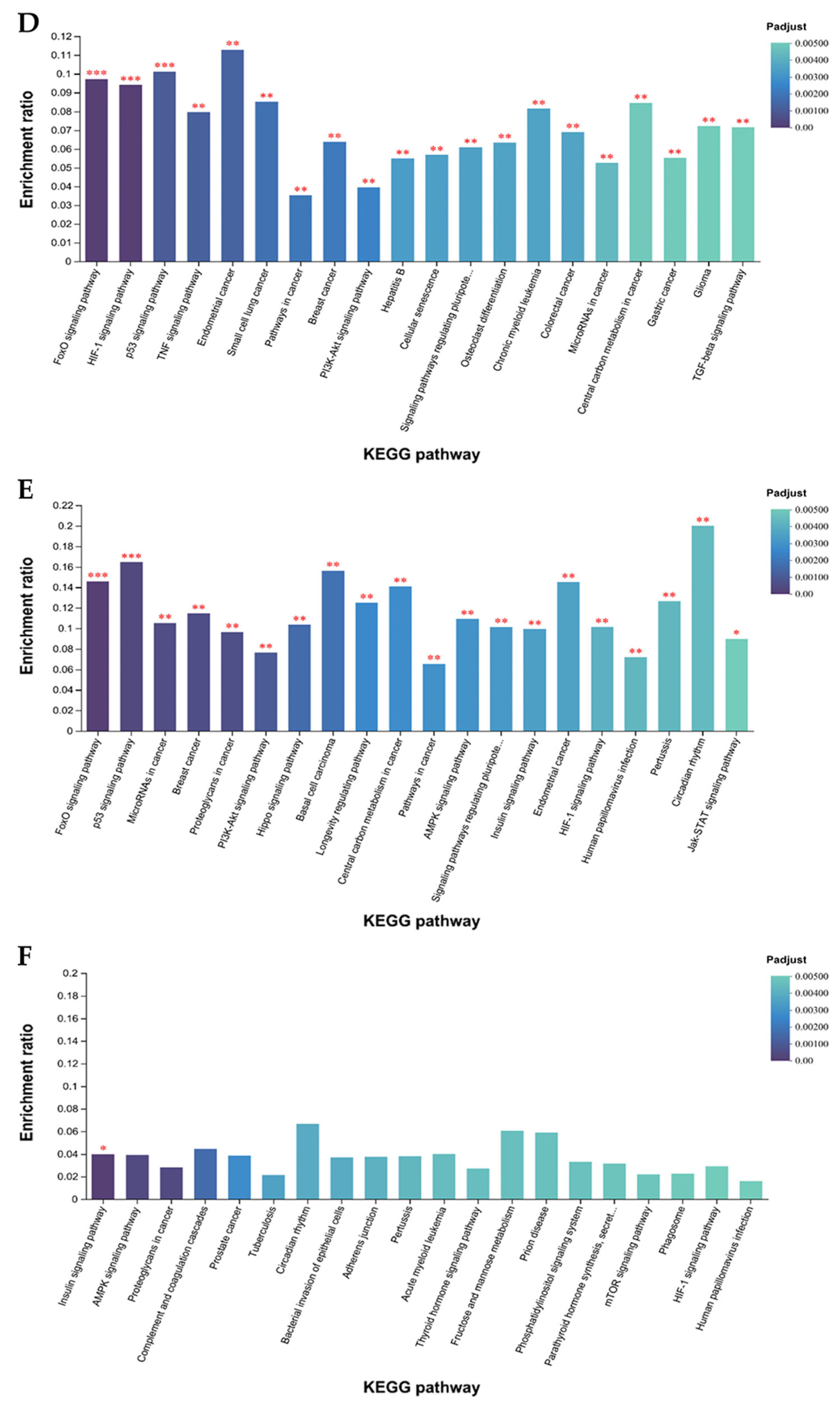
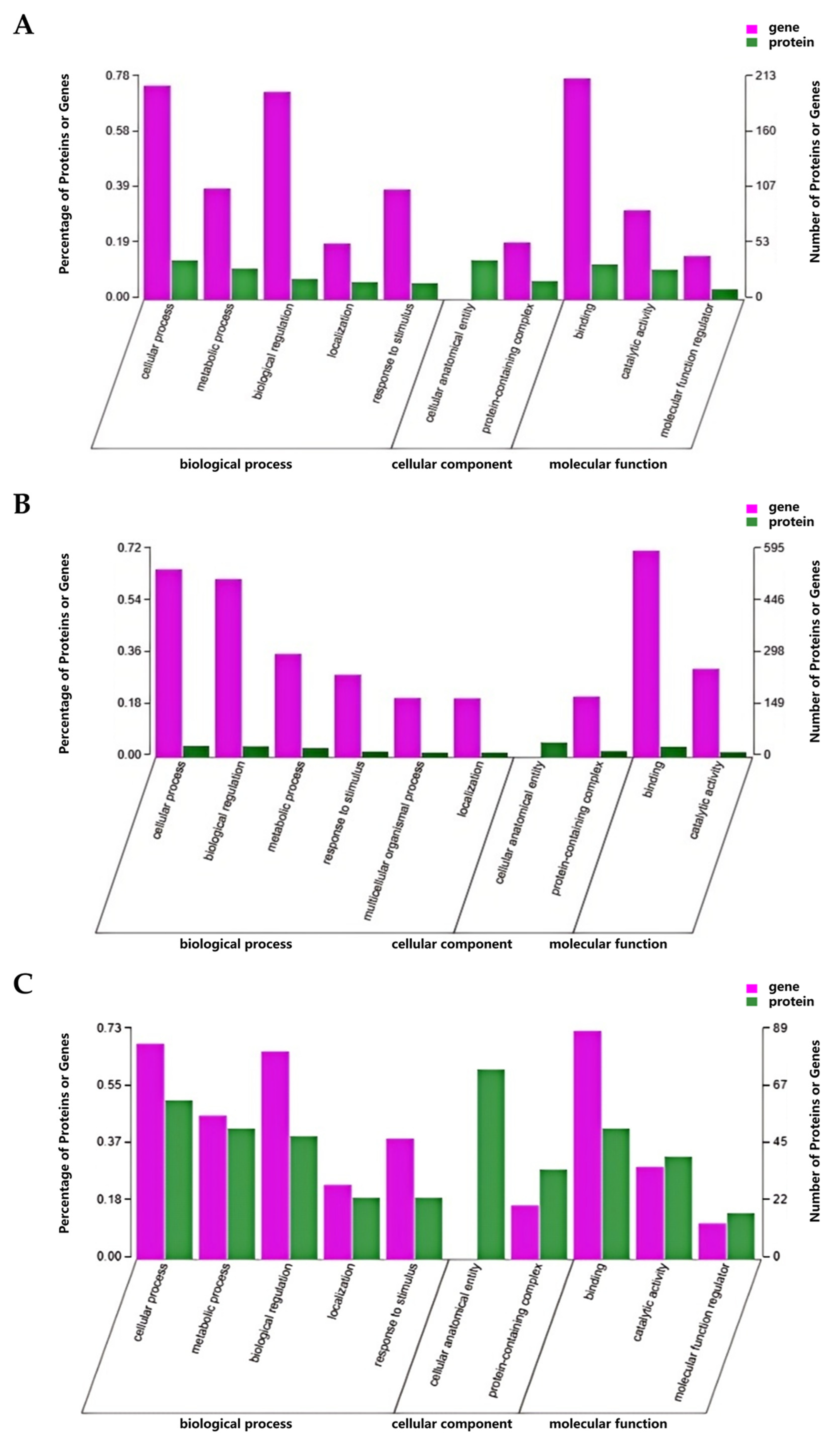
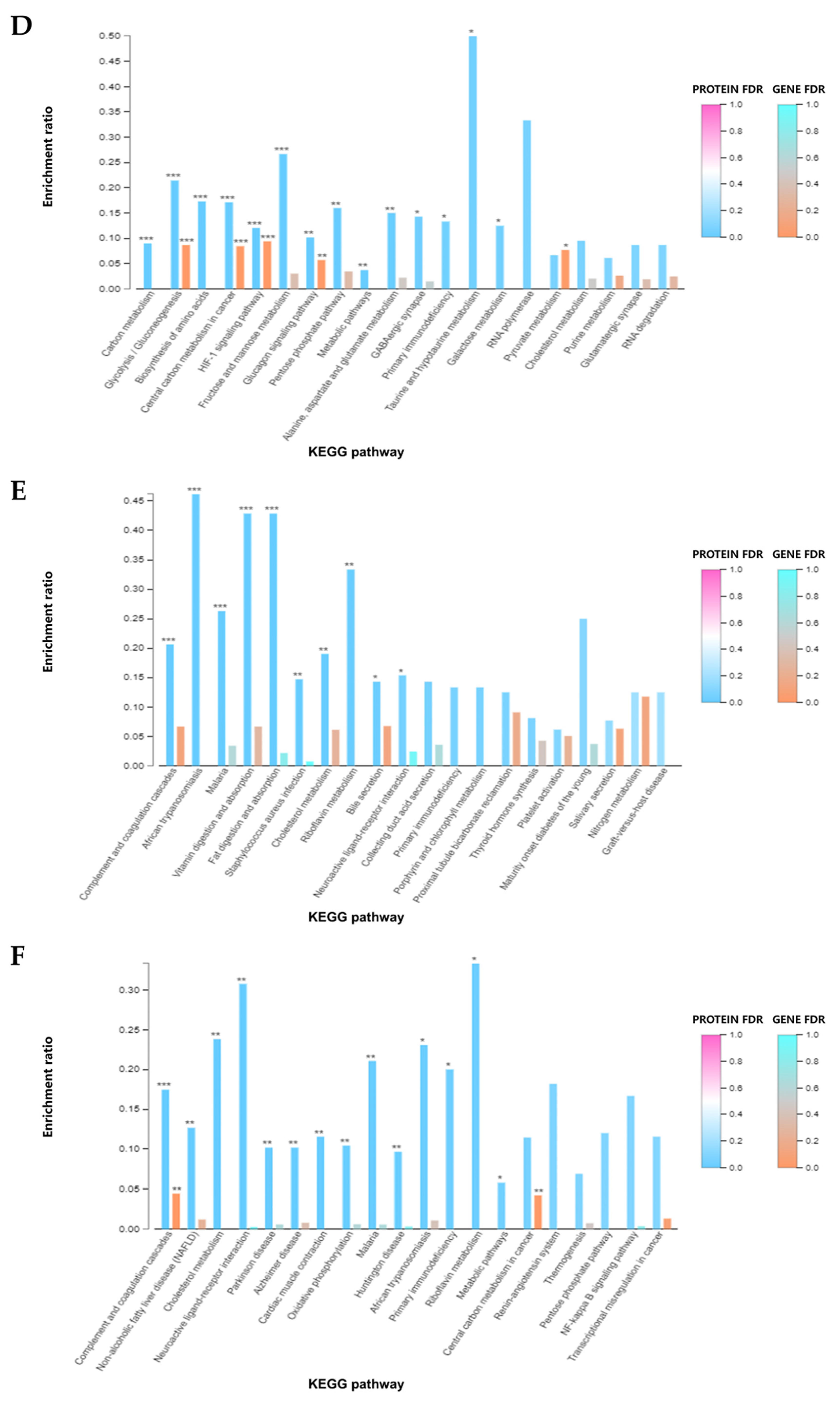
3.1.3. GO and KEGG Enrichment Analysis of DEGs
3.2. Proteome Changes in Muscles of Mongolian Horses Pre- and Post-Exercise
3.2.1. Comparative PCA of the Proteome Pre- and Post-Exercise
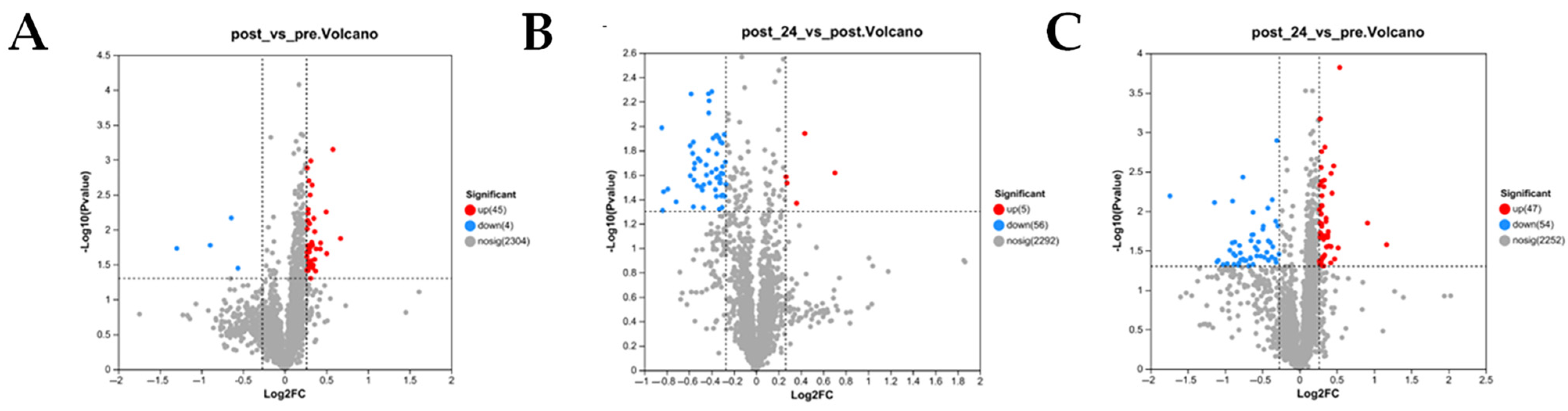
3.2.2. Differential Protein Expression (DEP) Analysis Pre- and Post-Exercise
3.2.3. GO and KEGG Enrichment Analysis of DEPs
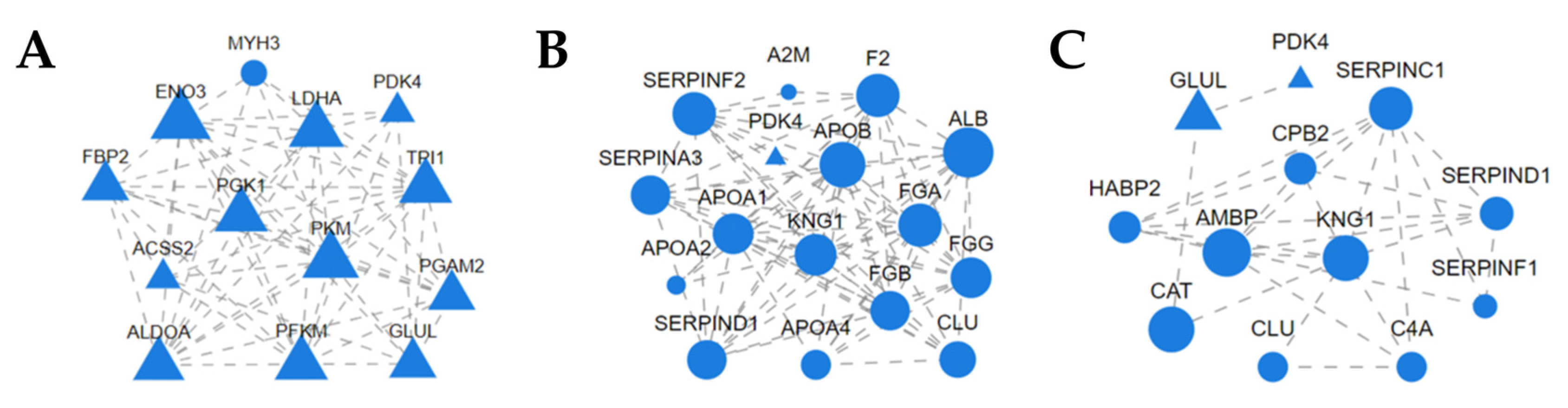
3.2.4. Protein–Protein Interaction Network Analysis of DEPs
3.3. Integrated Analysis of Muscle Transcriptome and Proteome in Mongolian Horses Pre- and Post-Exercise
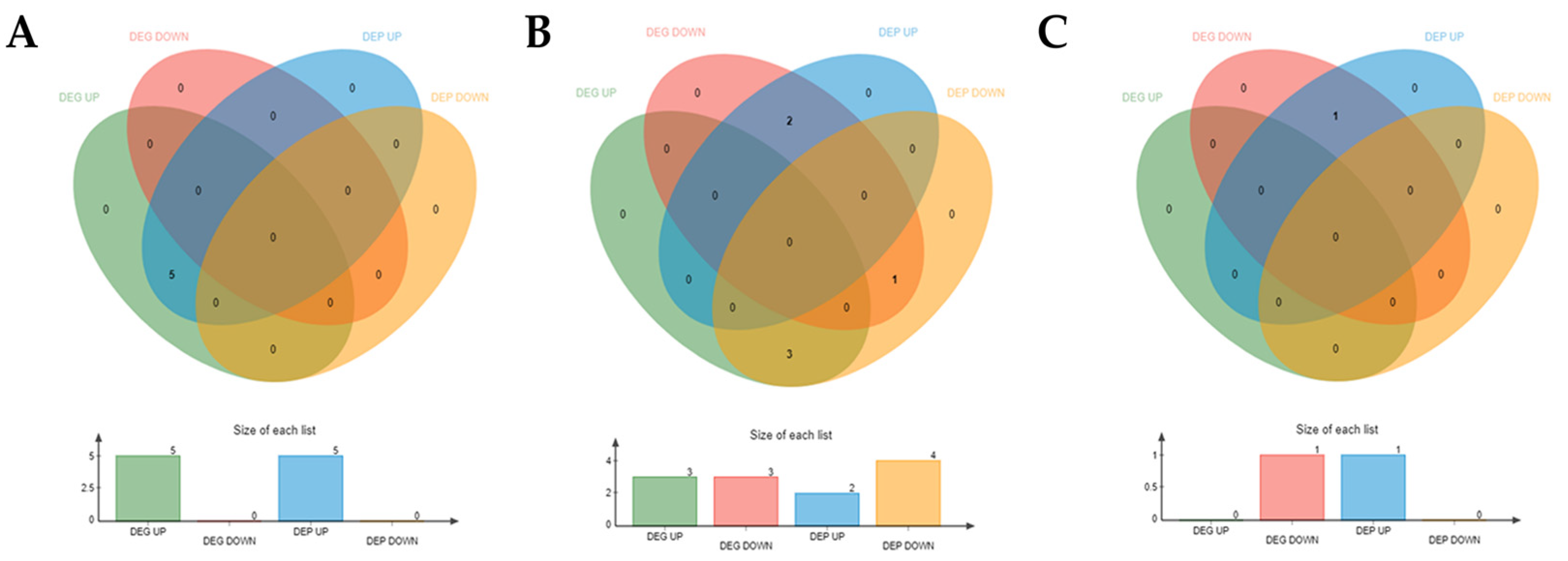
3.3.1. Integrated Analysis of DEGs and DEPs
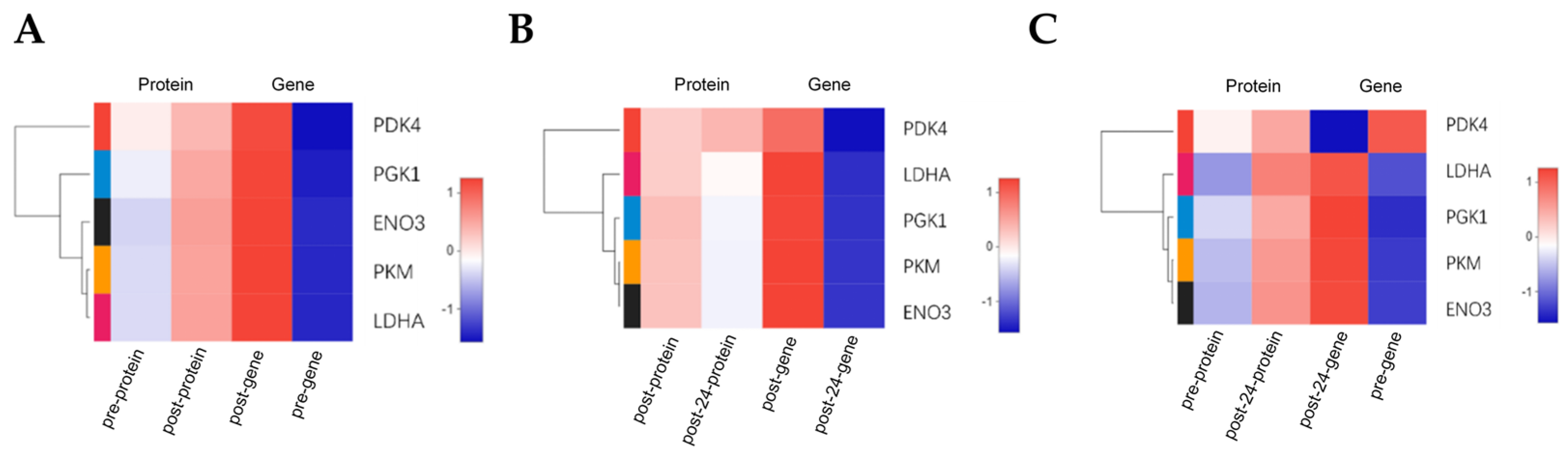
3.3.2. Cluster Analysis of Expression Levels in Correlated Data
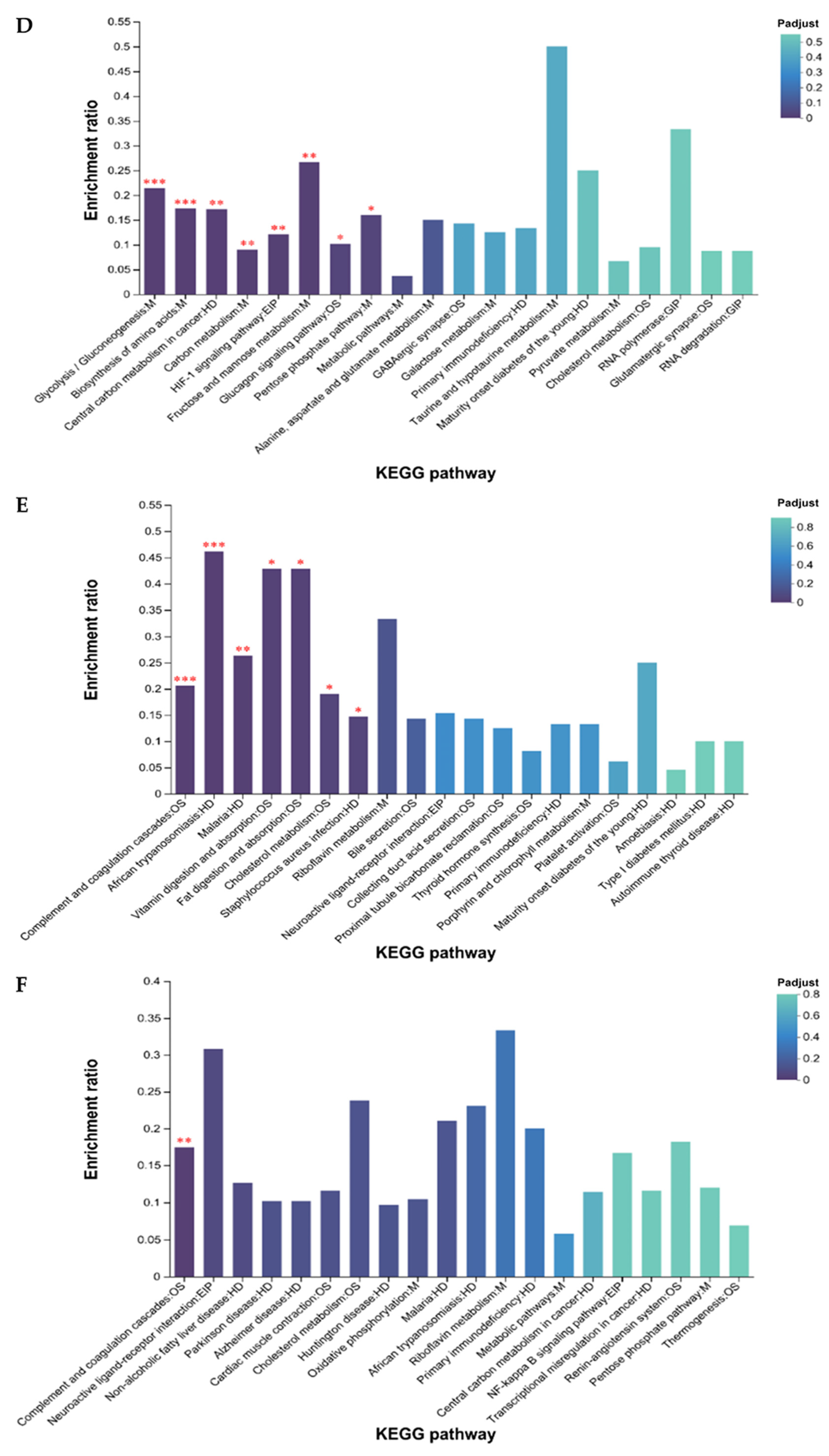
3.3.3. GO and KEGG Enrichment Analysis of DEGs and DEPs
4. Discussion
4.1. Transcriptome Changes in Muscles of Mongolian Horses Pre- and Post-Exercise
4.2. Proteome Changes in Muscles of Mongolian Horses Pre- and Post-Exercise
4.3. Integrated Analysis of Muscle Transcriptome and Proteome in Mongolian Horses Pre- and Post-Exercise
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, Y.; Sinjab, A.; Min, J.; Han, G.; Paradiso, F.; Zhang, Y.; Wang, R.; Pei, G.; Dai, Y.; Liu, Y.; et al. Conserved spatial subtypes and cellular neighborhoods of cancer-associated fibroblasts revealed by single-cell spatial multi-omics. Cancer Cell 2025, 43, 905–924. [Google Scholar] [CrossRef] [PubMed]
- Ayantoye, J.O.; Kolachi, H.A.; Zhang, X.; Shahzad, M.; Kandil, O.M.T.; Wan, P.; Zhao, X. Advances in Timed Artificial Insemination: Integrating Omics Technologies for Enhanced Reproductive Efficiency in Dairy Cattle. Animals 2025, 15, 816. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Li, P.; Huang, X.; Wang, C.; Zeng, Y.; Wang, J.; Yao, X.; Meng, J. Effects of Combined Transcriptome and Metabolome Analysis Training on Athletic Performance of 2-Year-Old Trot-Type Yili Horses. Genes 2025, 16, 197. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ren, W.; Li, Z.; Li, L.; Wang, R.; Ma, S.; Zeng, Y.; Meng, J.; Yao, X. Regulatory Mechanisms of Yili Horses During an 80 km Race Based on Transcriptomics and Metabolomics Analyses. Int. J. Mol. Sci. 2025, 26, 2426. [Google Scholar] [CrossRef]
- Wang, J.; Ren, W.; Sun, Z.; Han, Z.; Zeng, Y.; Meng, J.; Yao, X. Comparative transcriptome analysis of slow-twitch and fast-twitch muscles in Kazakh horses. Meat. Sci. 2024, 216, 109582. [Google Scholar] [CrossRef]
- Castiglione, G.M.; Chen, X.; Xu, Z.; Dbouk, N.H.; Bose, A.A.; Carmona-Berrio, D.; Chi, E.E.; Zhou, L.; Boronina, T.N.; Cole, R.N.; et al. Running a genetic stop sign accelerates oxygen metabolism and energy production in horses. Science 2025, 387, eadr8589. [Google Scholar] [CrossRef]
- Zhang, F.L.; Zhang, X.Y.; Zhao, J.X.; Zhu, K.X.; Liu, S.Q.; Zhang, T.; Sun, Y.J.; Wang, J.J.; Shen, W. Multispecies comparative analysis reveals transcriptional specificity during Mongolian horse testicular development. Reprod. Domest. Anim. 2022, 57, 1295–1306. [Google Scholar] [CrossRef]
- Bou, T.; Ding, W.; Ren, X.; Liu, H.; Gong, W.; Jia, Z.; Zhang, X.; Dugarjaviin, M.; Bai, D. Muscle fibre transition and transcriptional changes of horse skeletal muscles during traditional Mongolian endurance training. Equine Vet. J. 2024, 56, 178–192. [Google Scholar] [CrossRef]
- Shuken, S.R. An Introduction to Mass Spectrometry-Based Proteomics. J. Proteome Res. 2023, 22, 2151–2171. [Google Scholar] [CrossRef]
- MoTrPAC Study Group. Temporal dynamics of the multi-omic response to endurance exercise training. Nature 2024, 629, 174–183. [Google Scholar] [CrossRef]
- Noone, J.; Mucinski, J.M.; DeLany, J.P.; Sparks, L.M.; Goodpaster, B.H. Understanding the variation in exercise responses to guide personalized physical activity prescriptions. Cell Metab. 2024, 36, 702–724. [Google Scholar] [CrossRef]
- Zhao, L.; Yan, W.; Xiang, H.; Wang, X.; Qiao, H. Proteomic investigation of changes in rat skeletal muscle after exercise-induced fatigue. Biol. Res. 2012, 45, 75–80. [Google Scholar] [CrossRef]
- Ying, N.; Luo, H.; Li, B.; Gong, K.; Shu, Q.; Liang, F.; Gao, H.; Huang, T.; Zheng, H. Exercise Alleviates Behavioral Disorders but Shapes Brain Metabolism of APP/PS1 Mice in a Region- and Exercise-Specific Manner. J. Proteome Res. 2023, 22, 1649–1659. [Google Scholar] [CrossRef]
- Dai, W.; Chen, Q.; Wang, Q.; White, R.R.; Liu, J.; Liu, H. Complementary transcriptomic and proteomic analyses reveal regulatory mechanisms of milk protein production in dairy cows consuming different forages. Sci. Rep. 2017, 7, 44234. [Google Scholar] [CrossRef] [PubMed]
- Zeng, F.; Li, L.; Yang, J.; Liu, S.; Yuan, Y.; Zhao, C.; Wang, J. Transcriptomic and Proteomic Analyses Reveal New Insights into Regulatory Mechanisms of Strontium in Bovine Chondrocytes. Animals 2023, 13, 1301. [Google Scholar] [CrossRef] [PubMed]
- Xing, J.; Xie, L.; Qi, X.; Liu, G.; Akhtar, M.F.; Li, X.; Bou, G.; Bai, D.; Zhao, Y.; Dugarjaviin, M.; et al. Integrated analysis of transcriptome and proteome for exploring mechanism of promoting proliferation of equine satellite cells associated with leucine. Comp. Biochem. Physiol. Part D Genom. Proteom. 2023, 48, 101118. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, Y.; Li, L.; Ma, W.; Bai, D.; Dugarjaviin, M. Physiological and Metabolic Responses of Mongolian Horses to a 20 km Endurance Exercise and Screening for New Oxidative-Imbalance Biomarkers. Animals 2025, 15, 1350. [Google Scholar] [CrossRef]
- Li, B.; He, X.; Zhao, Y.; Bai, D.; Li, D.; Zhou, Z.; Manglai, D. Analysis of the miRNA transcriptome during testicular developmentand spermatogenesis of the Mongolian horse. Reprod. Fertil. Dev. 2020, 32, 582–593. [Google Scholar] [CrossRef]
- Bao, T.; Han, H.; Li, B.; Zhao, Y.; Bou, G.; Zhang, X.; Du, M.; Zhao, R.; Mongke, T.; Laxima; et al. The distinct transcriptomes of fast-twitch and slow-twitch muscles in Mongolian horses. Comp. Biochem. Physiol. Part D Genom. Proteom. 2020, 33, 100649. [Google Scholar] [CrossRef]
- Han, H.; McGivney, B.A.; Allen, L.; Bai, D.; Corduff, L.R.; Davaakhuu, G.; Davaasambuu, J.; Dorjgotov, D.; Hall, T.J.; Hemmings, A.J.; et al. Common protein-coding variants influence the racing phenotype in galloping racehorse breeds. Commun. Biol. 2022, 5, 1320. [Google Scholar] [CrossRef]
- Knierim, E.; Lucke, B.; Schwarz, J.M.; Schuelke, M.; Seelow, D. Systematic comparison of three methods for fragmentation of long-range PCR products for next generation sequencing. PLoS ONE 2011, 6, e28240. [Google Scholar] [CrossRef] [PubMed]
- Vantaggiato, L.; Landi, C.; Shaba, E.; Rossi, D.; Sorrentino, V.; Bini, L. Protein Extraction Methods Suitable for Muscle Tissue Proteomic Analysis. Proteomes 2024, 12, 27. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Yu, G.; Shi, C.; Liu, L.; Guo, Q.; Han, C.; Zhang, D.; Zhang, L.; Liu, B.; Gao, H.; et al. Majorbio Cloud: A one-stop, comprehensive bioinformatic platform for multiomics analyses. iMeta 2022, 1, e12. [Google Scholar] [CrossRef] [PubMed]
- Donovan, D.C.; Jackson, C.A.; Colahan, P.T.; Norton, N.; Hurley, D.J. Exercise-induced alterations in pro-inflammatory cytokines and prostaglandin F2alpha in horses. Vet. Immunol. Immunopathol. 2007, 118, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Purvis, D.; Gonsalves, S.; Deuster, P.A. Physiological and psychological fatigue in extreme conditions: Overtraining and elite athletes. PM R 2010, 2, 442–450. [Google Scholar] [CrossRef] [PubMed]
- Rennie, M.J.; Edwards, R.H.; Davies, C.T.; Krywawych, S.; Halliday, D.; Waterlow, J.C.; Millward, D.J. Protein and amino acid turnover during and after exercise. Biochem. Soc. Trans. 1980, 8, 499–501. [Google Scholar] [CrossRef]
- Rodriguez-Colman, M.J.; Dansen, T.B.; Burgering, B.M.T. FOXO transcription factors as mediators of stress adaptation. Nature reviews. Nat. Rev. Mol. Cell Biol. 2024, 25, 46–64. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Dong, H.H. FoxO integration of insulin signaling with glucose and lipid metabolism. J. Endocrinol. 2017, 233, R67–R79. [Google Scholar] [CrossRef]
- Link, W.; Ferreira, B.I. FOXO Transcription Factors: A Brief Overview. Methods Mol. Biol. 2025, 2871, 1–8. [Google Scholar]
- Guo, X.; Peng, K.; He, Y.; Xue, L. Mechanistic regulation of FOXO transcription factors in the nucleus. Biochim. Biophys. Acta Rev. Cancer 2024, 1879, 189083. [Google Scholar] [CrossRef]
- Solinas, G.; Becattini, B. PI3K and AKT at the Interface of Signaling and Metabolism. Curr. Top. Microbiol. Immunol. 2022, 436, 311–336. [Google Scholar] [PubMed]
- Shiau, J.P.; Chuang, Y.T.; Cheng, Y.B.; Tang, J.Y.; Hou, M.F.; Yen, C.Y.; Chang, H.W. Impacts of Oxidative Stress and PI3K/AKT/mTOR on Metabolism and the Future Direction of Investigating Fucoidan-Modulated Metabolism. Antioxidants 2022, 11, 911. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.S.; Cui, W. Proliferation, survival and metabolism: The role of PI3K/AKT/mTOR signalling in pluripotency and cell fate determination. Development 2016, 143, 3050–3060. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, N.; Heerding, D.A.; Duckett, D.R.; Eberwein, D.J.; Knick, V.B.; Lansing, T.J.; McConnell, R.T.; Gilmer, T.M.; Zhang, S.Y.; Robell, K.; et al. Characterization of an Akt kinase inhibitor with potent pharmacodynamic and antitumor activity. Cancer Res. 2008, 68, 2366–2374. [Google Scholar] [CrossRef]
- Hermida, M.A.; Dinesh Kumar, J.; Leslie, N.R. GSK3 and its interactions with the PI3K/AKT/mTOR signalling network. Adv. Biol. Regul. 2017, 65, 5–15. [Google Scholar] [CrossRef]
- Yamashita, S.I.; Sugiura, Y.; Matsuoka, Y.; Maeda, R.; Inoue, K.; Furukawa, K.; Fukuda, T.; Chan, D.C.; Kanki, T. Mitophagy mediated by BNIP3 and NIX protects against ferroptosis by downregulating mitochondrial reactive oxygen species. Cell Death Differ. 2024, 31, 651–661. [Google Scholar] [CrossRef]
- Semenza, G.L. HIF-1: Upstream and downstream of cancer metabolism. Curr. Opin. Genet. Dev. 2010, 20, 51–56. [Google Scholar] [CrossRef]
- Greijer, A.E.; van der Wall, E. The role of hypoxia inducible factor 1 (HIF-1) in hypoxia induced apoptosis. J. Clin. Pathol. 2004, 57, 1009–1014. [Google Scholar] [CrossRef]
- Majmundar, A.J.; Wong, W.J.; Simon, M.C. Hypoxia-inducible factors and the response to hypoxic stress. Mol. Cell. 2010, 40, 294–309. [Google Scholar] [CrossRef]
- Chen, Y.; Wu, G.; Li, M.; Hesse, M.; Ma, Y.; Chen, W.; Huang, H.; Liu, Y.; Xu, W.; Tang, Y.; et al. LDHA-mediated metabolic reprogramming promoted cardiomyocyte proliferation by alleviating ROS and inducing M2 macrophage polarization. Redox. Biol. 2022, 56, 102446. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, J.; Wang, J.; Zhang, T.; Xu, D.; Hu, W.; Feng, Z. The Interplay Between Tumor Suppressor p53 and Hypoxia Signaling Pathways in Cancer. Front. Cell Dev. Biol. 2021, 9, 648808. [Google Scholar] [CrossRef]
- Wu, D.; Prives, C. Relevance of the p53-MDM2 axis to aging. Cell Death Differ. 2018, 25, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Robertson, E.D.; Semenchenko, K.; Wasylyk, B. Crosstalk between Mdm2, p53 and HIF1-α: Distinct responses to oxygen stress and implications for tumour hypoxia. Subcell Biochem. 2014, 85, 199–214. [Google Scholar] [PubMed]
- Shi, T.; Dansen, T.B. Reactive Oxygen Species Induced p53 Activation: DNA Damage, Redox Signaling, or Both? Antioxid. Redox. Signal. 2020, 33, 839–859. [Google Scholar] [CrossRef] [PubMed]
- Karimian, A.; Ahmadi, Y.; Yousefi, B. Multiple functions of p21 in cell cycle, apoptosis and transcriptional regulation after DNA damage. DNA Repair 2016, 42, 63–71. [Google Scholar] [CrossRef]
- Franklin, S.H.; Van Erck-Westergren, E.; Bayly, W.M. Respiratory responses to exercise in the horse. Equine Vet. J. 2012, 44, 726–732. [Google Scholar] [CrossRef]
- Gastin, P.B. Energy system interaction and relative contribution during maximal exercise. Sports Med. 2001, 31, 725–741. [Google Scholar] [CrossRef]
- Pellegrino, J.K.; Anthony, T.G.; Gillies, P.; Arent, S.M. The exercise metabolome: Acute aerobic and anaerobic signatures. J. Int. Soc. Sports Nutr. 2022, 19, 603–622. [Google Scholar] [CrossRef]
- Gao, W.; Huang, M.; Chen, X.; Chen, J.; Zou, Z.; Li, L.; Ji, K.; Nie, Z.; Yang, B.; Wei, Z.; et al. The role of S-nitrosylation of PFKM in regulation of glycolysis in ovarian cancer cells. Cell Death Dis. 2021, 12, 408. [Google Scholar] [CrossRef]
- Wang, S.; Wu, X.; Bi, W.; Xu, J.; Hou, L.; Li, G.; Pan, Y.; Zhang, H.; Li, M.; Du, S.; et al. ROS-induced cytosolic release of mitochondrial PGAM5 promotes colorectal cancer progression by interacting with MST3. Nat. Commun. 2025, 16, 1406. [Google Scholar] [CrossRef]
- Li, S.; Wen, P.; Zhang, D.; Li, D.; Gao, Q.; Liu, H.; Di, Y. PGAM5 expression levels in heart failure and protection ROS-induced oxidative stress and ferroptosis by Keap1/Nrf2. Clin. Exp. Hypertens. 2023, 45, 2162537. [Google Scholar] [CrossRef] [PubMed]
- Nascentes Melo, L.M.; Cansiz, F.; Tasdogan, A. Aldolase A: The broker of glycolysis. Nat. Metab. 2025, 7, 242–244. [Google Scholar] [CrossRef]
- Bai, D.; Du, J.; Bu, X.; Cao, W.; Sun, T.; Zhao, J.; Zhao, Y.; Lu, N. ALDOA maintains NLRP3 inflammasome activation by controlling AMPK activation. Autophagy 2022, 18, 1673–1693. [Google Scholar] [CrossRef] [PubMed]
- Manio, M.C.; Inoue, K.; Fujitani, M.; Matsumura, S.; Fushiki, T. Combined pharmacological activation of AMPK and PPARδ potentiates the effects of exercise in trained mice. Physiol. Rep. 2016, 4, e12625. [Google Scholar] [CrossRef] [PubMed]
- Park, B.Y.; Jeon, J.H.; Go, Y.; Ham, H.J.; Kim, J.E.; Yoo, E.K.; Kwon, W.H.; Jeoung, N.H.; Jeon, Y.H.; Koo, S.H.; et al. PDK4 Deficiency Suppresses Hepatic Glucagon Signaling by Decreasing cAMP Levels. Diabetes 2018, 67, 2054–2068. [Google Scholar] [CrossRef]
- Digre, A.; Nan, J.; Frank, M.; Li, J.P. Heparin interactions with apoA1 and SAA in inflammation-associated HDL. Biochem. Biophys. Res. Commun. 2016, 474, 309–314. [Google Scholar] [CrossRef]
- Bayram, Ș.; Razzaque, Y.S.; Geisberger, S.; Pietzke, M.; Fürst, S.; Vechiatto, C.; Forbes, M.; Mastrobuoni, G.; Kempa, S. Investigating the role of GLUL as a survival factor in cellular adaptation to glutamine depletion via targeted stable isotope resolved metabolomics. Front. Mol. Biosci. 2022, 9, 859787. [Google Scholar] [CrossRef]
- Feng, T.; Sang, M.; Zhuang, H.; Xu, Z. In vitro and in vivo antioxidative and radioprotective capacities of polysaccharide isolated from Mesona Blumes gum. Starch-Stärke 2017, 69, 1700056. [Google Scholar] [CrossRef]
- Contreras-Zentella, M.L.; Villalobos-García, D.; Hernández-Muñoz, R. Ethanol Metabolism in the Liver, the Induction of Oxidant Stress, and the Antioxidant Defense System. Antioxidants 2022, 11, 1258. [Google Scholar] [CrossRef]
- Powers, S.K.; Deminice, R.; Ozdemir, M.; Yoshihara, T.; Bomkamp, M.P.; Hyatt, H. Exercise-induced oxidative stress: Friend or foe? J. Sport Health. Sci. 2020, 9, 415–425. [Google Scholar] [CrossRef]
- de Sousa, C.V.; Sales, M.M.; Rosa, T.S.; Lewis, J.E.; de Andrade, R.V.; Simões, H.G. The Antioxidant Effect of Exercise: A Systematic Review and Meta-Analysis. Sports Med. 2017, 47, 277–293. [Google Scholar] [CrossRef] [PubMed]
- Ungsudechachai, T.; Jittikoon, J.; Honsawek, S.; Udomsinprasert, W. Protective effect of clusterin against interleukin-1β-induced apoptosis and inflammation in human knee osteoarthritis chondrocytes. Clin. Transl. Sci. 2024, 17, e13881. [Google Scholar] [CrossRef] [PubMed]
- De Miguel, Z.; Khoury, N.; Betley, M.J.; Lehallier, B.; Willoughby, D.; Olsson, N.; Yang, A.C.; Hahn, O.; Lu, N.; Vest, R.T.; et al. Exercise plasma boosts memory and dampens brain inflammation via clusterin. Nature 2021, 600, 494–499. [Google Scholar] [CrossRef]
- Kostrzewa-Nowak, D.; Kubaszewska, J.; Nowakowska, A.; Nowak, R. Effect of Aerobic and Anaerobic Exercise on the Complement System of Proteins in Healthy Young Males. J. Clin. Med. 2020, 9, 2357. [Google Scholar] [CrossRef] [PubMed]
- Scoppetta, F.; Tartaglia, M.; Renzone, G.; Avellini, L.; Gaiti, A.; Scaloni, A.; Chiaradia, E. Plasma protein changes in horse after prolonged physical exercise: A proteomic study. J. Proteom. 2012, 75, 4494–4504. [Google Scholar] [CrossRef]
- Ritchie, M.D.; Holzinger, E.R.; Li, R.; Pendergrass, S.A.; Kim, D. Methods of integrating data to uncover genotype-phenotype interactions. Nat. Rev. Genet. 2015, 16, 85–97. [Google Scholar] [CrossRef]
- Muers, M. Gene expression: Transcriptome to proteome and back to genome. Nat. Rev. Genet. 2011, 12, 518. [Google Scholar] [CrossRef]
- Liao, H.Y.; Chien, C.C.; Tang, P.; Chen, C.C.; Chen, C.Y.; Chen, S.C. The integrated analysis of transcriptome and proteome for exploring the biodegradation mechanism of 2,4,6-trinitrotoluene by Citrobacter sp. J. Hazard. Mater. 2018, 349, 79–90. [Google Scholar] [CrossRef]
- Liu, M.; Xiong, L.B.; Tao, X.; Liu, Q.H.; Wang, F.Q.; Wei, D.Z. Integrated Transcriptome and Proteome Studies Reveal the Underlying Mechanisms for Sterol Catabolism and Steroid Production in Mycobacterium neoaurum. J. Agric. Food. Chem. 2018, 66, 9147–9157. [Google Scholar] [CrossRef]
- Tormos, K.V.; Chandel, N.S. Inter-connection between mitochondria and HIFs. J. Cell. Mol. Med. 2010, 14, 795–804. [Google Scholar] [CrossRef]
- Wang, Y.X.; Liu, H.B.; Li, P.S.; Yuan, W.X.; Liu, B.; Liu, S.T.; Qin, K.R. ROS and NO Dynamics in Endothelial Cells Exposed to Exercise-Induced Wall Shear Stress. Cell. Mol. Bioeng. 2018, 12, 107–120. [Google Scholar] [CrossRef] [PubMed]
- Sawada, Y.; Ichikawa, H.; Ebine, N.; Minamiyama, Y.; Alharbi, A.A.D.; Iwamoto, N.; Fukuoka, Y. Effects of High-Intensity Anaerobic Exercise on the Scavenging Activity of Various Reactive Oxygen Species and Free Radicals in Athletes. Nutrients 2023, 15, 222. [Google Scholar] [CrossRef] [PubMed]
- de Bont, C.M.; Boelens, W.C.; Pruijn, G.J.M. NETosis, complement, and coagulation: A triangular relationship. Cell Mol. Immunol. 2019, 16, 19–27. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Liu, Y.; Ma, W.; Li, L.; Bai, D.; Dugarjaviin, M. Integrated Transcriptomic and Proteomic Analysis Reveals Differential Gene and Protein Expression and Signaling Pathways During a 20 Km Endurance Exercise and Recovery in Mongolian Horses. Animals 2025, 15, 1981. https://doi.org/10.3390/ani15131981
Zhang X, Liu Y, Ma W, Li L, Bai D, Dugarjaviin M. Integrated Transcriptomic and Proteomic Analysis Reveals Differential Gene and Protein Expression and Signaling Pathways During a 20 Km Endurance Exercise and Recovery in Mongolian Horses. Animals. 2025; 15(13):1981. https://doi.org/10.3390/ani15131981
Chicago/Turabian StyleZhang, Xinzhuang, Yuanyi Liu, Wei Ma, Lianhao Li, Dongyi Bai, and Manglai Dugarjaviin. 2025. "Integrated Transcriptomic and Proteomic Analysis Reveals Differential Gene and Protein Expression and Signaling Pathways During a 20 Km Endurance Exercise and Recovery in Mongolian Horses" Animals 15, no. 13: 1981. https://doi.org/10.3390/ani15131981
APA StyleZhang, X., Liu, Y., Ma, W., Li, L., Bai, D., & Dugarjaviin, M. (2025). Integrated Transcriptomic and Proteomic Analysis Reveals Differential Gene and Protein Expression and Signaling Pathways During a 20 Km Endurance Exercise and Recovery in Mongolian Horses. Animals, 15(13), 1981. https://doi.org/10.3390/ani15131981




