Genomic Selection for Economically Important Traits in Dual-Purpose Simmental Cattle
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Phenotypic Data and Pedigree
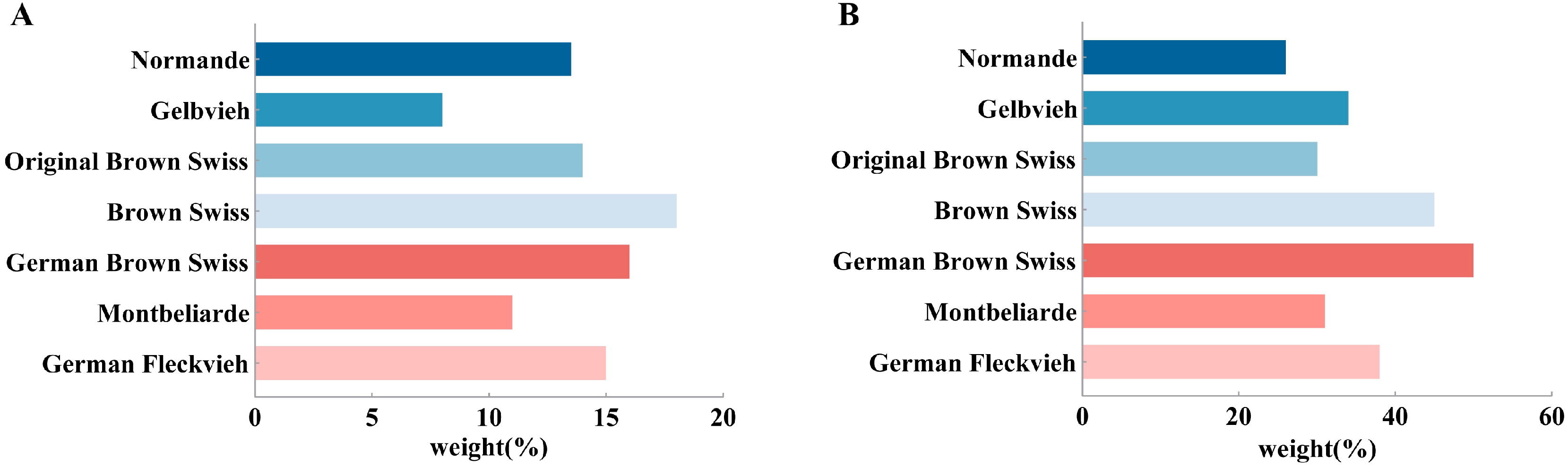
2.2. Genotype Data
2.3. Statistical Analysis
2.3.1. Significance Test of the Influence Factors
2.3.2. Single-Trait Animal Model
2.3.3. Two-Trait Animal Model
2.3.4. Reliability of Breeding Value Estimates
3. Results
3.1. Results of Fixed-Effects Analysis of Variance
3.1.1. Significance Testing of Factors Affecting Milk-Production Traits
3.1.2. Significance Testing of Factors Affecting Reproduction Traits
3.1.3. Significance Testing of Factors Affecting Growth Traits
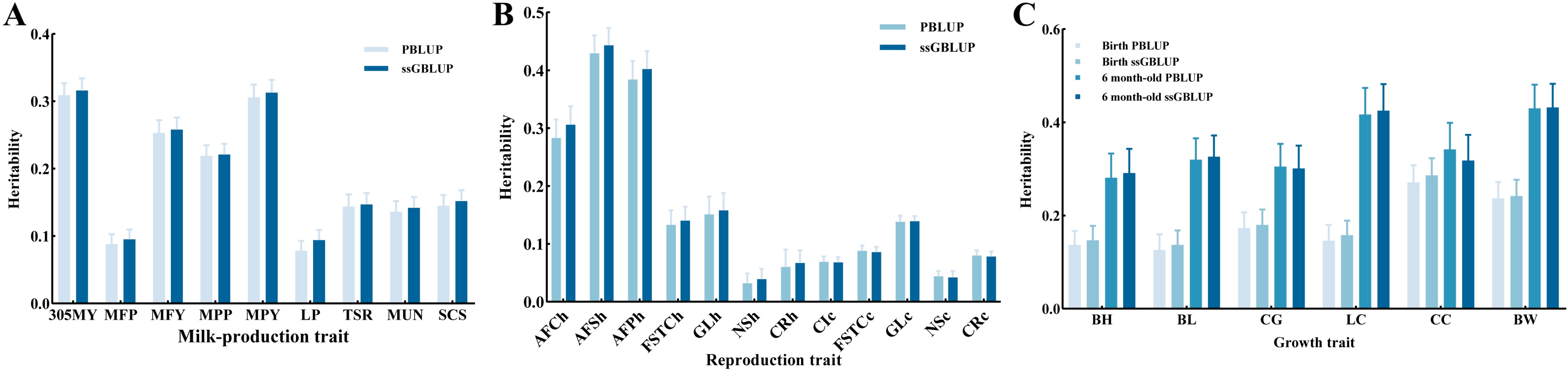
3.2. Estimation of the Variance Components and Heritability
3.2.1. Milk-Production Traits
3.2.2. Reproduction Traits
3.2.3. Growth Traits
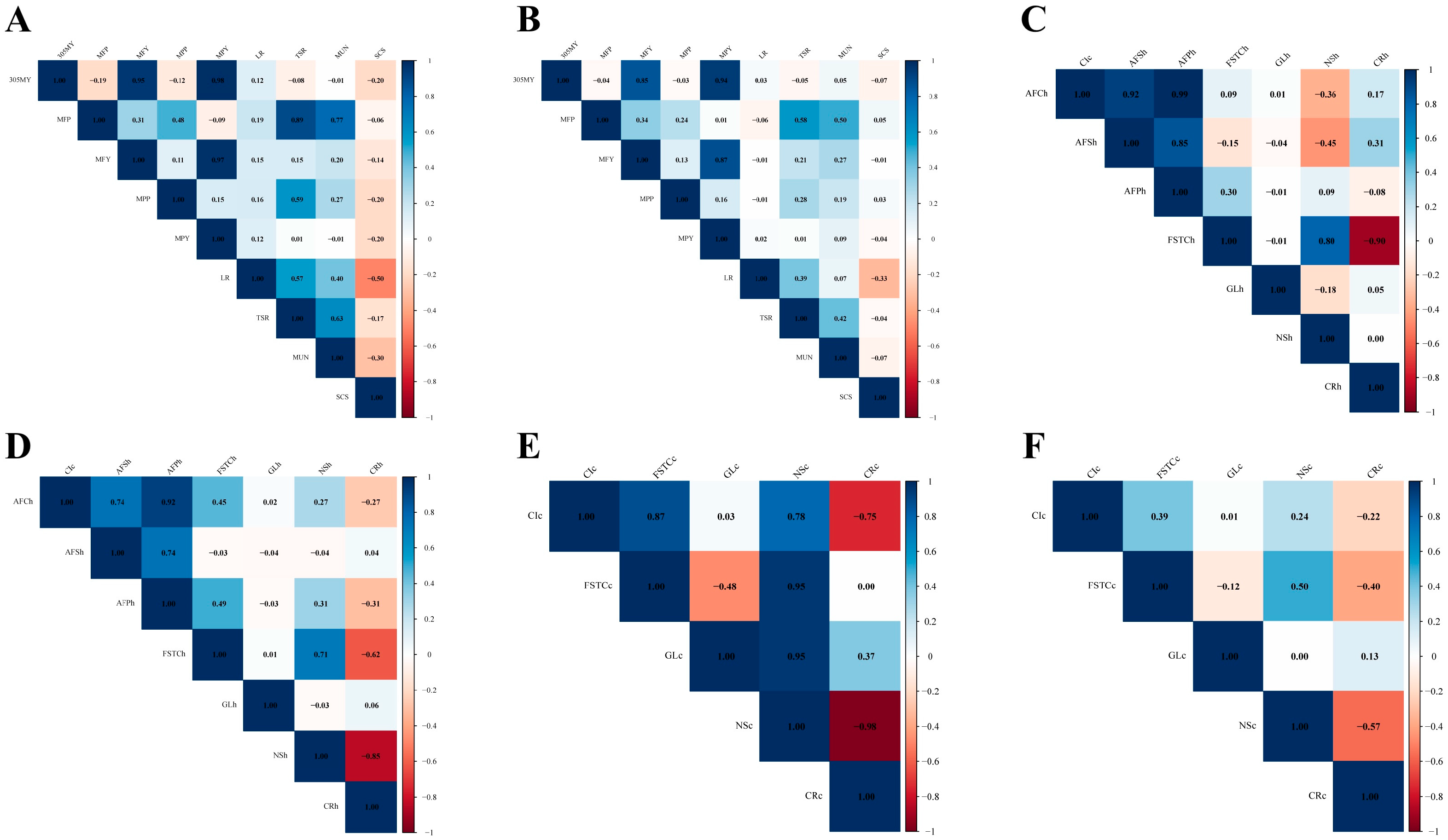
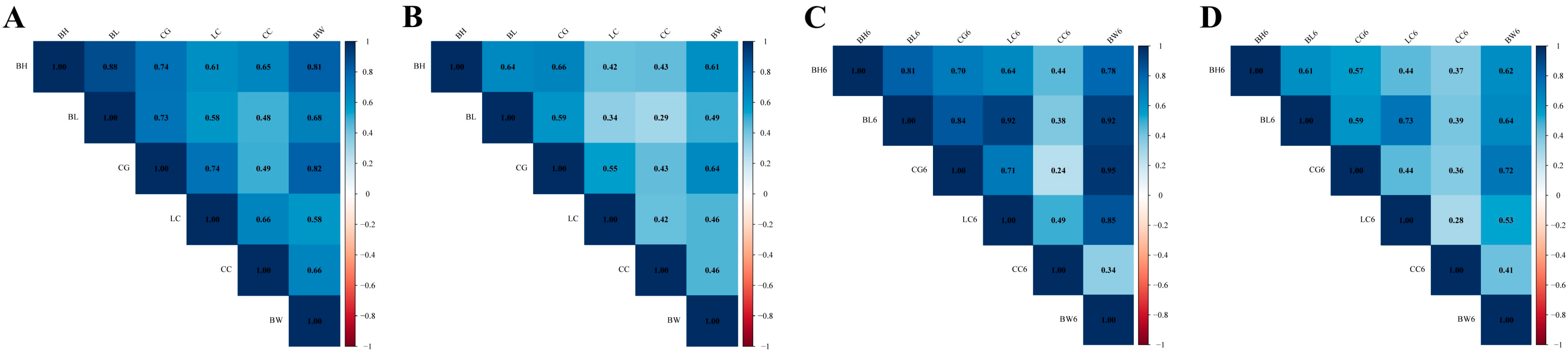
3.3. Genetic and Phenotypic Correlation
3.3.1. Genetic and Phenotypic Correlation of Milk-Production Traits
3.3.2. Genetic and Phenotypic Correlation of Reproduction Traits
3.3.3. Genetic and Phenotypic Correlation of Growth Traits
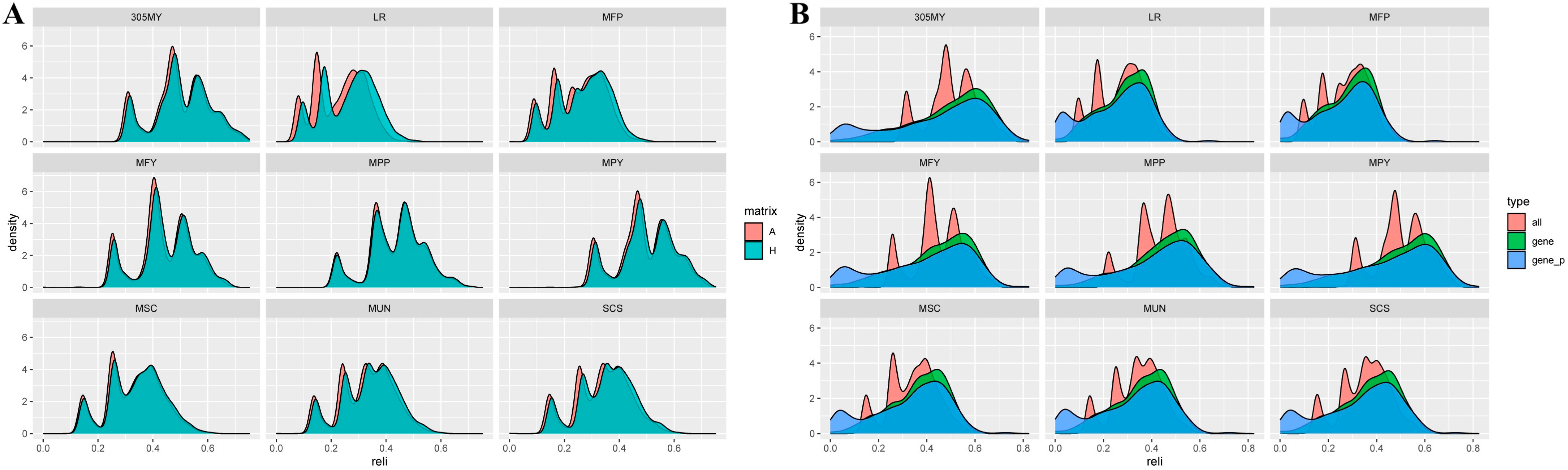
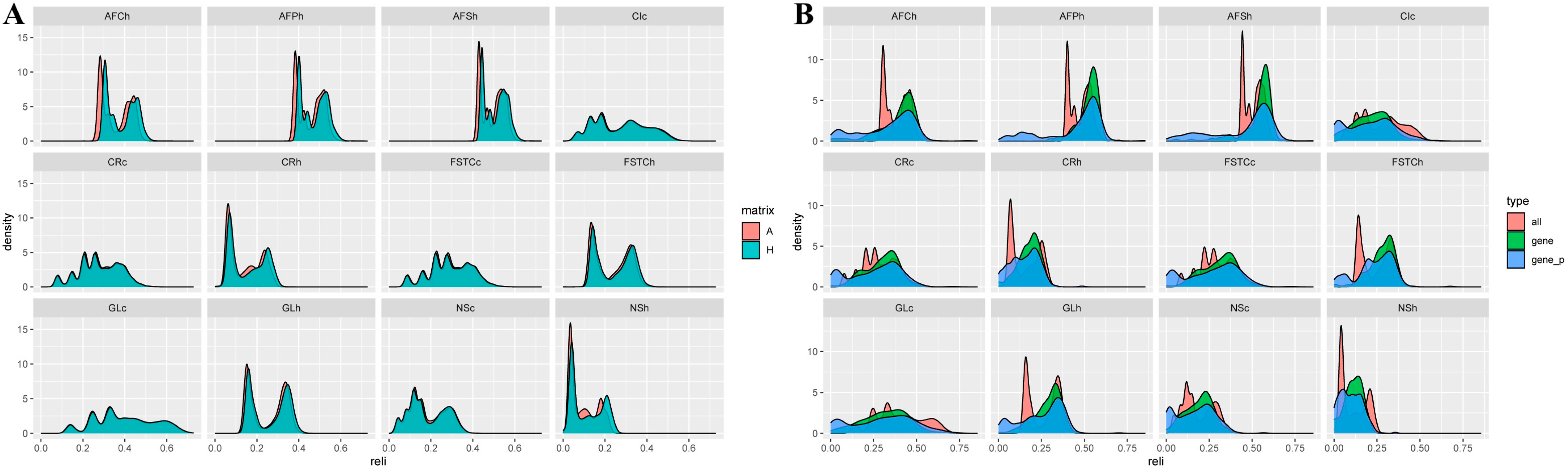
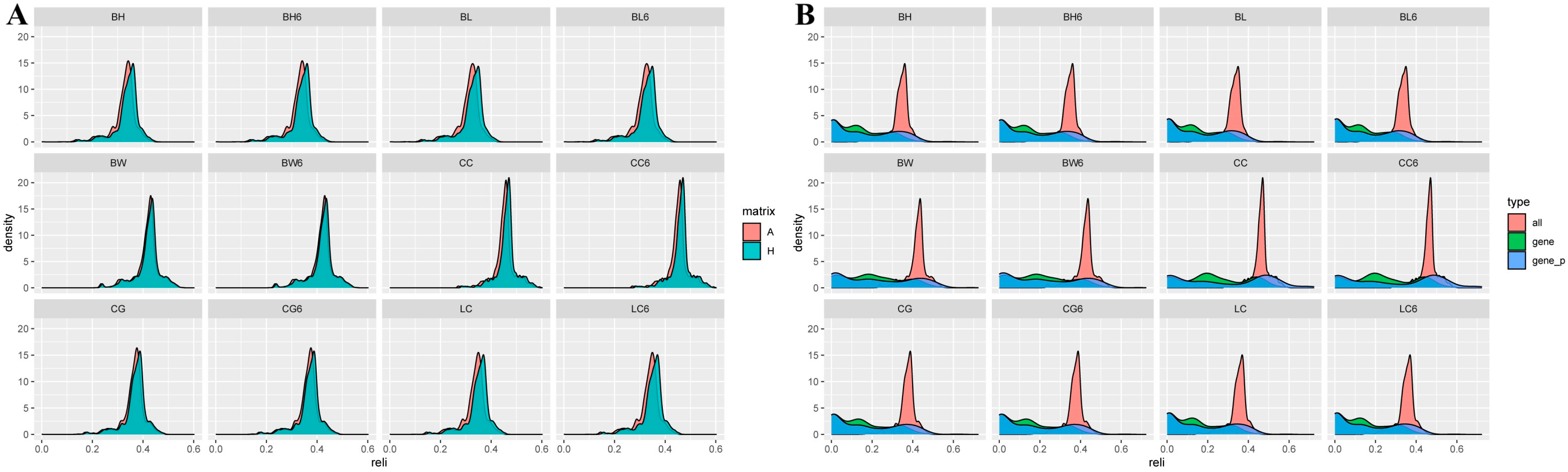
3.4. Reliability Prediction of Estimated Breeding Values (EBVs) and Genomic Estimated Breeding Values (GEBVs) Based on Different Matrices
3.4.1. Reliability Comparison of the Breeding Values for Milk-Production Traits
3.4.2. Reliability Comparison of the Breeding Values for Reproduction Traits
3.4.3. Reliability Comparison of the Breeding Values for Growth Traits
4. Discussion
4.1. Genetic Parameters of Traits in Dual-Purpose Simmental Cattle
4.1.1. Heritability of the Milk-Production Traits
4.1.2. Heritability of the Reproduction Traits
4.1.3. Heritability of the Growth Traits
4.2. Genetic and Phenotypic Correlations for Dual-Purpose Simmental Cattle
4.3. Reliability of EBV and GEBV for Each Trait in Dual-Purpose Simmental Cattle
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pfeiffer, C.; Fuerst, C.; Schwarzenbacher, H.; Fuerst-Waltl, B. Genotype by environment interaction in organic and conventional production systems and their consequences for breeding objectives in Austrian Fleckvieh cattle. Livest. Sci. 2016, 185, 50–55. [Google Scholar] [CrossRef]
- Byrne, T.J.; Santos, B.F.S.; Amer, P.R.; Martin-Collado, D.; Pryce, J.E.; Axford, M. New breeding objectives and selection indices for the Australian dairy industry. J. Dairy Sci. 2016, 99, 8146–8167. [Google Scholar] [CrossRef]
- Wahinya, P.K.; Jeyaruban, M.G.; Swan, A.A.; van der Werf, J.H.J. Breeding objectives for dairy cattle under low, medium and high production systems in the tropics. Animal 2022, 16, 100513. [Google Scholar] [CrossRef]
- Xue, X.; Hu, H.; Zhang, J.; Ma, Y.; Han, L.; Hao, F.; Jiang, Y.; Ma, Y. Estimation of Genetic Parameters for Conformation Traits and Milk Production Traits in Chinese Holsteins. Animals 2023, 13, 100. [Google Scholar] [CrossRef]
- Olasege, B.S.; Zhang, S.; Zhao, Q.; Liu, D.; Sun, H.; Wang, Q.; Ma, P.; Pan, Y. Genetic parameter estimates for body conformation traits using composite index, principal component, and factor analysis. J. Dairy Sci. 2019, 102, 5219–5229. [Google Scholar] [CrossRef]
- Xu, L.; Luo, H.; Zhang, X.; Lu, H.; Zhang, M.; Ge, J.; Zhang, T.; Yan, M.; Tan, X.; Huang, X.; et al. Factor Analysis of Genetic Parameters for Body Conformation Traits in Dual-Purpose Simmental Cattle. Animals 2022, 12, 2433. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, Z.; Lu, X.; Yang, Z. Effect of Dam Body Conformations on Birth Traits of Calves in Chinese Holsteins. Animals 2023, 13, 2253. [Google Scholar] [CrossRef]
- Mandel, A.; Reißmann, M.; Brockmann, G.A.; Korkuć, P. Whole-Genome Insights into the Genetic Basis of Conformation Traits in German Black Pied (DSN) Cattle. Genes 2025, 16, 445. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Brito, L.F.; Luo, H.; Shi, R.; Chang, Y.; Liu, L.; Guo, G.; Wang, Y. Genetic and Genomic Analyses of Service Sire Effect on Female Reproductive Traits in Holstein Cattle. Front. Genet. 2021, 12, 713575. [Google Scholar] [CrossRef] [PubMed]
- Pfeiffer, C.; Fuerst, C.; Ducrocq, V.; Fuerst-Waltl, B. Short communication: Genetic relationships between functional longevity and direct health traits in Austrian Fleckvieh cattle. J. Dairy Sci. 2015, 98, 7380–7383. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Liu, A.; Wang, Y.; Luo, H.; Yan, X.; Guo, X.; Li, X.; Liu, L.; Su, G. Genetic Parameters and Genome-Wide Association Studies of Eight Longevity Traits Representing Either Full or Partial Lifespan in Chinese Holsteins. Front. Genet. 2021, 12, 634986. [Google Scholar] [CrossRef]
- Soufleri, A.; Banos, G.; Panousis, N.; Tsiamadis, V.; Kougioumtzis, A.; Arsenos, G.; Valergakis, G.E. Estimated breeding values of dairy sires for cow colostrum and transfer of passive immunity traits. JDS Commun. 2025, 6, 74–78. [Google Scholar] [CrossRef] [PubMed]
- The Future Now Association. More on Normande Proofs and Pedigrees. Available online: https://normandegenetics.com/how-to-understand-our-proofs/ (accessed on 11 March 2025).
- Franken, Z.R. Zuchtprogramm für die Rasse Deutsches Gelbvieh. Available online: https://www.rzv-franken.de/fileadmin/user_upload/Zucht/Gelbviehzuchtprogramm_Doppelnutzung_RZV_Franken__ab_11-18_.pdf (accessed on 11 March 2025).
- Rust, M. Effizienz im Fokus. CHbraunvieh 2021, 10–11. Available online: https://homepage.braunvieh.ch/wp-content/uploads/2021/04/2021-01-CHbraunvieh.pdf (accessed on 11 March 2025).
- German Livestock Association. Brown Swiss. Available online: https://www.asr-rind.de/rinderrassen/deutsches-braunvieh (accessed on 11 March 2025).
- Montbeliarbe Associtation. Genetic: Selection Goals. Available online: https://www.montbeliarde.org/en-428 (accessed on 11 March 2025).
- German Livestock Association. Deutsches Fleckvieh. Available online: https://www.asr-rind.de/rinderrassen/deutsches-fleckvieh (accessed on 11 March 2025).
- Department of Seed Management; Ministry of Agriculture and Rural Affairs; National Animal Husbandry Service. 2024 Summary of Genetic Evaluation of Chinese Beef and Dairy-meat Dual-Purpose Breeding Bulls. Available online: https://www.nahs.org.cn/zt_10027/qgxqycgljh/ycpg/202412/t20241227_451505.htm (accessed on 11 March 2025).
- Aguilar, I.; Misztal, I.; Johnson, D.L.; Legarra, A.; Tsuruta, S.; Lawlor, T.J. Hot topic: A unified approach to utilize phenotypic, full pedigree, and genomic information for genetic evaluation of Holstein final score. J. Dairy Sci. 2010, 93, 743–752. [Google Scholar] [CrossRef]
- Christensen, O.F.; Lund, M.S. Genomic prediction when some animals are not genotyped. Genet. Sel. Evol. 2010, 42, 2. [Google Scholar] [CrossRef] [PubMed]
- Schaeffer, L.R. Strategy for applying genome-wide selection in dairy cattle. J. Anim. Breed. Genet. 2006, 123, 218–223. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Ranjan, S.; Gautam, A. Heritability Estimate. In Encyclopedia of Animal Cognition and Behavior; Vonk, J., Shackelford, T., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 1–4. [Google Scholar]
- Wei, C.; Luo, H.P.; Wang, Y.C.; Huang, X.X.; Zhang, M.H.; Zhang, X.X.; Wang, D.; Ge, J.J.; Xu, L.; Jiang, H.; et al. Analyses of the genetic relationships between lactose, somatic cell score, and growth traits in Simmental cattle. Animal 2021, 15, 100027. [Google Scholar] [CrossRef]
- Zhou, J.; Liu, L.; Chen, C.J.; Zhang, M.; Lu, X.; Zhang, Z.; Huang, X.; Shi, Y. Genome-wide association study of milk and reproductive traits in dual-purpose Xinjiang Brown cattle. BMC Genom. 2019, 20, 827. [Google Scholar] [CrossRef]
- Zhang, M.; Luo, H.; Xu, L.; Shi, Y.; Zhou, J.; Wang, D.; Zhang, X.; Huang, X.; Wang, Y. Genomic Selection for Milk Production Traits in Xinjiang Brown Cattle. Animals 2022, 12, 136. [Google Scholar] [CrossRef]
- Jayawardana, J.; Lopez-Villalobos, N.; McNaughton, L.R.; Hickson, R.E. Genomic Regions Associated with Milk Composition and Fertility Traits in Spring-Calved Dairy Cows in New Zealand. Genes 2023, 14, 860. [Google Scholar] [CrossRef]
- Su, M.; Lin, X.; Xiao, Z.; She, Y.; Deng, M.; Liu, G.; Sun, B.; Guo, Y.; Liu, D.; Li, Y. Genome-Wide Association Study of Lactation Traits in Chinese Holstein Cows in Southern China. Animals 2023, 13, 2545. [Google Scholar] [CrossRef]
- Wolf, M.J.; Neumann, G.B.; Kokuć, P.; Yin, T.; Brockmann, G.A.; König, S.; May, K. Genetic evaluations for endangered dual-purpose German Black Pied cattle using 50K SNPs, a breed-specific 200K chip, and whole-genome sequencing. J. Dairy Sci. 2023, 106, 3345–3358. [Google Scholar] [CrossRef] [PubMed]
- Laodim, T.; Elzo, M.A.; Koonawootrittriron, S.; Suwanasopee, T.; Jattawa, D. Genomic-polygenic and polygenic predictions for milk yield, fat yield, and age at first calving in Thai multibreed dairy population using genic and functional sets of genotypes. Livest. Sci. 2019, 219, 17–24. [Google Scholar] [CrossRef]
- Schneider, M.d.P.; Strandberg, E.; Ducrocq, V.; Roth, A. Survival Analysis Applied to Genetic Evaluation for Female Fertility in Dairy Cattle. J. Dairy Sci. 2005, 88, 2253–2259. [Google Scholar] [CrossRef]
- Maskal, J.M.; Pedrosa, V.B.; Rojas de Oliveira, H.; Brito, L.F. A comprehensive meta-analysis of genetic parameters for resilience and productivity indicator traits in Holstein cattle. J. Dairy Sci. 2024, 107, 3062–3079. [Google Scholar] [CrossRef]
- Wei, C.; Ge, J.; Abudureyimu, R.; Liu, L.; Huang, X.; Ma, G. Effect of Non-genetic Factors on reproduction traits of Simmental Cattle. Chin. J. Anim. Sci. 2017, 53, 39–43. [Google Scholar] [CrossRef]
- Lu, X. Estimation of Genetic Parameter and Genome-Wide Association Study for Female Reproductive in Xinjiang Brown Cattle. Master’s Thesis, Ningxia University, Yinchuan, China, 2020. [Google Scholar]
- Xu, L. Estimates of Genetic Parameters and Total Perfoemance Index Construction of Xinjiang Brown Cattle. Master’s Thesis, Xinjiang Agricultural University, Urumqi, China, 2020. [Google Scholar]
- Dong, G.; Zhang, X.; Wang, Y.; Wu, H.; Liu, A.; Zhang, Y.; Wang, D.; Cui, J. Genetic Evaluation of Reproduction Traits in Sanhe Cattle. China Anim. Husb. Vet. Med. 2015, 42, 1547–1552. [Google Scholar] [CrossRef]
- Kgari, R.D.; Muller, C.; Dzama, K.; Makgahlela, M.L. Estimation of Genetic Parameters for Heifer and Cow Fertility Traits Derived from On-Farm AI Service Records of South African Holstein Cattle. Animals 2022, 12, 2023. [Google Scholar] [CrossRef]
- Haque, M.A.; Lee, Y.-M.; Ha, J.-J.; Jin, S.; Park, B.; Kim, N.-Y.; Won, J.-I.; Kim, J.-J. Genomic Predictions in Korean Hanwoo Cows: A Comparative Analysis of Genomic BLUP and Bayesian Methods for Reproductive Traits. Animals 2024, 14, 27. [Google Scholar] [CrossRef]
- Weller, J.I.; Ezra, E.; Gershoni, M. Genetic and genomic analysis of age at first insemination in Israeli dairy cattle. J. Dairy Sci. 2022, 105, 5192–5205. [Google Scholar] [CrossRef]
- Roy, I.; Rahman, M.; Karunakaran, M.; Gayari, I.; Baneh, H.; Mandal, A. Genetic relationships between reproductive and production traits in Jersey crossbred cattle. Gene 2024, 894, 147982. [Google Scholar] [CrossRef] [PubMed]
- Sbardella, A.P.; Watanabe, R.N.; da Costa, R.M.; Bernardes, P.A.; Braga, L.G.; Baldi Rey, F.S.; Lôbo, R.B.; Munari, D.P. Genome-Wide Association Study Provides Insights into Important Genes for Reproductive Traits in Nelore Cattle. Animals 2021, 11, 1386. [Google Scholar] [CrossRef]
- Jiménez, J.M.; Morales, R.M.; Demyda-Peyrás, S.; Molina, A. Genetic relationships between male and female reproductive traits in Retinta beef cattle. Livest. Sci. 2025, 291, 105610. [Google Scholar] [CrossRef]
- Canaza-Cayo, A.W.; Cobuci, J.A.; Lopes, P.S.; de Almeida Torres, R.; Martins, M.F.; dos Santos Daltro, D.; Barbosa da Silva, M.V.G. Genetic trend estimates for milk yield production and fertility traits of the Girolando cattle in Brazil. Livest. Sci. 2016, 190, 113–122. [Google Scholar] [CrossRef]
- Chen, Z.; Shi, R.; Luo, H.; Tian, J.; Wei, C.; Zhang, W.; Li, W.; Wen, W.; Wang, Y.; Wang, Y. Estimation of genetic parameters of reproduction traits of Holstein in Ningxia. Acta Vet. Et Zootech. Sin. 2021, 52, 344–351. [Google Scholar] [CrossRef]
- Berry, D.P.; Wall, E.; Pryce, J.E. Genetics and genomics of reproductive performance in dairy and beef cattle. Animal 2014, 8, 105–121. [Google Scholar] [CrossRef] [PubMed]
- Caetano, S.L.; Savegnago, R.P.; Boligon, A.A.; Ramos, S.B.; Chud, T.C.S.; Lôbo, R.B.; Munari, D.P. Estimates of genetic parameters for carcass, growth and reproductive traits in Nellore cattle. Livest. Sci. 2013, 155, 1–7. [Google Scholar] [CrossRef]
- do Amaral Grossi, D.; Grupioni, N.V.; Buzanskas, M.E.; de Paz, C.C.P.; de Almeida Regitano, L.C.; de Alencar, M.M.; Schenkel, F.S.; Munari, D.P. Allele substitution effects of IGF1, GH and PIT1 markers on estimated breeding values for weight and reproduction traits in Canchim beef cattle. Livest. Sci. 2015, 180, 78–83. [Google Scholar] [CrossRef]
- Lopez, B.I.; Son, J.-H.; Seo, K.; Lim, D. Estimation of Genetic Parameters for Reproductive Traits in Hanwoo (Korean Cattle). Animals. 2019, 9, 715. [Google Scholar] [CrossRef]
- Zhu, K.; Li, T.; Liu, D.; Wang, S.; Wang, S.; Wang, Q.; Pan, Y.; Zan, L.; Ma, P. Estimation of genetic parameters for fertility traits in Chinese Holstein of south China. Front. Genet. 2023, 14, 1288375. [Google Scholar] [CrossRef]
- Kluska, S.; Olivieri, B.F.; Bonamy, M.; Chiaia, H.L.J.; Feitosa, F.L.B.; Berton, M.P.; Peripolli, E.; Lemos, M.V.A.; Tonussi, R.L.; Lôbo, R.B.; et al. Estimates of genetic parameters for growth, reproductive, and carcass traits in Nelore cattle using the single step genomic BLUP procedure. Livest. Sci. 2018, 216, 203–209. [Google Scholar] [CrossRef]
- Aponte, P.F.C.; Carneiro, P.L.S.; Araujo, A.C.; Pedrosa, V.B.; Fotso-Kenmogne, P.R.; Silva, D.A.; Miglior, F.; Schenkel, F.S.; Brito, L.F. Investigating the genomic background of calving-related traits in Canadian Jersey cattle. J. Dairy Sci. 2024, 107, 11195–11213. [Google Scholar] [CrossRef]
- Jourdain, J.; Capitan, A.; Saintilan, R.; Hozé, C.; Fouéré, C.; Fritz, S.; Boichard, D.; Barbat, A. Genetic parameters, genome-wide association study, and selection perspective on gestation length in 16 French cattle breeds. J. Dairy Sci. 2024, 107, 8157–8169. [Google Scholar] [CrossRef]
- Shi, R.; Brito, L.F.; Li, S.; Han, L.; Guo, G.; Wen, W.; Yan, Q.; Chen, S.; Wang, Y. Genomic prediction and validation strategies for reproductive traits in Holstein cattle across different Chinese regions and climatic conditions. J. Dairy Sci. 2025, 108, 707–725. [Google Scholar] [CrossRef] [PubMed]
- Boonkum, W.; Chankitisakul, V.; Duangjinda, M.; Buaban, S.; Sumreddee, P.; Sungkhapreecha, P. Genomic Selection Using Single-Step Genomic BLUP on the Number of Services per Conception Trait in Thai–Holstein Crossbreeds. Animals 2023, 13, 3609. [Google Scholar] [CrossRef]
- Ma, L.; Cole, J.B.; Da, Y.; VanRaden, P.M. Symposium review: Genetics, genome-wide association study, and genetic improvement of dairy fertility traits. J. Dairy Sci. 2019, 102, 3735–3743. [Google Scholar] [CrossRef] [PubMed]
- Cochran, S.D.; Cole, J.B.; Null, D.J.; Hansen, P.J. Discovery of single nucleotide polymorphisms in candidate genes associated with fertility and production traits in Holstein cattle. BMC Genet. 2013, 14, 49. [Google Scholar] [CrossRef]
- An, B.; Xu, L.; Xia, J.; Wang, X.; Miao, J.; Chang, T.; Song, M.; Ni, J.; Xu, L.; Zhang, L.; et al. Multiple association analysis of loci and candidate genes that regulate body size at three growth stages in Simmental beef cattle. BMC Genet. 2020, 21, 32. [Google Scholar] [CrossRef]
- Duan, X.; An, B.; Du, L.; Chang, T.; Liang, M.; Yang, B.-G.; Xu, L.; Zhang, L.; Li, J.; E, G.; et al. Genome-Wide Association Analysis of Growth Curve Parameters in Chinese Simmental Beef Cattle. Animals 2021, 11, 192. [Google Scholar] [CrossRef]
- Guo, M.; Meng, Y.; Wang, H.; Wang, Y.; Che, L.; Shen, L.; Wang, X. Estimation of genetic parameters of body weight and body size traits in Jinnan cattle at different growth stages. Acta Vet. Zootechn. Sin. 2023, 54, 1452–1464. [Google Scholar]
- Kamprasert, N.; Duijvesteijn, N.; Van der Werf, J.H.J. Estimation of genetic parameters for BW and body measurements in Brahman cattle. Animal 2019, 13, 1576–1582. [Google Scholar] [CrossRef]
- Ma, K.; Zhang, X.; Li, X.; Wang, W.; Wang, D.; Xu, L.; Zhang, M.; Ge, J.; He, J.; Huang, X. Estimation of Genetic Parameters for Body Weight Traits in Chinese Simmental Cattle from Xinjiang Changji Region. Chin. J. Anim. Sci. 2024, 60, 232–238. [Google Scholar] [CrossRef]
- Ren, X.; Zhang, X.; Wang, Y.; Wu, H.; Liu, A.; Zhang, Y.; Wang, D.; Cui, J.; Dou, T.; Yuan, P.; et al. Genetic parameter estiamtion for body measurements and weight at birth in sanhe cattle. Sci. Agric. Sin. 2013, 46, 5020–5025. [Google Scholar] [CrossRef]
- Xu, L.; Zhang, M.; Wang, D.; Zhang, X.; Fan, S.; Liu, J.; Liu, J.; Yan, C.; Huang, X.; Wang, Y. Estimation of genetic parameters of body size and body weight in Xinjiang brown cattle at different growth stages. Chin. Anim. Husb. Vet. Med. 2021, 48, 4105–4114. [Google Scholar] [CrossRef]
- Zhang, X.; Li, Y.; Zhao, J.; Zhao, F.; Chu, G.; Mamatiniyaz, M.; Wang, D.; Wei, C.; You, Z.; Huang, X.; et al. Analysis on growth and development pattern of simmental calves in xinjiang area. Chin. J. Anim. Sci. 2018, 54, 35–39. [Google Scholar] [CrossRef]
- Brito, L.C.; Peixoto, M.; Carrara, E.R.; Fonseca, E.S.F.; Ventura, H.T.; Bruneli, F.A.T.; Lopes, P.S. Genetic parameters for milk, growth, and reproductive traits in Guzerá cattle under tropical conditions. Trop. Anim. Health Prod. 2020, 52, 2251–2257. [Google Scholar] [CrossRef] [PubMed]
- Vargas, G.; Buzanskas, M.E.; Guidolin, D.G.; Grossi Ddo, A.; Bonifácio Ada, S.; Lôbo, R.B.; da Fonseca, R.; Oliveira, J.A.; Munari, D.P. Genetic parameter estimation for pre- and post-weaning traits in Brahman cattle in Brazil. Trop. Anim. Health Prod. 2014, 46, 1271–1278. [Google Scholar] [CrossRef]
- Afolayan, R.A.; Pitchford, W.S.; Deland, M.P.B.; McKiernan, W.A. Breed variation and genetic parameters for growth and body development in diverse beef cattle genotypes. Animal 2007, 1, 13–20. [Google Scholar] [CrossRef]
- Lembeyea, F.; Lopez-Villalobosa, N.; Burke, J.L.; Davis, S.R. Estimation of genetic parameters for milk traits in cows milked once- or twice-daily in New Zealand. Livest. Sci. 2016, 18, 142–147. [Google Scholar] [CrossRef]
- Samoré, A.B.; Rizzi, R.; Rossoni, A.; Bagnato1, A. Genetic parameters for functional longevity, type traits, somatic cell scores, milk flow and production in the Italian Brown Swiss. Ital. J. Anim. Sci. 2010, 9, 145–152. Available online: https://www.tandfonline.com/doi/full/10.4081/ijas.2010.e28 (accessed on 26 April 2025). [CrossRef]
- Frigo, E.; BSamorè, A.; Vicario, D.; Pedron, A.B.O. Heritabilities and Genetic Correlations of Body Condition Score and Muscularity with Productive Traits and their Trend Functions in Italian Simmental Cattle. Ital. J. Anim. Sci. 2013, 40, 240–246. [Google Scholar] [CrossRef]
- Mazza, S.; Guzzo, N.; Sartori, C.; Mantovani, R. Genetic correlations between type and test-day milk yield in small dual-purpose cattle populations: The Aosta Red Pied breed as a case study. J. Dairy Sci. 2016, 99, 8127–8136. [Google Scholar] [CrossRef]
- Sartori, C.; Guzzo, N.; Mazza, S.; Mantovani, R. Genetic correlations among milk yield, morphology, performance test traits and somatic cells in dual-purpose Rendena breed. Animal 2018, 12, 906–914. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, T.; Coffey, M.; Mrode, R.; Wall, E. Genetic parameters for production, health, fertility and longevity traits in dairy cows. Animal 2013, 7, 34–46. [Google Scholar] [CrossRef]
- Petersson, K.J.; Strandberg, E.; Gustafsson, H.; Royal, M.D.; Berglund, B. Detection of delayed cyclicity in dairy cows based on progesterone content in monthly milk samples. Prev. Vet. Med. 2008, 86, 153–163. [Google Scholar] [CrossRef]
- Royal, M.; Mann, G.E.; Flint, A.P.F. Strategies for Reversing the Trend Towards Subfertility in Dairy Cattle. Vet. J. 2000, 160, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Naserkheil, M.; Mehrban, H.; Lee, D.; Park, M.N. Evaluation of Genome-Enabled Prediction for Carcass Primal Cut Yields Using Single-Step Genomic Best Linear Unbiased Prediction in Hanwoo Cattle. Genes 2021, 12, 1886. [Google Scholar] [CrossRef]
- Peripolli, E.; Temp, L.B.; Fukumasu, H.; Pereira, A.S.C.; Fabricio, E.C.; Ferraz, J.B.S.; Baldi, F. Identification of candidate genes and genomic prediction for early heifer pregnancy in Nelore beef cattle. Livest. Sci. 2024, 289, 105582. [Google Scholar] [CrossRef]
- Bolormaa, S.; Pryce, J.E.; Kemper, K.; Savin, K.; Hayes, B.J.; Barendse, W.; Zhang, Y.; Reich, C.M.; Mason, B.A.; Bunch, R.J.; et al. Accuracy of prediction of genomic breeding values for residual feed intake and carcass and meat quality traits in Bos taurus, Bos indicus, and composite beef cattle. J. Anim. Sci. 2013, 91, 3088–3104. [Google Scholar] [CrossRef]
- Lund, M.S.; de Roos, A.P.W.; de Vries, A.G.; Druet, T.; Ducrocq, V.; Fritz, S.; Guillaume, F.; Guldbrandtsen, B.; Liu, Z.; Reents, R.; et al. A common reference population from four European Holstein populations increases reliability of genomic predictions. Genet. Sel. Evol. 2011, 43, 43. [Google Scholar] [CrossRef]
- Li, X.; Lund, M.S.; Zhang, Q.; Costa, C.N.; Ducrocq, V.; Su, G. Short communication: Improving accuracy of predicting breeding values in Brazilian Holstein population by adding data from Nordic and French Holstein populations. J. Dairy Sci. 2016, 99, 4574–4579. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Xu, L.; Lu, H.; Luo, H.; Zhou, J.; Wang, D.; Zhang, X.; Huang, X.; Wang, Y. Genomic prediction based on a joint reference population for the Xinjiang Brown cattle. Front. Genet. 2024, 15, 1394636. [Google Scholar] [CrossRef] [PubMed]
| Trait | Herd | Parity | Calving Year | Calving Season | Lactation |
|---|---|---|---|---|---|
| 305MY | 1923.31 ** | 2.31 * | 10.90 ** | 2.94 * | - |
| MFP | 1157.58 ** | 14.18 ** | 6.62 ** | 103.52 ** | 97.97 ** |
| MFY | 1745.86 ** | 3.02 * | 36.99 ** | 0.03 * | - |
| MPP | 18.99 ** | 7.84 ** | 86.97 ** | 74.86 ** | 5.22 ** |
| MPY | 1326.81 ** | 4.66 ** | 13.79 ** | 5.31 ** | - |
| LR | 132.82 ** | 63.76 ** | 39.14 ** | 1.32 ns | 39.57 ** |
| TSR | 50.37 ** | 15.57 ** | 64.13 ** | 6.39 ** | 2.18 ns |
| MUN | 2533.45 ** | 4.77 ** | 206.25 ** | 139.86 ** | 17.08 ** |
| SCS | 1263.25 ** | 5.52 ** | 18.31 ** | 2.10 ns | 9.91 ** |
| Trait | Herd | Breeding Technician | Service Year | Service Season | Parity | |||
|---|---|---|---|---|---|---|---|---|
| First | Last | First | Last | First | Last | |||
| AFCh | - | - | 69.28 ** | - | - | 60.63 ** | 5.53 ** | - |
| AFSh | 518.64 ** | - | - | 547.84 ** | 74.18 ** | - | - | - |
| AFPh | - | - | 29.26 ** | - | - | 103.03 ** | 1.10 ns | - |
| FSTCh | - | - | 35.24 ** | - | - | 27.80 ** | 7.18 ** | - |
| GLh | 267.56 ** | - | - | - | - | 38.75 ** | 34.19 ** | - |
| NSh | - | 18.71 ** | - | 8.71 ** | 0.78 ns | - | - | - |
| CRh | - | 25.64 ** | - | 7.13 ** | 0.63 ns | - | - | - |
| CIc | 656.62 ** | - | - | - | - | 5.27 ** | 0.25 ns | 15.64 ** |
| FSTCc | - | - | 68.61 ** | - | - | 19.80 ** | 20.08 ** | 10.23 ** |
| GLc | 687.59 ** | - | - | - | 67.72 ** | 91.00 ** | 4.51 ** | |
| NSc | - | - | 23.64 ** | - | - | 31.46 ** | 38.00 ** | 12.12 ** |
| CRc | - | 22.30 ** | 16.59 ** | 13.41 ** | - | - | - | 16.78 ** |
| Trait | Newborn | Six-Month-Old | ||||
|---|---|---|---|---|---|---|
| Birth Year | Birth Month | Sex | Birth Year | Birth Month | Sex | |
| BH | 52.99 ** | 1.42 ns | 119.12 ** | 85.71 ** | 6.21 ** | 12.49 ** |
| BL | 88.93 ** | 1.79 ns | 83.85 ** | 167.47 ** | 13.30 ** | 25.09 ** |
| CG | 22.61 ** | 2.74 ** | 129.63 ** | 193.72 ** | 4.81 ** | 26.40 ** |
| LC | 32.03 ** | 3.23 ** | 93.85 ** | 74.47 ** | 1.55 ** | 4.26 ** |
| CC | 42.34 ** | 5.90 ** | 181.18 ** | 176.00 ** | 21.12 ** | 176.48 ** |
| BW | 13.04 ** | 2.30 ** | 351.82 ** | 165.55 ** | 8.77 ** | 28.60 ** |
| Traits | Whole Population | Genotyped Subpopulation | ||||||
|---|---|---|---|---|---|---|---|---|
| PBLUP | ssGBLUP | (%) | Correlation | PBLUP | ssGBLUP | (%) | Correlation | |
| 305MY | 0.496 | 0.506 | 1 | 0.989 ** | 0.549 | 0.566 | 1.7 | 0.966 ** |
| MFP | 0.254 | 0.271 | 1.7 | 0.975 ** | 0.294 | 0.319 | 2.5 | 0.881 ** |
| MFY | 0.440 | 0.450 | 1 | 0.985 ** | 0.487 | 0.505 | 1.8 | 0.957 ** |
| MPP | 0.432 | 0.438 | 0.6 | 0.986 ** | 0.480 | 0.496 | 1.6 | 0.945 ** |
| MPY | 0.491 | 0.502 | 1.1 | 0.990 ** | 0.538 | 0.557 | 1.9 | 0.965 ** |
| LR | 0.233 | 0.265 | 3.2 | 0.958 ** | 0.275 | 0.315 | 4 | 0.846 ** |
| TSR | 0.332 | 0.341 | 0.9 | 0.979 ** | 0.387 | 0.404 | 1.7 | 0.916 ** |
| MUN | 0.329 | 0.342 | 1.3 | 0.973 ** | 0.375 | 0.397 | 2.2 | 0.873 ** |
| SCS | 0.339 | 0.354 | 1.5 | 0.974 ** | 0.389 | 0.413 | 2.4 | 0.909 ** |
| Traits | Whole Population | Genotyped Subpopulation | ||||||
|---|---|---|---|---|---|---|---|---|
| PBLUP | ssGBLUP | (%) | Correlation | PBLUP | ssGBLUP | (%) | Correlation | |
| AFCh | 0.359 | 0.383 | 2.4 | 0.986 ** | 0.393 | 0.429 | 3.6 | 0.929 ** |
| AFSh | 0.494 | 0.509 | 1.5 | 0.991 ** | 0.527 | 0.555 | 2.8 | 0.969 ** |
| AFPh | 0.456 | 0.474 | 1.8 | 0.992 ** | 0.503 | 0.534 | 3.1 | 0.945 ** |
| FSTCh | 0.233 | 0.244 | 1.1 | 0.975 ** | 0.268 | 0.287 | 1.9 | 0.882 ** |
| GLh | 0.251 | 0.262 | 1.1 | 0.983 ** | 0.280 | 0.303 | 2.3 | 0.902 ** |
| NSh | 0.095 | 0.113 | 1.8 | 0.954 ** | 0.099 | 0.117 | 1.8 | 0.718 ** |
| CRh | 0.143 | 0.157 | 1.4 | 0.970 ** | 0.163 | 0.181 | 1.8 | 0.820 ** |
| CIc | 0.275 | 0.275 | 0 | 0.971 ** | 0.242 | 0.254 | 1.2 | 0.814 ** |
| FSTCc | 0.293 | 0.292 | −0.1 | 0.979 ** | 0.313 | 0.325 | 1.2 | 0.895 ** |
| GLc | 0.398 | 0.400 | 0.2 | 0.989 ** | 0.359 | 0.377 | 1.8 | 0.889 ** |
| NSc | 0.186 | 0.185 | −0.1 | 0.978 ** | 0.199 | 0.203 | 0.4 | 0.882 ** |
| CRc | 0.280 | 0.280 | 0 | 0.987 ** | 0.301 | 0.313 | 1.2 | 0.883 ** |
| Traits | Whole Population | Genotyped Subpopulation | ||||||
|---|---|---|---|---|---|---|---|---|
| PBLUP | ssGBLUP | (%) | Correlation | PBLUP | ssGBLUP | (%) | Correlation | |
| Newborn | ||||||||
| BH | 0.325 | 0.338 | 1.3 | 0.988 ** | 0.302 | 0.164 | −13.8 | 0.842 ** |
| BL | 0.310 | 0.325 | 1.5 | 0.987 ** | 0.289 | 0.158 | −13.1 | 0.857 ** |
| CG | 0.361 | 0.369 | 0.8 | 0.996 ** | 0.337 | 0.179 | −15.8 | 0.891 ** |
| LC | 0.334 | 0.349 | 1.5 | 0.994 ** | 0.313 | 0.169 | −14.4 | 0.909 ** |
| CC | 0.451 | 0.463 | 1.2 | 0.996 ** | 0.433 | 0.262 | −17.1 | 0.909 ** |
| BW | 0.415 | 0.420 | 0.5 | 0.996 ** | 0.351 | 0.222 | −12.9 | 0.916 ** |
| Six months | ||||||||
| BH | 0.325 | 0.338 | 1.3 | 0.978 ** | 0.302 | 0.164 | −13.8 | 0.906 ** |
| BL | 0.310 | 0.325 | 1.5 | 0.974 ** | 0.289 | 0.158 | −13.1 | 0.922 ** |
| CG | 0.361 | 0.369 | 0.8 | 0.981 ** | 0.337 | 0.179 | −15.8 | 0.915 ** |
| LC | 0.334 | 0.349 | 1.5 | 0.998 ** | 0.313 | 0.169 | −14.4 | 0.964 ** |
| CC | 0.451 | 0.463 | 1.2 | 0.973 ** | 0.433 | 0.262 | −17.1 | 0.916 ** |
| BW | 0.415 | 0.420 | 0.5 | 0.986 ** | 0.351 | 0.222 | −12.9 | 0.944 ** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Wang, D.; Zhang, M.; Xu, L.; Huang, X.; Wang, Y. Genomic Selection for Economically Important Traits in Dual-Purpose Simmental Cattle. Animals 2025, 15, 1960. https://doi.org/10.3390/ani15131960
Zhang X, Wang D, Zhang M, Xu L, Huang X, Wang Y. Genomic Selection for Economically Important Traits in Dual-Purpose Simmental Cattle. Animals. 2025; 15(13):1960. https://doi.org/10.3390/ani15131960
Chicago/Turabian StyleZhang, Xiaoxue, Dan Wang, Menghua Zhang, Lei Xu, Xixia Huang, and Yachun Wang. 2025. "Genomic Selection for Economically Important Traits in Dual-Purpose Simmental Cattle" Animals 15, no. 13: 1960. https://doi.org/10.3390/ani15131960
APA StyleZhang, X., Wang, D., Zhang, M., Xu, L., Huang, X., & Wang, Y. (2025). Genomic Selection for Economically Important Traits in Dual-Purpose Simmental Cattle. Animals, 15(13), 1960. https://doi.org/10.3390/ani15131960






