Research on the Historical Dynamics of Baicheng Oil Chicken Populations
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Samples
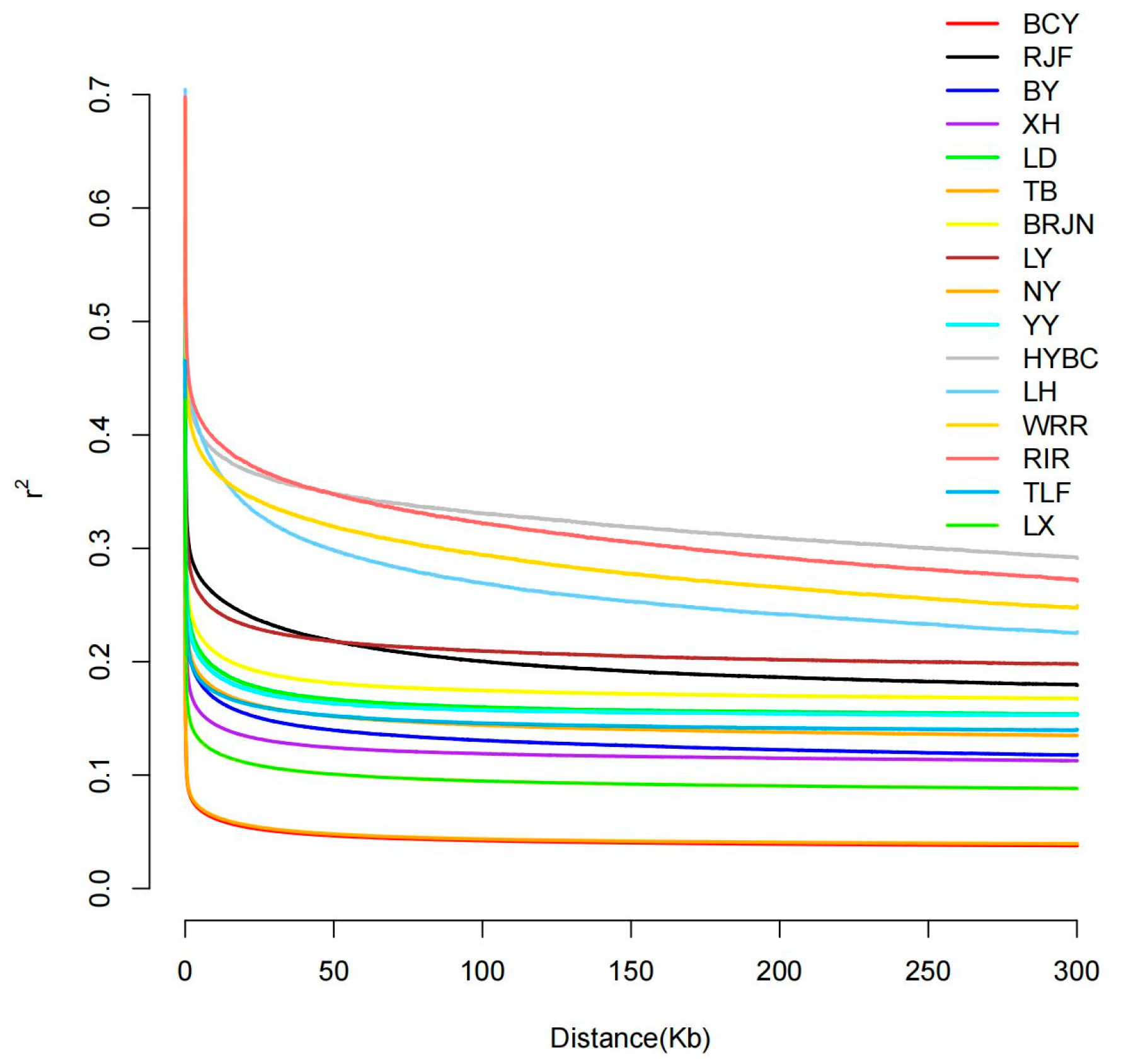
2.2. LD Decay Linkage Disequilibrium
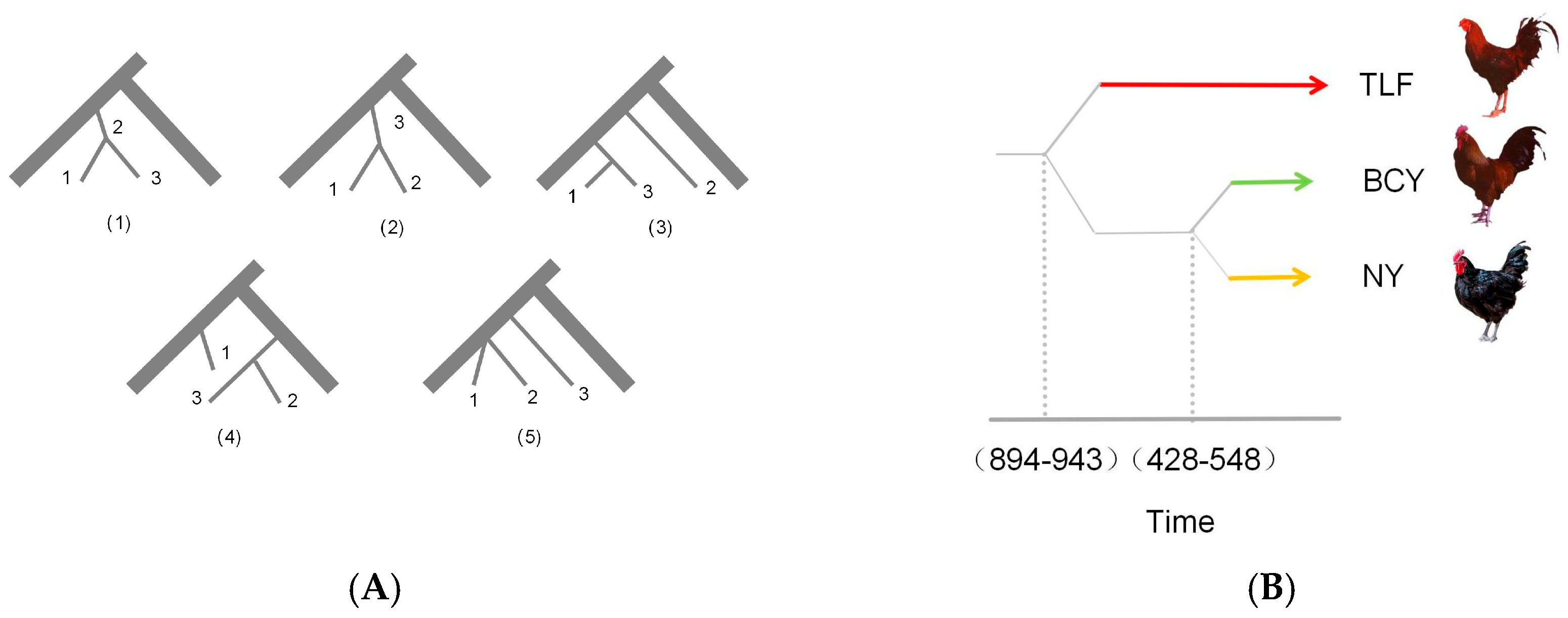
2.3. Estimation of Historical Effective Population Size and Divergence Time
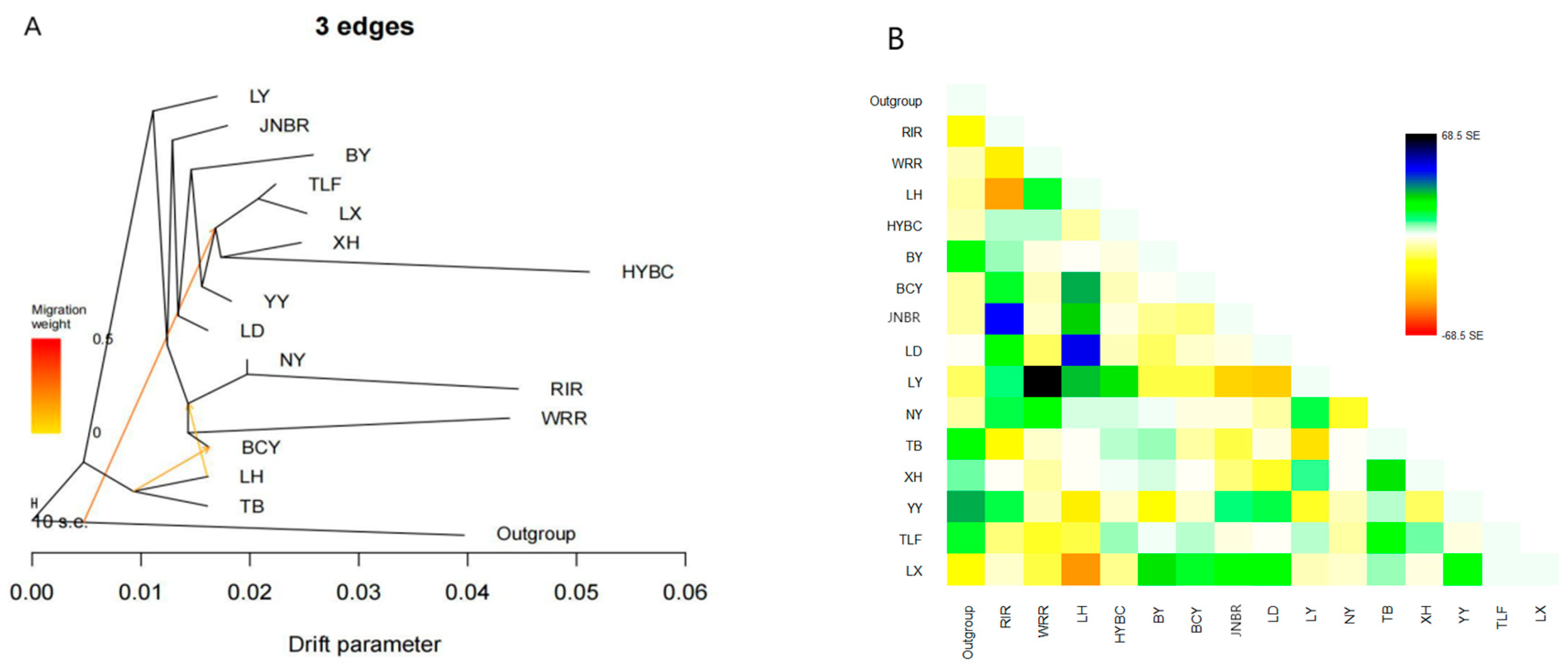
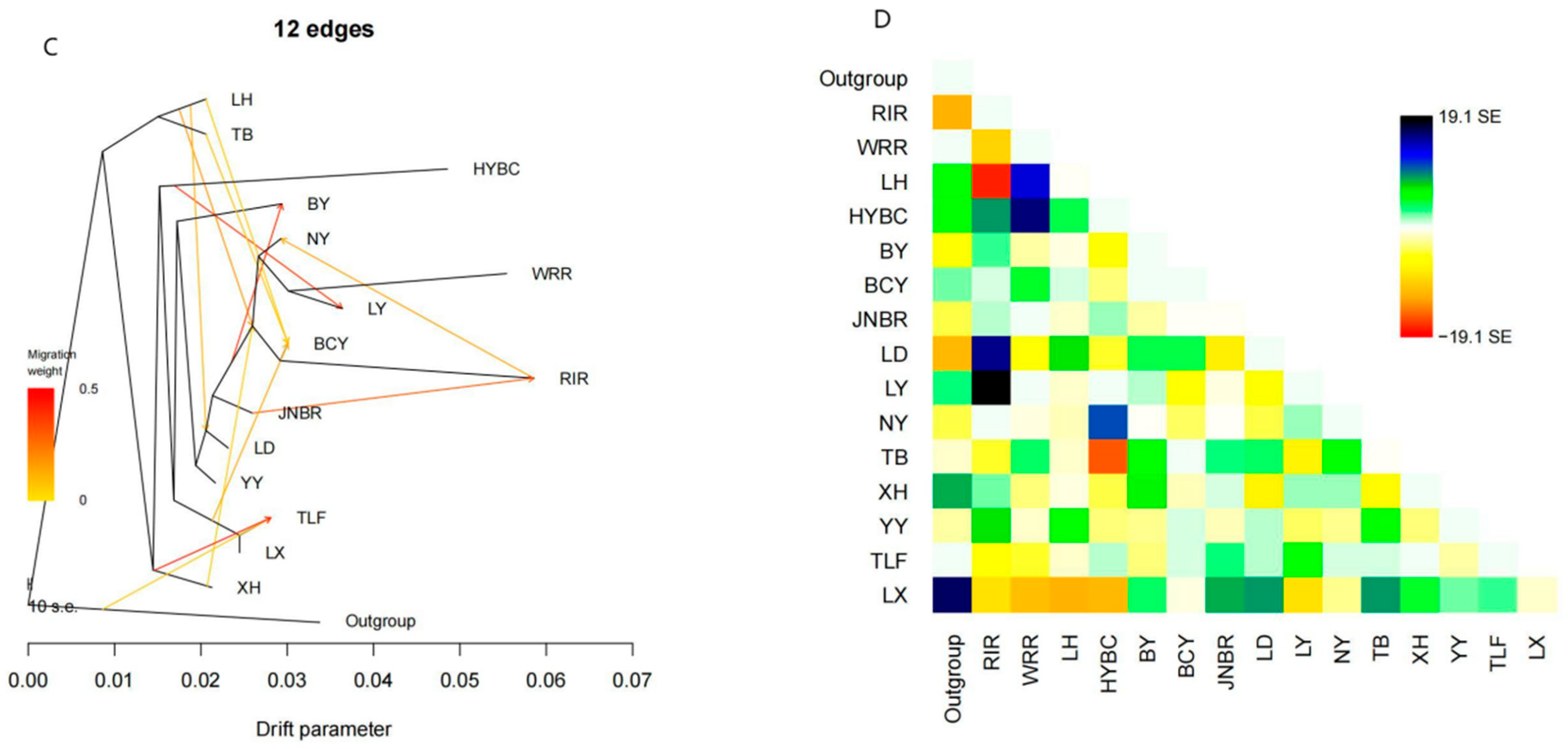
2.4. TreeMix Analysis
2.5. D Statistical Genetic Infiltration Analysis
2.6. Identification of Candidate Regions for Gene Infiltration
3. Result
3.1. LD Decay
3.2. Effective Population Size of Ancestors and Divergence Time
3.3. Gene Flow Between Populations
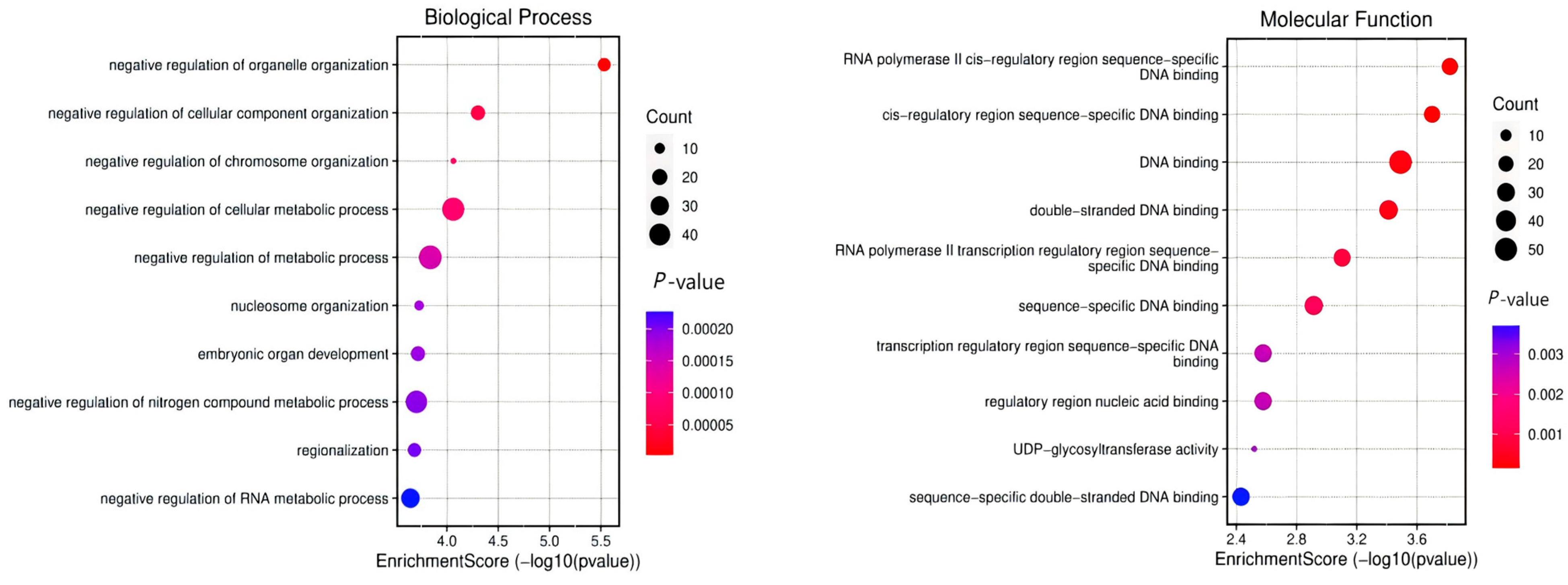
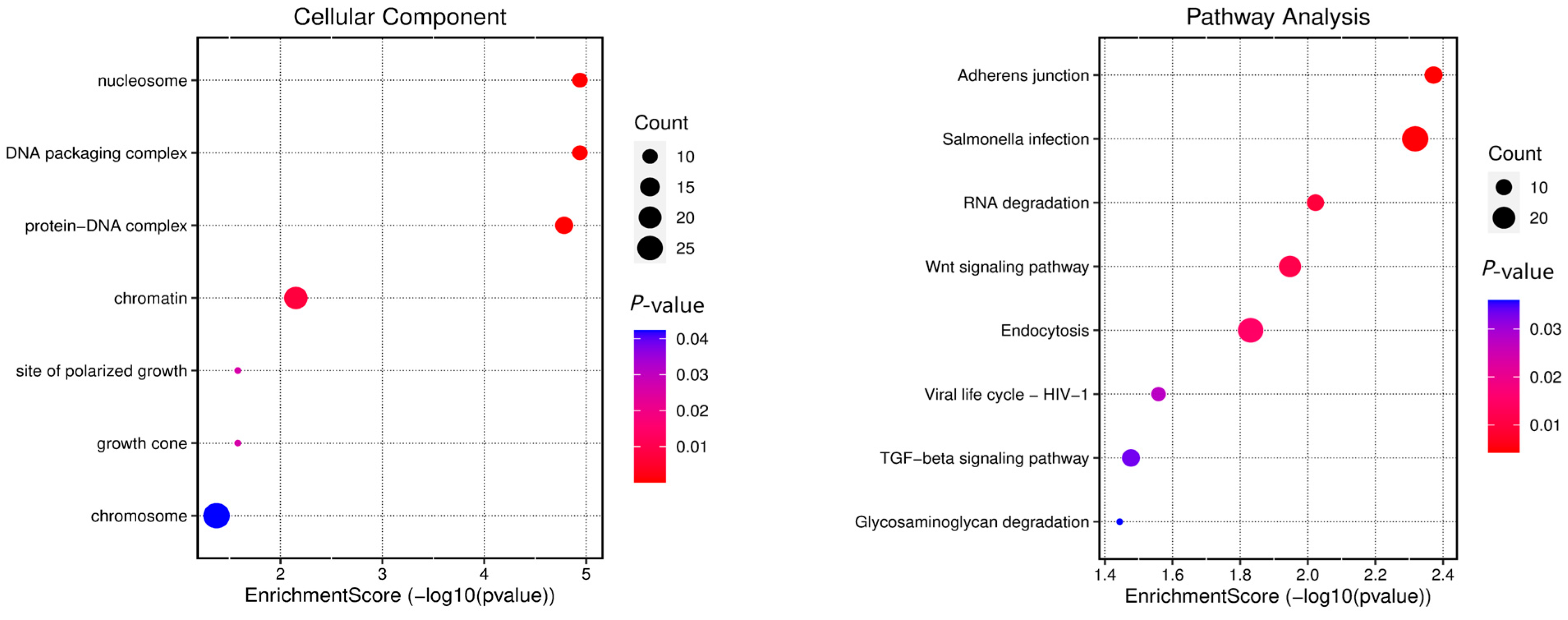
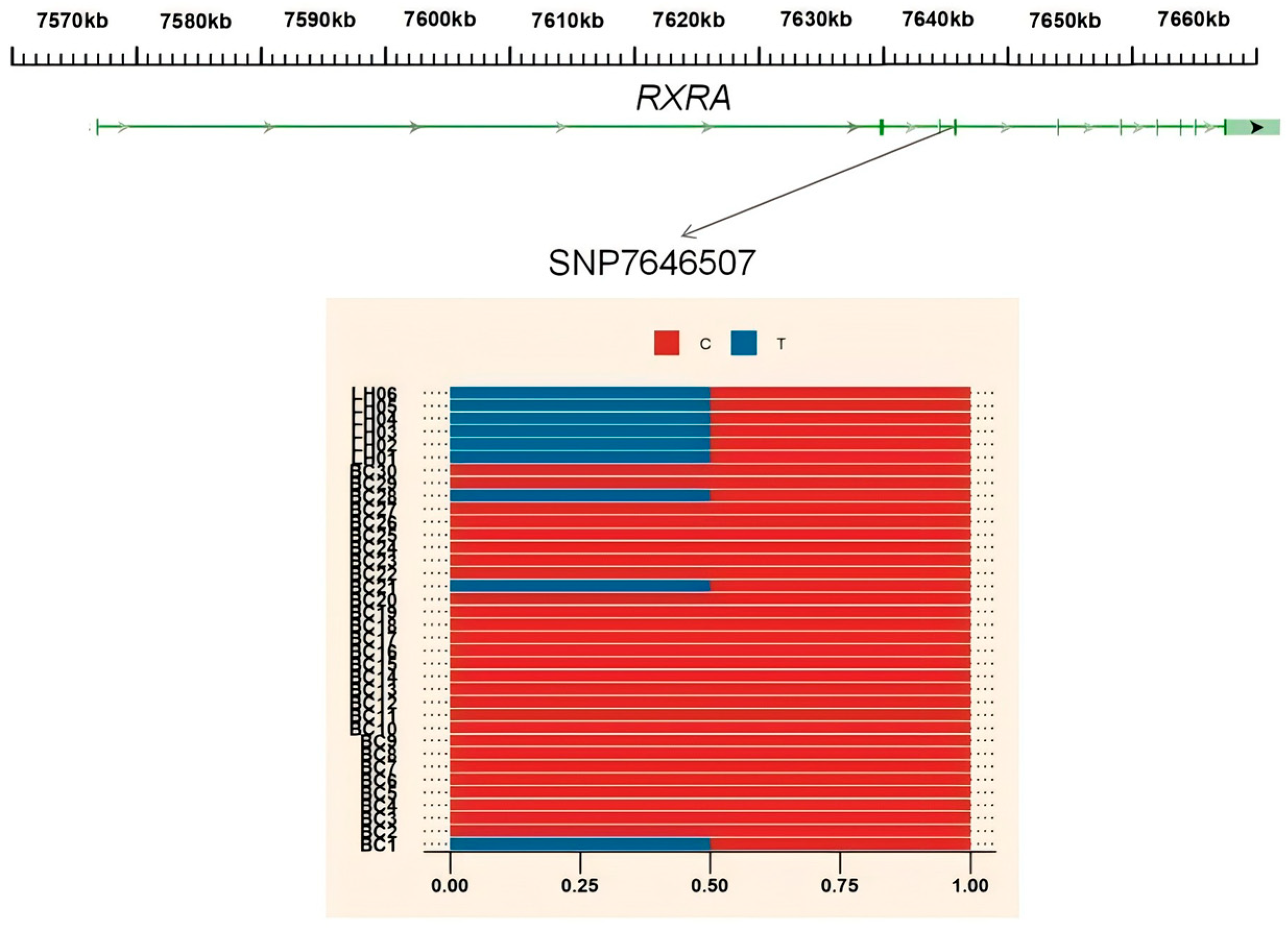
3.4. Gene Annotation of the BCY Infiltration Area by LH
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Du, J.; Qin, C.; He, Z.; Wang, J.; Wang, W.; Dong, W. Research on the Current Status and Countermeasures of the Bayingol Chicken Industry. Chin. J. Livest. Poult. Breed. 2023, 19, 18–21. [Google Scholar]
- Wright, S. Evolution in Mendelian Populations. Genetics 1931, 16, 97–159. [Google Scholar] [CrossRef]
- Kimura, M. The neutral theory of molecular evolution. Sci. Am. 1979, 241, 98–100, 102, 108. [Google Scholar] [CrossRef]
- Elbeltagy, A.R.; Bertolini, F.; Fleming, D.S.; Van Goor, A.; Ashwell, C.M.; Schmidt, C.J.; Kugonza, D.R.; Lamont, S.J.; Rothschild, M.F. Natural Selection Footprints Among African Chicken Breeds and Village Ecotypes. Front. Genet. 2019, 10, 376. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Guan, Z.; Li, H.; Wen, J.; Zhao, X.; Wang, G.; Zhang, X.; Wang, H.; Zhang, L.; Yu, F.; et al. Extensive intra- and inter-genetic admixture of Chinese gamecock and other indigenous chicken breeds revealed by genomic data. Poult. Sci. 2023, 102, 102766. [Google Scholar] [CrossRef] [PubMed]
- Lv, F.; Cao, Y.; Liu, G.; Luo, L.-Y.; Lu, R.; Liu, M.-J.; Li, W.-R.; Zhou, P.; Wang, X.-H.; Shen, M.; et al. Whole-Genome Resequencing of Worldwide Wild and Domestic Sheep Elucidates Genetic Diversity, Introgression, and Agronomically Important Loci. Mol. Biol. Evol. 2022, 39, msab353. [Google Scholar] [CrossRef]
- Maiorano, A.M.; Lourenco, D.L.; Tsuruta, S.; Toro Ospina, A.M.; Stafuzza, N.B.; Masuda, Y.; Filho, A.E.V.; Dos Santos Goncalves Cyrillo, J.N.; Curi, R.A.; De Vasconcelos Silva, J.A. Assessing genetic architecture and signatures of selection of dual purpose Gir cattle populations using genomic information. PLoS ONE 2018, 13, e200694. [Google Scholar] [CrossRef]
- Wang, H.; Zhao, X.; Wen, J.; Wang, C.; Zhang, X.; Ren, X.; Zhang, J.; Li, H.; Muhatai, G.; Qu, L. Comparative population genomics analysis uncovers genomic footprints and genes influencing body weight trait in Chinese indigenous chicken. Poult. Sci. 2023, 102, 103031. [Google Scholar] [CrossRef]
- Zhang, C.; Dong, S.S.; Xu, J.Y.; He, W.-M.; Yang, T.-L. PopLDdecay: A fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics 2019, 35, 1786–1788. [Google Scholar] [CrossRef]
- Terhorst, J.; Kamm, J.A.; Song, Y.S. Robust and scalable inference of population history from hundreds of unphased whole genomes. Nat. Genet. 2017, 49, 303–309. [Google Scholar] [CrossRef]
- Excoffier, L.; Marchi, N.; Marques, D.A.; Matthey-Doret, R.; Gouy, A.; Sousa, V.C. fastsimcoal2: Demographic inference under complex evolutionary scenarios. Bioinformatics 2021, 37, 4882–4885. [Google Scholar] [CrossRef] [PubMed]
- Pickrell, J.K.; Pritchard, J.K. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 2012, 8, e1002967. [Google Scholar] [CrossRef] [PubMed]
- Malinsky, M.; Matschiner, M.; Svardal, H. Dsuite—FastD-statistics and related admixture evidence from VCF files. Mol. Ecol. Resour. 2021, 21, 584–595. [Google Scholar] [CrossRef] [PubMed]
- Palstra, F.P.; Ruzzante, D.E. Genetic estimates of contemporary effective population size: What can they tell us about the importance of genetic stochasticity for wild population persistence? Mol. Ecol. 2008, 17, 3428–3447. [Google Scholar] [CrossRef]
- Rubin, C.; Zody, M.C.; Eriksson, J.; Meadows, J.R.S.; Sherwood, E.; Webster, M.T.; Jiang, L.; Ingman, M.; Sharpe, T.; Ka, S.; et al. Whole-genome resequencing reveals loci under selection during chicken domestication. Nature 2010, 464, 587–591. [Google Scholar] [CrossRef]
- Kardos, M.; Armstrong, E.E.; Fitzpatrick, S.W.; Hauser, S.; Hedrick, P.W.; Miller, J.M.; Tallmon, D.A.; Funk, W.C. The crucial role of genome-wide genetic variation in conservation. Proc. Natl. Acad. Sci. USA 2021, 118, e2104642118. [Google Scholar] [CrossRef]
- Pouyet, F.; Aeschbacher, S.; Thiery, A.; Excoffier, L. Background selection and biased gene conversion affect more than 95% of the human genome and bias demographic inferences. eLife 2018, 7, e36317. [Google Scholar] [CrossRef]
- Armstrong, E.E.; Khan, A.; Taylor, R.W.; Gouy, A.; Greenbaum, G.; Thiéry, A.; Kang, J.T.; A Redondo, S.; Prost, S.; Barsh, G.; et al. Recent Evolutionary History of Tigers Highlights Contrasting Roles of Genetic Drift and Selection. Mol. Biol. Evol. 2021, 38, 2366–2379. [Google Scholar] [CrossRef]
- Lu, K.; Wei, L.; Li, X.; Wang, Y.; Wu, J.; Liu, M.; Zhang, C.; Chen, Z.; Xiao, Z.; Jian, H.; et al. Whole-genome resequencing reveals Brassica napus origin and genetic loci involved in its improvement. Nat. Commun. 2019, 10, 1154. [Google Scholar] [CrossRef]
- Vazquez-Rosas-Landa, M.; Ponce-Soto, G.Y.; Aguirre-Liguori, J.A.; Thakur, S.; Scheinvar, E.; Barrera-Redondo, J.; Ibarra-Laclette, E.; Guttman, D.S.; Eguiarte, L.E.; Souza, V. Population genomics of Vibrionaceae isolated from an endangered oasis reveals local adaptation after an environmental perturbation. BMC Genom. 2020, 21, 418. [Google Scholar] [CrossRef]
- Wu, D. A Historical Study of Cockfighting (Second Report on Henan Cockfighting). J. Sci. Res. West. Henan Agric. Coll. 1983, 49–50+63. [Google Scholar]
- Shi, S.; Shao, D.; Yang, L.; Liang, Q.; Han, W.; Xue, Q.; Qu, L.; Leng, L.; Li, Y.; Zhao, X.; et al. Whole genome analyses reveal novel genes associated with chicken adaptation to tropical and frigid environments. J. Adv. Res. 2023, 47, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z. Lingnan Omnivorous Poem; Zhonghua Book Company: Beijing, China, 1985. [Google Scholar]
- Zhong, Y.; Zhong, Z.; Li, H. Conservation of Genetic Resources and Breeding Improvement of “Bayingol Chicken” in Xinjiang. In Proceedings of the 14th National Symposium on Poultry Science, Harbin, China, 1 July 2009. [Google Scholar]
- Hu, J.N. The State of Jingjue in the Western Regions. Tutoring Middle Sch. Stud. 2013, 13, 42–44. [Google Scholar]
- Zhang, C.; Lin, D.; Wang, Y.; Peng, D.; Li, H.; Fei, J.; Chen, K.; Yang, N.; Hu, X.; Zhao, Y.; et al. Widespread introgression in Chinese indigenous chicken breeds from commercial broiler. Evol. Appl. 2018, 12, 610–621. [Google Scholar] [CrossRef] [PubMed]
- Tepekoy, F.; Akkoyunlu, G.; Demir, R. The role of Wnt signaling members in the uterus and embryo during pre-implantation and implantation. J. Assist. Reprod. Genet. 2015, 32, 337–346. [Google Scholar] [CrossRef]
- Ricken, A.; Lochhead, P.; Kontogiannea, M.; Farookhi, R. Wnt signaling in the ovary: Identification and compartmentalized expression of wnt-2, wnt-2b, and frizzled-4 mRNAs. Endocrinology 2002, 143, 2741–2749. [Google Scholar] [CrossRef]
- Wolc, A.; Lisowski, M.; Szwaczkowski, T. Heritability of egg production in laying hens under different environmental conditions. Czech J. Anim. Sci. 2007, 52, 254–259. [Google Scholar] [CrossRef]
- Braunschweig, U.; Barbosa-Morais, N.L.; Pan, Q.; Nachman, E.N.; Alipanahi, B.; Gonatopoulos-Pournatzis, T.; Frey, B.; Irimia, M.; Blencowe, B.J. Widespread intron retention in mammals functionally tunes transcriptomes. Genome Res. 2014, 24, 1774–1786. [Google Scholar] [CrossRef]
- Monteuuis, G.; Wong, J.J.L.; Bailey, C.G.; Schmitz, U.; Rasko, J.E. The changing paradigm of intron retention: Regulation, ramifications and recipes. Nucleic Acids Res. 2019, 47, 11497–11513. [Google Scholar] [CrossRef]
- Johnson, A.L. Ovarian follicle selection and granulosa cell differentiation. Poult. Sci. 2015, 94, 781–785. [Google Scholar] [CrossRef]
- Nguyen, T.M.; Duittoz, A.; Praud, C.; Combarnous, Y.; Blesbois, E. Calcium channels in chicken sperm regulate motility and the acrosome reaction. FEBS J. 2016, 283, 1902–1920. [Google Scholar] [CrossRef]
- Vaishnav, S.; Chauhan, A.; Ajay, A.; Saini, B.L.; Kumar, S.; Kumar, A.; Bhushan, B.; Gaur, G.K. Allelic to genome wide perspectives of swine genetic variation to litter size and its component traits. Mol. Biol. Rep. 2023, 50, 3705–3721. [Google Scholar] [CrossRef] [PubMed]
- Pang, P.; Li, Z.; Hu, H.; Wang, L.; Sun, H.; Mei, S.; Li, F. Genetic effect and combined genotype effect of ESR, FSHbeta, CTNNAL1 and miR-27a loci on litter size in a Large White population. Anim. Biotechnol. 2019, 30, 287–292. [Google Scholar] [CrossRef] [PubMed]
- Chawla, A.; Repa, J.J.; Evans, R.M.; Mangelsdorf, D.J. Nuclear receptors and lipid physiology: Opening the X-files. Science 2001, 294, 1866–1870. [Google Scholar] [CrossRef] [PubMed]
- Bastien, J.; Rochette-Egly, C. Nuclear retinoid receptors and the transcription of retinoid-target genes. Gene 2004, 328, 1–16. [Google Scholar] [CrossRef]
- Park, B.; Nguyen, N.T.; Dutt, P.; Merdek, K.D.; Bashar, M.; Sterpetti, P.; Tosolini, A.; Testa, J.R.; Toksoz, D. Association of Lbc Rho guanine nucleotide exchange factor with alpha-catenin-related protein, alpha-catulin/CTNNAL1, supports serum response factor activation. J. Biol. Chem. 2002, 277, 45361–45370. [Google Scholar] [CrossRef]


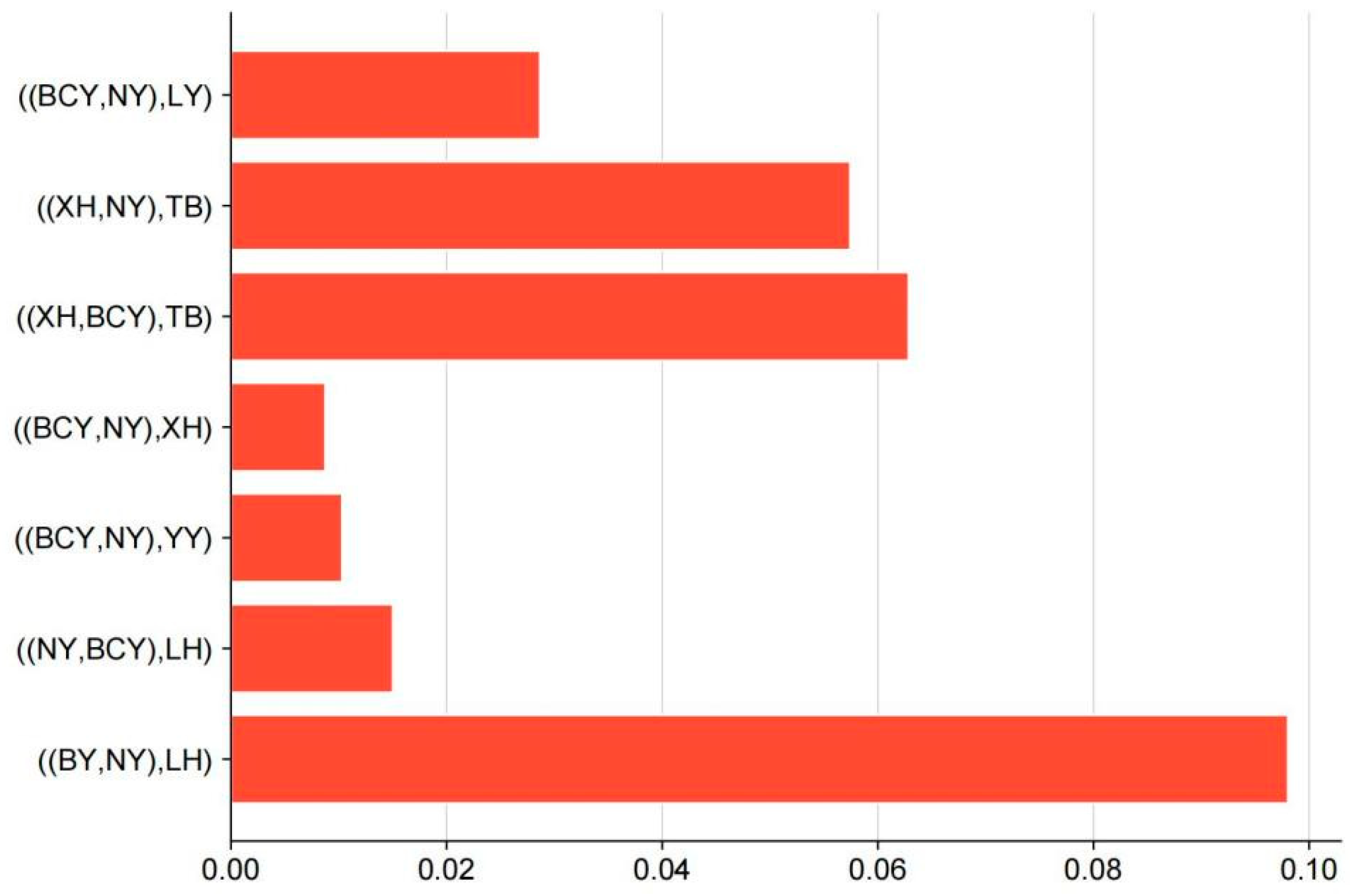

| ((P1,P2),P3) | chr | Window Start | Window End | D Value | f_d | f_dM | d_f |
|---|---|---|---|---|---|---|---|
| ((NY,BCY),LH) | chr17 | 6738123 | 9797958 | 0.074077 | 0.060236 | 0.042896 | 0.039979 |
| chr22 | 2015588 | 4127909 | 0.0738 | 0.063796 | 0.044762 | 0.037595 | |
| chr1 | 20420138 | 53163584 | 0.070559 | 0.063676 | 0.042755 | 0.038425 | |
| chr2 | 47984993 | 92026288 | 0.069768 | 0.044631 | 0.034249 | 0.037573 | |
| chr2 | 58104921 | 100875426 | 0.069497 | 0.052234 | 0.038317 | 0.035799 | |
| ((XH,BCY),TB) | chr5 | 24005918 | 49859805 | 0.141173 | 0.23659 | 0.114761 | 0.078218 |
| chr5 | 16171926 | 38768130 | 0.137803 | 0.236848 | 0.111738 | 0.078419 |
| CHROM | BIN_START | BIN_END | GENE |
|---|---|---|---|
| chr1 | 33820001 | 33830000 | RASSF3 |
| chr17 | 7640001 | 7650000 | RXRA |
| chr2 | 49130001 | 49140000 | No gene |
| chr2 | 54155001 | 54165000 | CNTNAP2 |
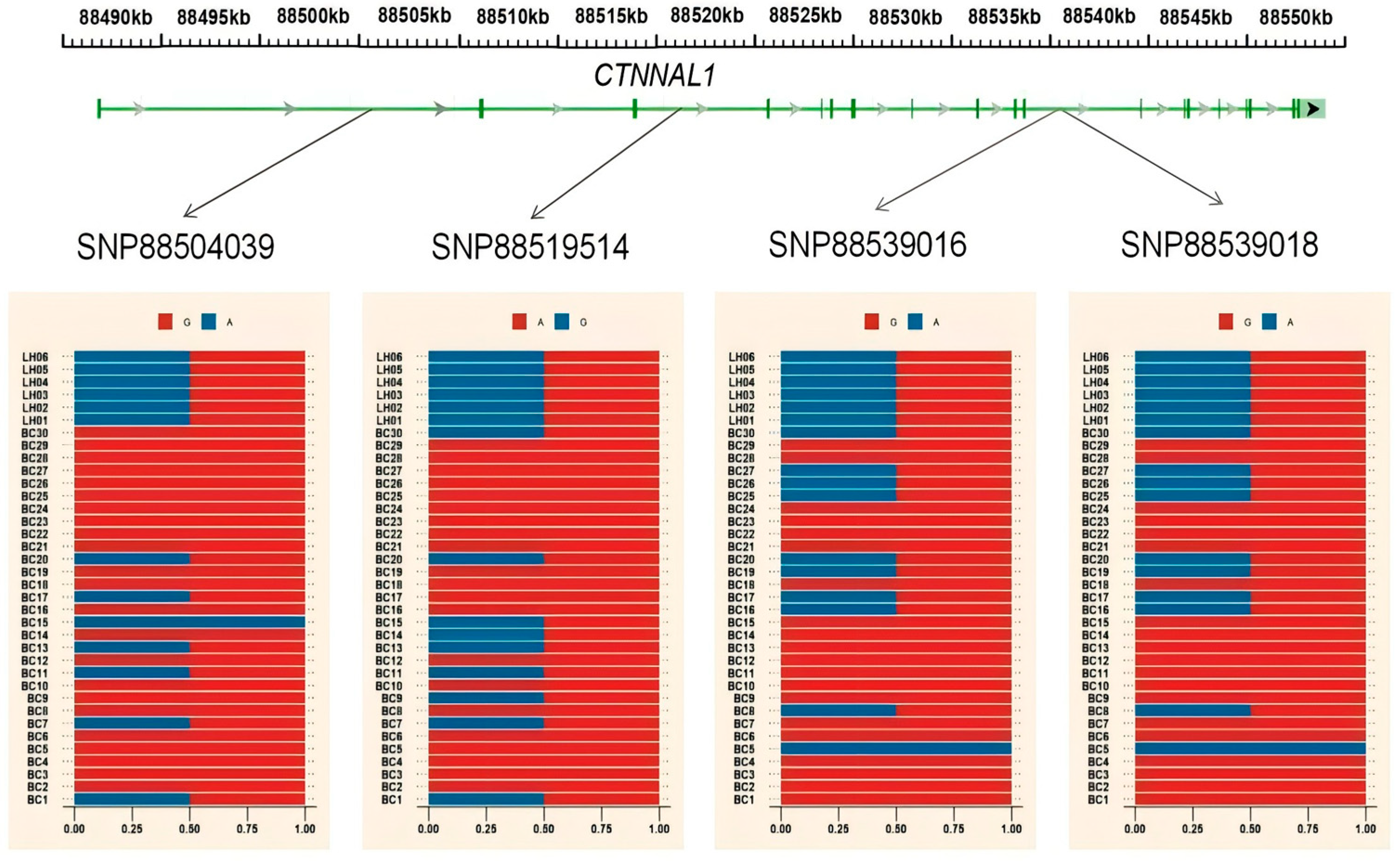
| chr2 | 88495001 | 88555000 | CTNNAL1 |
| chr2 | 88550001 | 88560000 | FAM206A |
| chr2 | 88625001 | 88650000 | ENSGALT00000104496 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Liu, T.; Wang, G.; Zhao, X.; Wang, C.; Li, F.; Muhatai, G.; Qu, L. Research on the Historical Dynamics of Baicheng Oil Chicken Populations. Animals 2025, 15, 1952. https://doi.org/10.3390/ani15131952
Wang H, Liu T, Wang G, Zhao X, Wang C, Li F, Muhatai G, Qu L. Research on the Historical Dynamics of Baicheng Oil Chicken Populations. Animals. 2025; 15(13):1952. https://doi.org/10.3390/ani15131952
Chicago/Turabian StyleWang, Huie, Tianci Liu, Gang Wang, Xiurong Zhao, Chengqian Wang, Fugui Li, Gemingguli Muhatai, and Lujiang Qu. 2025. "Research on the Historical Dynamics of Baicheng Oil Chicken Populations" Animals 15, no. 13: 1952. https://doi.org/10.3390/ani15131952
APA StyleWang, H., Liu, T., Wang, G., Zhao, X., Wang, C., Li, F., Muhatai, G., & Qu, L. (2025). Research on the Historical Dynamics of Baicheng Oil Chicken Populations. Animals, 15(13), 1952. https://doi.org/10.3390/ani15131952





