Metabolomics and Transcriptomics Reveal Age-Dependent Development of Meat Quality Traits in Jingyuan Chicken
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Animal and Sample Collection
2.3. Meat Quality Traits Determination
2.4. Metabolomics Analysis by Untargeted HPLC-HRMS
2.5. Transcriptomics Analysis
2.6. Combined Analysis of Differential Genes and Differential Metabolites
2.7. Statistical Analysis
3. Results
3.1. Meat Quality Traits Performance of Jingyuan Hens at Different Developmental Stages
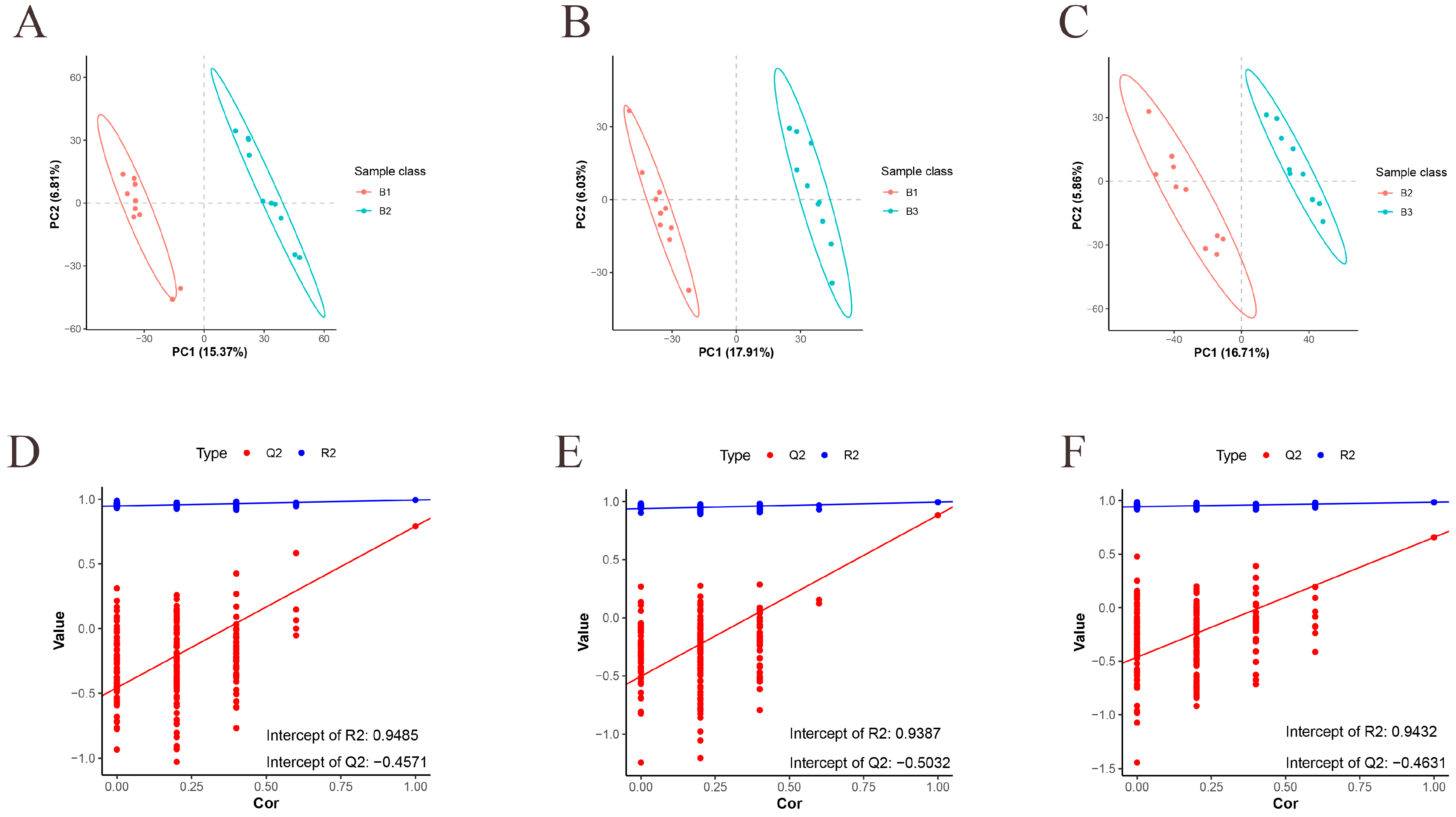
3.2. Metabolome QC and OPLS-DA Analysis
3.3. Differential Metabolite Analysis
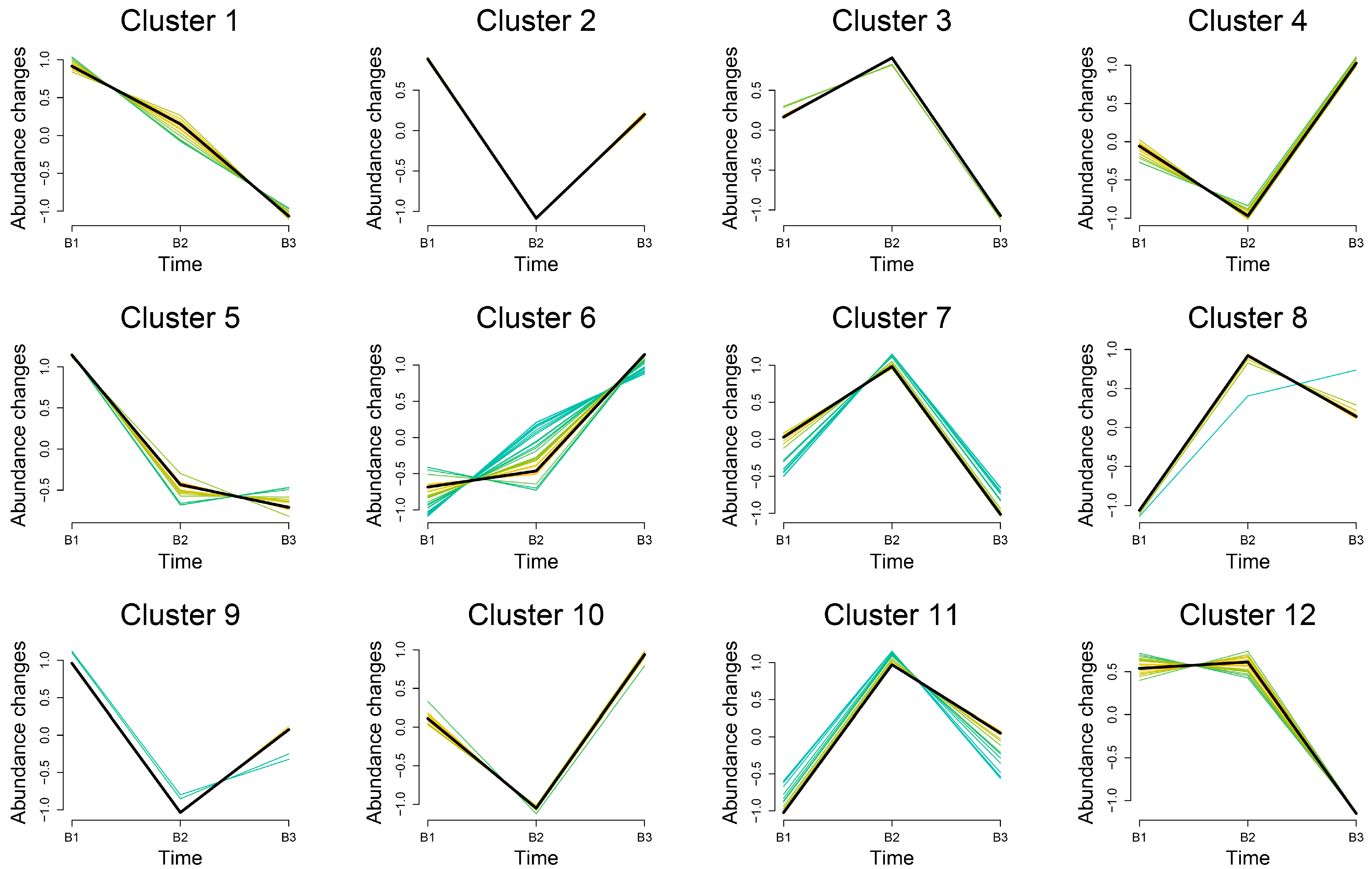
3.4. Trend Analysis of Differential Metabolite Expression
3.5. KEGG Pathway Enrichment Analysis
3.6. Joint Analysis of Transcriptome and Metabolome
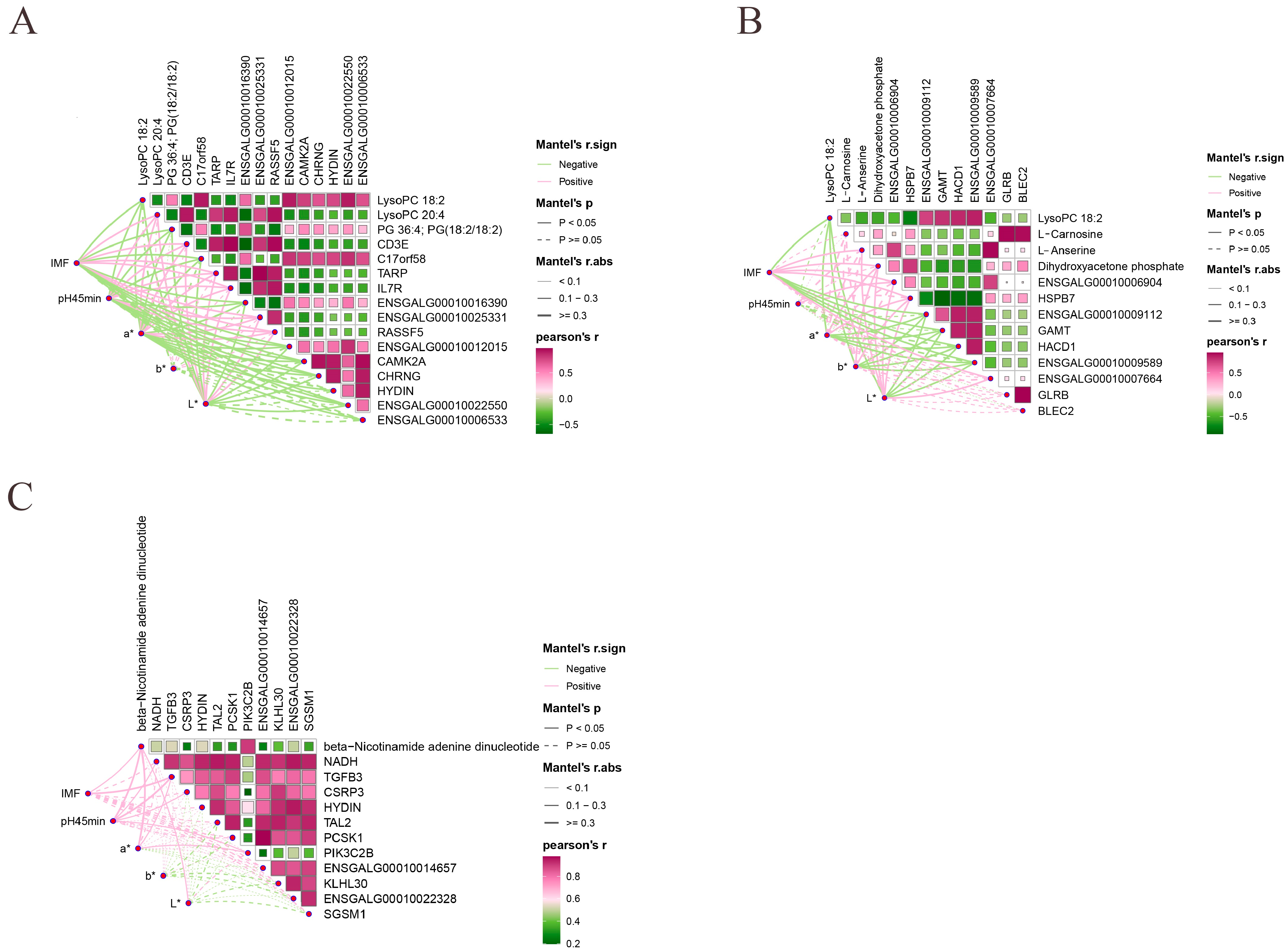
3.7. Identification of Meat Quality Traits Related Candidate Genes and Metabolites
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MLC | Muscle lipid content. |
| DEMs | Directory of open access journals. |
| DEGs | Three-letter acronym. |
| OPLS-DA | Linear dichroism. |
| PCA | Principal component analysis. |
| HMDB | Human Metabolome Database. |
| FC | Fold change. |
| dT | Dynabeads Oligo. |
| QC | Quality traits control |
References
- Zhao, W.; Hu, J.; Li, L.; Xue, L.; Tian, J.; Zhang, T.; Yang, L.; Gu, Y.; Zhang, J. Integrating lipidomics and metabolomics to reveal biomarkers of fat deposition in chicken meat. Food Chem. 2025, 464 Pt 2, 141732. [Google Scholar] [CrossRef]
- Burke, M.; Martini, L.; Blake, C.; Younginer, N.; Draper, C.; Bell, B.; Liese, A.; Jones, S. Stretching Food and Being Creative: Caregiver Responses to Child Food Insecurity. J. Nutr. Educ. Behav. 2017, 49, 296–303.e1. [Google Scholar] [CrossRef]
- Yu, B.; Cai, Z.; Liu, J.; Zhao, W.; Fu, X.; Gu, Y.; Zhang, J. Transcriptome and co-expression network analysis reveals the molecular mechanism of inosine monophosphate-specific deposition in chicken muscle. Front. Physiol. 2023, 14, 1199311. [Google Scholar] [CrossRef] [PubMed]
- Lu, P.; Li, D.; Yin, J.; Zhang, L.; Wang, Z. Flavour differences of cooked longissimus muscle from chinese indigenous pig breeds and hybrid pig breed (duroc × landrace × large white). Food Chem. 2008, 107, 1529–1537. [Google Scholar] [CrossRef]
- Qiu, F.; Xie, L.; Ma, J.E.; Luo, W.; Zhang, L.; Chao, Z.; Chen, S.; Nie, Q.; Lin, Z.; Zhang, X. Lower Expression of SLC27A1 Enhances Intramuscular Fat Deposition in Chicken via Down-Regulated Fatty Acid Oxidation Mediated by CPT1A. Front. Physiol. 2017, 8, 449. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Li, W.; Chen, C.; Yu, H.; Huang, J.; Tian, H. Novel cinnamon essential oil-bacterial cellulose microcapsules for enhanced preservation of prefabricated meat. Int. J. Biol. Macromol. 2024, 282 Pt 1, 136851. [Google Scholar] [CrossRef]
- Mottram, D.-S. Flavour formation in meat and meat products: A review. Food Chem. 1998, 62, 415–424. [Google Scholar] [CrossRef]
- Huang, Z.; Zhang, J.; Gu, Y.; Cai, Z.; Wei, D.; Feng, X.; Yang, C. Analysis of the molecular mechanism of inosine monophosphate deposition in Jingyuan chicken muscles using a proteomic approach. Poult. Sci. 2022, 101, 101741. [Google Scholar] [CrossRef]
- Beauclercq, S.; Mignon-Grasteau, S.; Petit, A.; Berger, Q.; Lefèvre, A.; Métayer-Coustard, S.; Tesseraud, S.; Emond, P.; Berri, C.; Le Bihan-Duval, E. A Divergent Selection on Breast Meat Ultimate pH, a Key Factor for Chicken Meat Quality traits, is Associated With Different Circulating Lipid Profiles. Front. Physiol. 2022, 13, 935868. [Google Scholar] [CrossRef]
- Liu, Y.; Liang, S.; Wang, K.; Zi, X.; Zhang, R.; Wang, G.; Kang, J.; Li, Z.; Dou, T.; Ge, C. Physicochemical, Nutritional Properties and Metabolomics Analysis Fat Deposition Mechanism of Chahua Chicken No. 2 and Yao Chicken. Genes 2022, 13, 1358. [Google Scholar] [CrossRef]
- Beauclercq, S.; Nadal-Desbarats, L.; Hennequet-Antier, C.; Collin, A.; Tesseraud, S.; Bourin, M.; Le Bihan-Duval, E.; Berri, C. Serum and Muscle Metabolomics for the Prediction of Ultimate pH, a Key Factor for Chicken-Meat Quality traits. J. Proteome Res. 2016, 15, 1168–1178. [Google Scholar] [CrossRef] [PubMed]
- Hirai, M.Y.; Yano, M.; Goodenowe, D.B.; Kanaya, S.; Kimura, T.; Awazuhara, M.; Arita, M.; Fujiwara, T.; Saito, K. Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2004, 101, 10205–10210. [Google Scholar] [CrossRef]
- Xiao, C.; Sun, T.; Yang, Z.; Xu, W.; Wang, J.; Zeng, L.; Deng, J.; Yang, X. Transcriptome landscapes of differentially expressed genes related to fat deposits in Nandan-Yao chicken. Funct. Integr. Genom. 2021, 21, 113–124. [Google Scholar] [CrossRef]
- Liu, L.; Liu, X.; Cui, H.; Liu, R.; Zhao, G.; Wen, J. Transcriptional insights into key genes and pathways controlling muscle lipid metabolism in broiler chickens. BMC Genom. 2019, 20, 863. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Liu, J.; Cai, Z.; Mu, T.; Gu, Y.; Xin, G.; Zhang, J. miRNA-mRNA associations with inosine monophosphate specific deposition in the muscle of Jingyuan chicken. Br. Poult. Sci. 2022, 63, 821–832. [Google Scholar] [CrossRef]
- GB 5009.6-2016; National Standard for Food Safety Determination of Fat in Food. State Food and Drug Administration: Beijing, China, 2017.
- Warwick, B.D.; David, B.; Paul, B.; Eva, Z.; Sue, F.-M.; Nadine, A.; Marie, B.; Joshua, K.; Antony, H.; John, N.H.; et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2011, 6, 1060–1083. [Google Scholar]
- Wen, B.; Mei, Z.; Zeng, C.; Liu, S. metaX: A flexible and comprehensive software for processing metabolomics data. BMC Bioinform. 2017, 18, 183. [Google Scholar] [CrossRef]
- Hu, J.H. Sereening of Key Genes and Key Metabolites Associated with Intramuscular Fat Deposition in Jingyuan Chicken; Ningxia University: Yinchuan, China, 2024. [Google Scholar]
- Oliver, T.; Ferdinand von, M.; Zoë, H.; John, A.; Andrew, W.; Richard, W.; Siân, G.; Felix, K.; Simon, A.; Ivana, B.; et al. Low rates of mutation in clinical grade human pluripotent stem cells under different culture conditions. Nat. Commun. 2020, 11, 1528. [Google Scholar]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Sayed Mohammad Ebrahim, S.; Marghoob, M.; Robert, S.; Hagen, T.; Pegah Tootoonchi, A.; Kin Fai, A.; Narges Bani, A.; Mark, G.; Wing Hung, W.; Snyder, M.; et al. Gaining comprehensive biological insight into the transcriptome by performing a broad-spectrum RNA-seq analysis. Nat. Commun. 2017, 8, 59. [Google Scholar]
- Nguyen, D.V.; Nguyen, O.C.; Malau-Aduli, A.E.O. Main regulatory factors of marbling level in beef cattle. Vet. Anim. Sci. 2021, 14, 100219. [Google Scholar] [CrossRef] [PubMed]
- Griffin, H.D.; Guo, K.; Windsor, D.; Butterwith, S.C. Adipose tissue lipogenesis and fat deposition in leaner broiler chickens. J. Nutr. 1992, 122, 363–368. [Google Scholar] [CrossRef] [PubMed]
- Kanaley, J.A.; Shadid, S.; Sheehan, M.T.; Guo, Z.; Jensen, M.D. Relationship between plasma free fatty acid, intramyocellular triglycerides and long-chain acylcarnitines in resting humans. J Physiol. 2009, 587 Pt 24, 5939–5950. [Google Scholar] [CrossRef] [PubMed]
- Heo, K.; Hong, E.; Kim, C.; Kim, H.; Lee, M.; Choo, H.; Choi, H.; Mushtaq, M.; Parvin, R.; Kim, J. The quality traits characteristics of Korean native chickenby the age. Asian-Australas J. Anim. Sci. 2015, 28, 382–390. [Google Scholar] [CrossRef]
- Moeller, S.J.; Rahn, M.; Schneider, F.T. Effect of different phosphate preparations on consistency and sensory of cooked sausages. Fleischwirtschaft 2001, 81, 101–103. [Google Scholar]
- Honikel, K.O.; Kim, C.J.; Hamm, R.; Roncales, P. Sarcomere shortening of prerigor muscles and its influence on drip loss. Meat Sci. 1986, 16, 267–282. [Google Scholar] [CrossRef]
- Cerretelli, P.; Samaja, M. Acid-base balance at exercise in normoxia and in chronic hypoxia. Revisiting the “lactate paradox”. Eur. J. Appl. Physiol. 2003, 90, 431–448. [Google Scholar] [CrossRef]
- Northcutt, J.K.; Pringle, T.D.; Dickens, J.A.; Buhr, R.J.; Young, L.L. Effects of age and tissue type on the calpain proteolytic system in turkey skeletal muscle. Poult. Sci. 1998, 77, 367–372. [Google Scholar] [CrossRef]
- Moore, G.E.; Goldspink, G. The effect of reduced activity on the enzymatic development of phasic and tonic muscles in the chicken. J. Dev. Physiol. 1985, 7, 381–386. [Google Scholar]
- Gáspárdy, A.; Bélley, R.; Barta, I. Meat-Producing Ability of Two Autochthonous Chicken Breeds Under Traditional and Semi-Intensive Conditions. Agriculture 2025, 15, 21. [Google Scholar] [CrossRef]
- Hughes, J.M.; Oiseth, S.K.; Purslow, P.P.; Warner, R.D. A structural approach to understanding the interactions between colour, water-holding capacity and tenderness. Meat Sci. 2014, 98, 520–532. [Google Scholar] [CrossRef]
- Canto, A.C.; Suman, S.P.; Nair, M.N.; Li, S.; Rentfrow, G.; Beach, C.M.; Silva, T.J.; Wheeler, T.L.; Shackelford, S.D.; Grayson, A.; et al. Differential abundance of sarcoplasmic proteome explains animal effect on beef Longissimus lumborum color stability. Meat Sci. 2015, 102, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Skotland, T.; Sandvig, K. The role of PS 18:0/18:1 in membrane function. Nat. Commun. 2019, 10, 2752. [Google Scholar] [CrossRef] [PubMed]
- Héliès-Toussaint, C.; Gambert, S.; Roller, P.; Tricot, S.; Lacour, B.; Grynberg, A. Lipid metabolism in human endothelial cells. Biochim. Biophys. Acta. 2006, 1761, 765–774. [Google Scholar] [CrossRef] [PubMed]
- Kaser, S.; Moschen, A.; Cayon, A.; Kaser, A.; Crespo, J.; Pons-Romero, F.; Ebenbichler, C.F.; Patsch, J.R.; Tilg, H. Adiponectin and its receptors in non-alcoholic steatohepatitis. Gut 2005, 54, 117–121. [Google Scholar] [CrossRef]
- Sprick, M.R.; Walczak, H. The interplay between the Bcl-2 family and death receptor-mediated apoptosis. Biochim. Et. Biophys. Acta 2004, 1644, 125–132. [Google Scholar] [CrossRef]
- Xu, Z.R.; Wang, M.Q.; Mao, H.X.; Zhan, X.A.; Hu, C.H. Effects of L-carnitine on growth performance, carcass composition, and metabolism of lipids in male broilers. Poult. Sci. 2003, 82, 408–413. [Google Scholar] [CrossRef]
- Rovira Gonzalez, Y.I.; Moyer, A.L.; LeTexier, N.J.; Bratti, A.D.; Feng, S.; Sun, C.; Liu, T.; Mula, J.; Jha, P.; Iyer, S.R.; et al. Mss51 deletion enhances muscle metabolism and glucose homeostasis in mice. JCI Insight 2019, 4, e122247. [Google Scholar] [CrossRef]
- Chiang, J.Y.L.; Ferrell, J.M. Bile Acid Metabolism in Liver Pathobiology. Gene Expr. 2018, 18, 71–87. [Google Scholar] [CrossRef]
- Nikopoulou, C.; Kleinenkuhnen, N.; Parekh, S.; Sandoval, T.; Ziegenhain, C.; Schneider, F.; Giavalisco, P.; Donahue, K.F.; Vesting, A.J.; Kirchner, M.; et al. Spatial and single-cell profiling of the metabolome, transcriptome and epigenome of the aging mouse liver. Nat. Aging 2023, 3, 1430–1445. [Google Scholar] [CrossRef]
- Fan, S.; Yuan, P.; Li, S.; Li, H.; Zhai, B.; Li, Y.; Zhang, H.; Gu, J.; Li, H.; Tian, Y.; et al. Genetic architecture and key regulatory genes of fatty acid composition in Gushi chicken breast muscle determined by GWAS and WGCNA. BMC Genom. 2023, 24, 434. [Google Scholar] [CrossRef]
- Kumar, P.; Liu, C.; Suliburk, J.; Hsu, J.W.; Muthupillai, R.; Jahoor, F.; Minard, C.G.; Taffet, G.E.; Sekhar, R.V. Supplementing Glycine and N-Acetylcysteine (GlyNAC) in Older Adults Improves Glutathione Deficiency, Oxidative Stress, Mitochondrial Dysfunction, Inflammation, Physical Function, and Aging Hallmarks: A Randomized Clinical Trial. J. Gerontol. A Biol. Sci. Med. Sci. 2023, 78, 75–89. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Yang, P.; Liang, Y.; Xia, N.; Li, Y.; Su, H.; Pan, H. Effects of liraglutide on lipolysis and the AC3/PKA/HSL pathway. Diabetes Metab. Syndr. Obes. 2019, 12, 1697–1703. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Wang, J.; Li, H.; Gao, L.; Geng, J.; Ma, Z.; Liu, J.; Zhang, J.; Xie, P.; Chen, L. Combined transcriptome and proteome analyses reveal differences in the longissimus dorsi muscle between Kazakh cattle and Xinjiang brown cattle. Anim. Biosci. 2021, 34, 1439–1450. [Google Scholar] [CrossRef]
- Zhang, R.; Yang, M.; Hou, X.; Hou, R.; Wang, L.; Shi, L.; Zhao, F.; Liu, X.; Meng, Q.; Wang, L.; et al. Characterization and difference of lipids and metabolites from Jianhe White Xiang and Large White pork by high-performance liquid chromatography-tandem mass spectrometry. Food Res Int. 2022, 162 Pt A, 111946. [Google Scholar] [CrossRef]
- Wang, J.; Xiang, H.; Lu, Y.; Wu, T.; Ji, G. The role and therapeutic implication of CPTs in fatty acid oxidation and cancers progression. Am. J. Cancer Res. 2021, 11, 2477–2494. [Google Scholar] [PubMed]
- Kim, D.Y.; Kim, J.M. Multi-omics integration strategies for animal epigenetic studies—A review. Anim. Biosci. 2021, 34, 1271–1282. [Google Scholar] [CrossRef] [PubMed]
- Spiegel, S.; Merrill, A.H., Jr. Sphingolipid metabolism and cell growth regulation. FASEB J. 1996, 10, 1388–1397. [Google Scholar] [CrossRef] [PubMed]
- Murase, T.; Aoki, M.; Wakisaka, T.; Hase, T.; Tokimitsu, I. Anti-obesity effect of dietary diacylglycerol in C57BL/6J mice: Dietary diacylglycerol stimulates intestinal lipid metabolism. J. Lipid Res. 2002, 43, 1312–1319. [Google Scholar] [CrossRef]
- Li, H.; Wang, H.; Yu, L.; Wang, M.; Liu, S.; Sun, L.; Chen, Q. Effects of supplementation of rumen-protected choline on growth performance, meat quality traits and gene expression in longissimus dorsi muscle of lambs. Arch. Anim. Nutr. 2015, 69, 340–350. [Google Scholar] [CrossRef]
- Hollenbeck, C.B. An introduction to the nutrition and metabolism of choline. Cent. Nerv. Syst. Agents Med. Chem. 2012, 12, 100–113. [Google Scholar] [CrossRef] [PubMed]
- Bricotte, L.; Chougrani, K.; Alard, V.; Ladmiral, V.; Caillol, S. Dihydroxyacetone: A User Guide for a Challenging Bio-Based Synthon. Molecules 2023, 28, 2724. [Google Scholar] [CrossRef] [PubMed]





| Metabolites | Class | FC | VIP | p Value |
|---|---|---|---|---|
| Acylcarnitine 9:0 | Fatty acyls | 0.16 | 3.48 | 0.01 |
| Caffeoylcholine | 0.23 | 3.19 | 0.01 | |
| Acylcarnitine 20:4 | Fatty acyls | 0.22 | 2.66 | 0.01 |
| Dihydroxyacetone phosphate | Organooxygen compounds | 0.24 | 2.76 | 0.01 |
| 2,6-Dihydroxybenzoic acid | Benzene and substituted derivatives | 0.35 | 2.47 | 0.01 |
| 2-Piperidinone | Piperidines | 0.44 | 2.24 | 0.01 |
| Gallic acid | Benzene and substituted derivatives | 0.24 | 2.67 | 0.01 |
| L-Glutathione, reduced | Carboxylic acids and derivatives | 0.55 | 1.76 | 0.01 |
| Linoleoylcarnitine | Fatty acyls | 0.27 | 2.46 | 0.01 |
| Acylcarnitine 18:3 | Fatty acyls | 0.20 | 2.84 | 0.01 |
| N-Undecanoylglycine | Carboxylic acids and derivatives | 0.48 | 1.61 | 0.01 |
| 4-Hydroxyquinoline | Quinolines and derivatives | 0.18 | 2.85 | 0.01 |
| 3-Methylglutarylcarnitine | Fatty acyls | 0.34 | 2.13 | 0.01 |
| Adenosine 5′-monophosphate | Purine nucleotides | 0.54 | 1.76 | 0.01 |
| D-Mannose-6-phosphate | Organooxygen compounds | 0.30 | 2.08 | 0.01 |
| Lauroyl-L-carnitine | Fatty acyls | 0.38 | 2.19 | 0.01 |
| D-Mannose-6-phosphate | Organooxygen compounds | 0.39 | 1.85 | 0.01 |
| (2R)-3-Hydroxyisovaleroylcarnitine | Fatty acyls | 0.62 | 1.66 | 0.01 |
| (Z)-14-Methyl-6-pentadecenoic acid | Purine nucleosides | 0.55 | 1.42 | 0.01 |
| Clofentezine | 0.63 | 1.70 | 0.01 | |
| N-Acetylhistamine | Carboxylic acids and derivatives | 0.45 | 1.50 | 0.01 |
| Eremopetasinorol | Alcohols and polyols | 0.56 | 1.39 | 0.01 |
| L-Carnosine | Peptidomimetics | 0.58 | 1.77 | 0.01 |
| 2-Hydroxy-2-methylbutyric acid | Fatty acyls | 0.52 | 1.53 | 0.01 |
| 2-Fluoromethamphetamine | 0.39 | 1.96 | 0.01 | |
| Dehydro-beta-Ionone | Prenol lipids | 0.62 | 1.12 | 0.01 |
| Xanthine | Carboxylic acids and derivatives | 0.61 | 1.56 | 0.01 |
| D-2-Phosphoglyceric acid | 0.14 | 2.05 | 0.01 | |
| Succinic acid | Prenol lipids | 0.41 | 1.69 | 0.01 |
| Metabolites | Class | FC | VIP | p Value |
|---|---|---|---|---|
| Taurodeoxycholic acid | Carboxylic acids and derivatives | 2.27 | 1.59 | 0.03 |
| Choline | Fatty acyls | 1.75 | 1.51 | 0.01 |
| 1,2,5,6-Tetrahydro-4H-pyrrolo [3,2,1-ij] quinolin-4-one | Fatty acyls | 2.18 | 1.78 | 0.01 |
| Prolyl-Valine | Purine nucleotides | 1.63 | 1.62 | 0.01 |
| Methylglutaric acid | Fatty acyls | 2.24 | 2.06 | 0.01 |
| 5,8,11-Eicosatrienoic acid | Fatty acyls | 2.12 | 2.23 | 0.01 |
| 8Z,11Z-eicosadienoic acid | Fatty acyls | 2.04 | 2.07 | 0.01 |
| Threonine | Carboxylic acids and derivatives | 2.08 | 2.03 | 0.01 |
| L-Propionylcarnitine | Fatty acyls | 3.72 | 2.20 | 0.01 |
| LysoPC 20:3 | Glycerophospholipids | 2.50 | 2.44 | 0.01 |
| Groups | Kegg Pathway | Metabolites | FC | p Value | VIP | Regulate |
|---|---|---|---|---|---|---|
| 42 and 126 days old | Glycerophospholipid metabolism (map00564) | LysoPC 16:0 | 0.57 | 0.02 | 1.34 | down |
| LysoPC 18:2 | 2.32 | 0.00 | 2.33 | up | ||
| LysoPC 20:4 | 0.49 | 0.00 | 2.29 | down | ||
| PG 36:4; PG(18:2/18:2) | 1.86 | 0.00 | 2.08 | up | ||
| PG 36:3; PG(18:1/18:2) | 1.61 | 0.00 | 1.73 | up | ||
| PI 38:4; PI(18:0/20:4) | 0.56 | 0.04 | 1.12 | down | ||
| Choline | 1.75 | 0.01 | 1.51 | up | ||
| LysoPC 22:4 | 0.47 | 0.01 | 1.73 | down | ||
| Longevity-regulating pathway (map04211) | AMP | 1.78 | 0.04 | 1.44 | up | |
| beta-Nicotinamide adenine dinucleotide | 4.57 | 0.00 | 3.20 | up | ||
| NADH | 4.39 | 0.00 | 3.09 | up | ||
| 42 and 180 days old | Glycerophospholipid metabolism (map00564) | Dihydroxyacetone phosphate | 0.24 | 0.00 | 2.76 | down |
| LysoPS 18:1 | 0.41 | 0.02 | 1.40 | down | ||
| LysoPI 20:3 | 3.00 | 0.00 | 2.58 | up | ||
| PG 36:4; PG(18:2/18:2) | 1.87 | 0.00 | 1.83 | up | ||
| PG 36:3; PG(18:1/18:2) | 1.67 | 0.00 | 1.62 | up | ||
| LysoPE 18:2 | 1.57 | 0.02 | 1.66 | up | ||
| LysoPC 18:2 | 3.34 | 0.00 | 2.93 | up | ||
| LysoPC 20:3 | 2.50 | 0.00 | 2.44 | up | ||
| beta-Alanine metabolism (map00410) | L-Carnosine | 0.53 | 0.03 | 1.24 | down | |
| Pantothenic acid | 0.64 | 0.02 | 1.12 | down | ||
| L-Anserine | 0.63 | 0.00 | 1.35 | down | ||
| 126 and 180 days old | Oxidative phosphorylation (map00190) | Succinic acid | 0.41 | 0.04 | 1.69 | down |
| beta-Nicotinamide adenine dinucleotide | 0.29 | 0.00 | 2.62 | down | ||
| NADH | 0.18 | 0.00 | 3.36 | down | ||
| Longevity-regulating pathway (map04212) | AMP | 0.44 | 0.00 | 2.09 | down | |
| beta-Nicotinamide adenine dinucleotide | 0.29 | 0.00 | 2.62 | down |
| Metabolite | Groups | Regulate | Kegg Pathway | Genes |
|---|---|---|---|---|
| LysoPC 18:2 | 42 and 126 days old | up | Glycerophospholipid metabolism | CHRNG, ENSGALG00010006533, HYDIN, CAMK2A, ENSGALG00010012015, ENSGALG00010022550, C17orf58 |
| PG 36:4; PG(18:2/18:2) | up | ENSGALG00010016390 | ||
| LysoPC 22:4 | CD3E, IL7R, RASSF5, ENSGALG00010025331, TARP | |||
| LysoPC 20:4 | down | ENSGALG00010025331, TARP, IL7R, CD3E, RASSF5 | ||
| LysoPC 18:2 | 42 and 180 days old | up | Glycerophospholipid metabolism | ENSGALG00010009589, HACD1, GAMT, HSPB7, ENSGALG00010009112 |
| L-Carnosine | down | beta-Alanine metabolism | GLRB, BLEC2 | |
| L-Anserine | down | ENSGALG00010007664, ENSGALG00010006904 | ||
| Dihydroxyacetone phosphate | down | HSPB7 | ||
| NADH | 126 and 180 days old | down | Oxidative phosphorylation | ENSGALG00010022328, TAL2, PCSK1, SGSM1, HYDIN, ENSGALG00010014657, KLHL30, TGFB3, CSRP3 |
| beta-Nicotinamide adenine dinucleotide | down | PIK3C2B |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, J.; Zhao, W.; Zhao, J.; Tian, J.; Yang, L.; Wang, H.; Chen, S.; Ma, R.; Gu, Y.; Wei, D.; et al. Metabolomics and Transcriptomics Reveal Age-Dependent Development of Meat Quality Traits in Jingyuan Chicken. Animals 2025, 15, 1938. https://doi.org/10.3390/ani15131938
Hu J, Zhao W, Zhao J, Tian J, Yang L, Wang H, Chen S, Ma R, Gu Y, Wei D, et al. Metabolomics and Transcriptomics Reveal Age-Dependent Development of Meat Quality Traits in Jingyuan Chicken. Animals. 2025; 15(13):1938. https://doi.org/10.3390/ani15131938
Chicago/Turabian StyleHu, Jiahuan, Wei Zhao, Jinyan Zhao, Jinli Tian, Lijuan Yang, Hua Wang, Siyu Chen, Ruimin Ma, Yaling Gu, Dawei Wei, and et al. 2025. "Metabolomics and Transcriptomics Reveal Age-Dependent Development of Meat Quality Traits in Jingyuan Chicken" Animals 15, no. 13: 1938. https://doi.org/10.3390/ani15131938
APA StyleHu, J., Zhao, W., Zhao, J., Tian, J., Yang, L., Wang, H., Chen, S., Ma, R., Gu, Y., Wei, D., & Zhang, J. (2025). Metabolomics and Transcriptomics Reveal Age-Dependent Development of Meat Quality Traits in Jingyuan Chicken. Animals, 15(13), 1938. https://doi.org/10.3390/ani15131938







