ASIP, AHCY and ITCH Genes Are Associated with the Coat Color of Local Goats (Capra hircus) of Southwestern China
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data and Quality Control
2.2. Genome-Wide Association Study
2.3. Haplotype Analysis
2.4. Gene Function Annotation and Pathway Analysis
3. Results
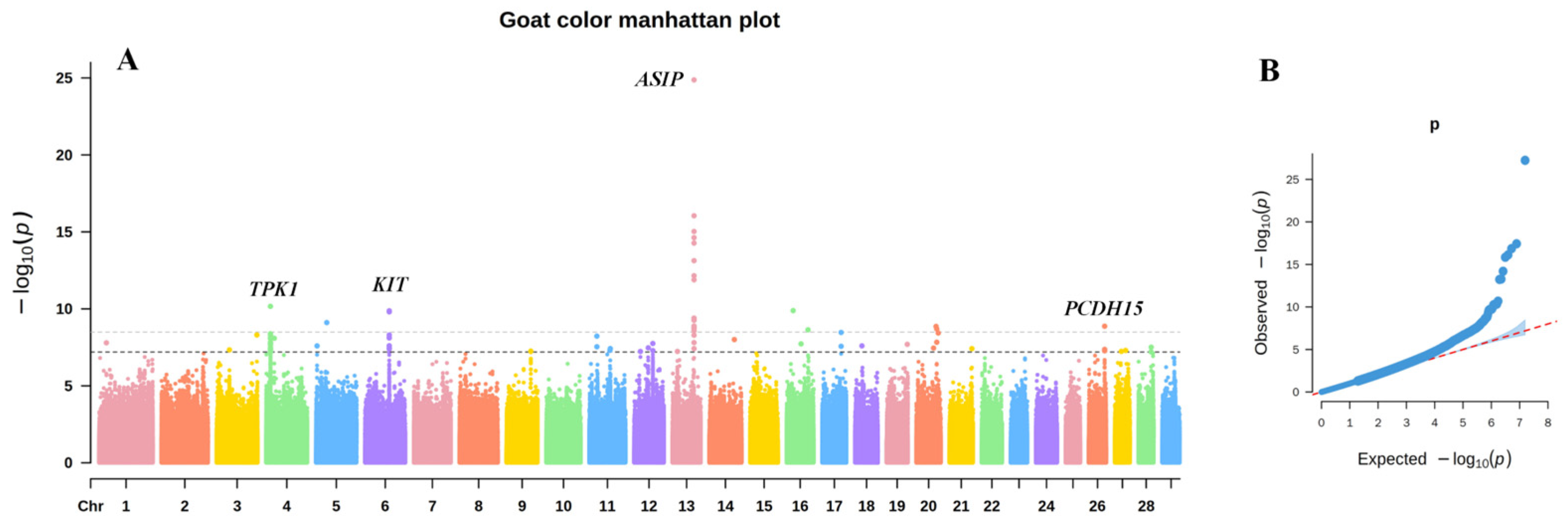
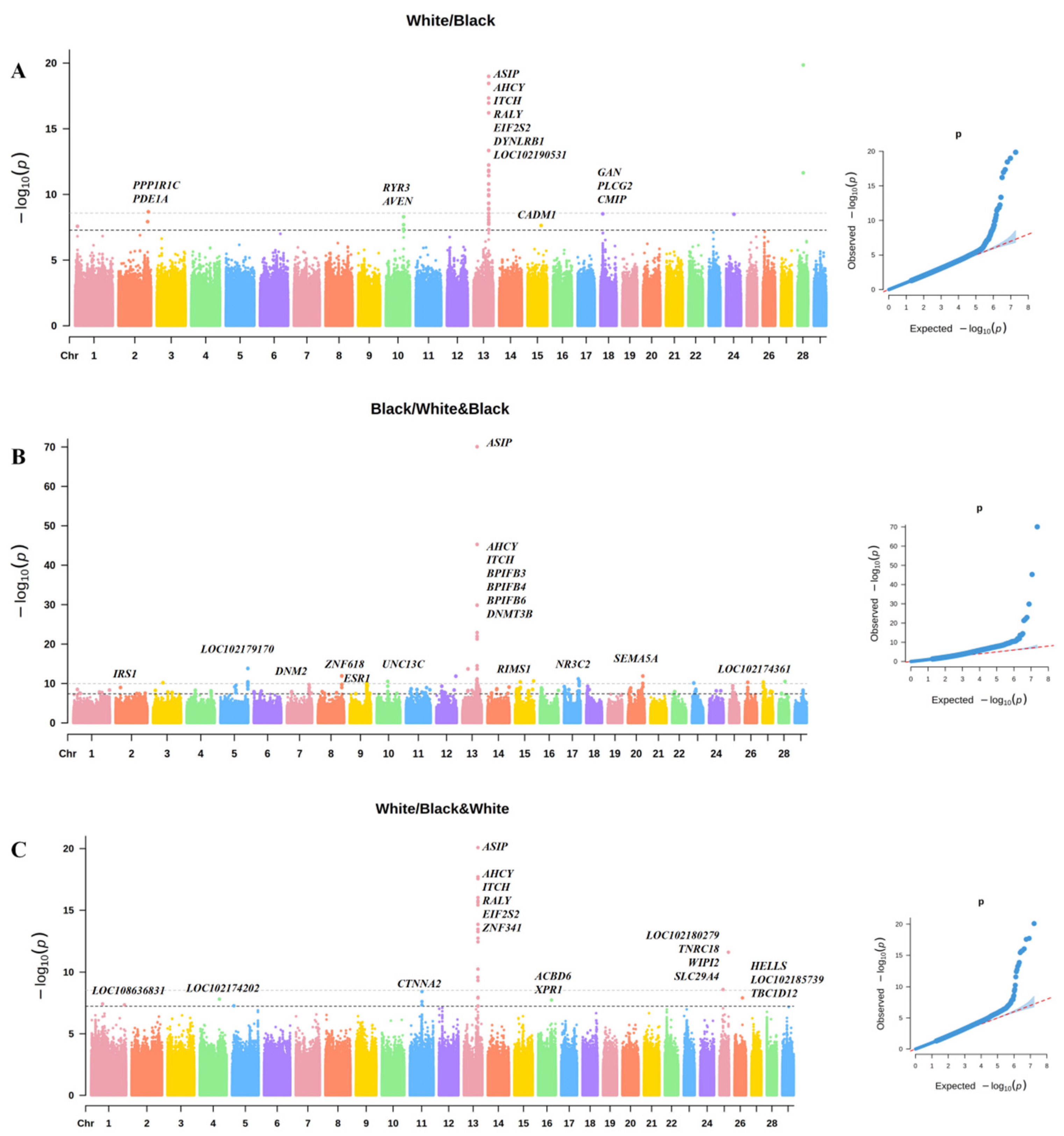
3.1. GWAS of Coat Color in Chongqing Local Goats
3.2. Haplotype Analysis of Coat Color Significant SNP Region
3.3. Results of Gene Function Annotation and Pathway Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cieslak, M.; Reissmann, M.; Hofreiter, M.; Ludwig, A. Colours of domestication. Biol. Rev. Camb. Philos. Soc. 2011, 86, 885–899. [Google Scholar] [CrossRef] [PubMed]
- Bhat, B.; Singh, A.; Iqbal, Z.; Kaushik, J.; Rao, A.; Ahmad, S.; Bhat, H.; Ayaz, A.; Sheikh, F.; Kalra, S.; et al. Comparative transcriptome analysis reveals the genetic basis of coat color variation in Pashmina goat. Sci. Rep. 2019, 9, 6361. [Google Scholar] [CrossRef] [PubMed]
- Henkel, J.; Saif, R.; Jagannathan, V.; Schmocker, C.; Zeindler, F.; Bangerter, E.; Herren, U.; Posantzis, D.; Bulut, Z.; Ammann, P.; et al. Selection signatures in goats reveal copy number variants underlying breed-defining coat color phenotypes. PLoS Genet. 2019, 15, e1008536. [Google Scholar] [CrossRef]
- Arenas-Báez, P.; Torres-Hernández, G.; Castillo-Hernández, G.; Hernández-Rodríguez, M.; Sánchez-Gutiérrez, R.; Vargas-Lόpez, S.; González-Maldonado, J.; Domínguez-Martínez, P.; Granados-Rivera, L.; Maldonado-Jáquez, J. Coat Color in Local Goats: Influence on Environmental Adaptation and Productivity, and Use as a Selection Criterion. Biology 2023, 12, 929. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Zhao, W.; Ma, Q.; Wang, J.; Zhao, Z.; Zhang, J.; Gu, Y. Genome-Wide Association Study of Birth Wool Length, Birth Weight, and Head Color in Chinese Tan Sheep Through Whole-Genome Re-Sequencing. Animals 2024, 14, 3495. [Google Scholar] [CrossRef] [PubMed]
- Bush, W.; Moore, J. Chapter 11: Genome-wide association studies. PLoS Comput. Biol. 2012, 8, e1002822. [Google Scholar] [CrossRef]
- Pavan, S.; Delvento, C.; Ricciardi, L.; Lotti, C.; Ciani, E.; D’Agostino, N. Recommendations for Choosing the Genotyping Method and Best Practices for Quality Control in Crop Genome-Wide Association Studies. Front. Genet. 2020, 11, 447. [Google Scholar] [CrossRef]
- Zhang, L.; Duan, Y.; Zhao, S.; Yu, H.; Zhang, J.; Xu, N.; Zhao, Y. Genome-wide association studies reveal novel loci associated with carcass and body measures in goats (Capra hircus). bioRxiv 2025. [Google Scholar] [CrossRef]
- Chang, C.; Chow, C.; Tellier, L.; Vattikuti, S.; Purcell, S.; Lee, J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. GigaScience 2015, 4, 7. [Google Scholar] [CrossRef]
- Zhou, X.; Matthew, S. Efficient multivariate linear mixed model algorithms for genome-wide association studies. Nat. Methods 2014, 11, 407–409. [Google Scholar] [CrossRef]
- Stephen, D.T. qqman: An R package for visualizing GWAS results using Q-Q and manhattan plots. J. Open Source Softw. 2018, 3, 731. [Google Scholar] [CrossRef]
- Yin, L. CMplot: Circle Manhattan Plot. R Package Version 4.5.0. 2023. Available online: https://CRAN.R-project.org/package=CMplot (accessed on 12 September 2024).
- Browning, B.L.; Tian, X.; Zhou, Y.; Browning, S.R. Fast two-stage phasing of large-scale sequence data. Am. J. Hum. Genet. 2021, 108, 1880–1890. [Google Scholar] [CrossRef]
- Paradis, E. pegas: An R package for population genetics with an integrated-modular approach. Bioinformatics 2010, 26, 419–420. [Google Scholar] [CrossRef] [PubMed]
- Paradis, E.; Schliep, K. ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 2019, 35, 526–528. [Google Scholar] [CrossRef]
- Bandelt, H.; Forster, P.; Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- Peng, Y.; Wang, Y.; Zhou, C.; Mei, W.; Zeng, C. PI3K/Akt/mTOR Pathway and Its Role in Cancer Therapeutics: Are We Making Headway? Front. Oncol. 2022, 12, 819128. [Google Scholar] [CrossRef]
- Videira, I.F.; Moura, D.F.; Magina, S. Mechanisms regulating melanogenesis. An. Bras. Dermatol. 2013, 88, 76–83. [Google Scholar] [CrossRef]
- Slominski, A.; Tobin, D.J.; Shibahara, S.; Wortsman, J. Melanin pigmentation in mammalian skin and its hormonal regulation. Physiol. Rev. 2004, 84, 1155–1228. [Google Scholar] [CrossRef] [PubMed]
- Hearing, V.J. Determination of melanin synthetic pathways. J. Investig. Dermatol. 2011, 131, E8–E11. [Google Scholar] [CrossRef]
- Tarricone, S.; Schlosserová, N.; Bruno, S.; Sardina, M.T.; Landi, V.; Giannico, F.; Colonna, M.A.; Sarti, F.M.; Lasagna, E.; Ceccobelli, S.; et al. Selection Signatures in Italian Goat Populations Sharing the “facciuto” Phenotype. Genes 2025, 16, 390. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, M.N.; Raudsepp, T.; Alshanbari, F.; Gutiérrez, G.; Ponce, F.A. Chromosomal Localization of Candidate Genes for Fiber Growth and Color in Alpaca (Vicugna pacos). Front. Genet. 2019, 10, 583. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Lyu, Y.; Zhang, D.; Reddi, K.K.; Sun, F.; Yi, J.; Liu, C.; Li, H.; Yao, H.; Dai, J.; et al. Genomic Characteristics and Selection Signatures in Indigenous Chongming White Goat (Capra hircus). Front. Genet. 2020, 11, 901. [Google Scholar] [CrossRef] [PubMed]
- Wan, X.; Jing, J.N.; Wang, D.F.; Lv, F.H. Whole-genome selective scans detect genes associated with important phenotypic traits in goat (Capra hircus). Front. Genet. 2023, 14, 1173017. [Google Scholar] [CrossRef]
- Wu, D.; Fan, J.; Pang, Y.; Wen, B.; Li, W.; Yang, G.; Cheng, H.; Shi, J.; Wang, T.; Hu, S.; et al. Identification and Expression Patterns of Critical Genes Related to Coat Color in Cashmere Goats. Genes 2025, 16, 222. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Zhao, S.; Yu, H.; Duan, Y.; Zhang, J.; Xu, N.; Zhao, Y. ASIP, AHCY and ITCH Genes Are Associated with the Coat Color of Local Goats (Capra hircus) of Southwestern China. Animals 2025, 15, 1849. https://doi.org/10.3390/ani15131849
Zhang L, Zhao S, Yu H, Duan Y, Zhang J, Xu N, Zhao Y. ASIP, AHCY and ITCH Genes Are Associated with the Coat Color of Local Goats (Capra hircus) of Southwestern China. Animals. 2025; 15(13):1849. https://doi.org/10.3390/ani15131849
Chicago/Turabian StyleZhang, Linyun, Shengnan Zhao, Houmo Yu, Yixin Duan, Jipan Zhang, Naiyi Xu, and Yongju Zhao. 2025. "ASIP, AHCY and ITCH Genes Are Associated with the Coat Color of Local Goats (Capra hircus) of Southwestern China" Animals 15, no. 13: 1849. https://doi.org/10.3390/ani15131849
APA StyleZhang, L., Zhao, S., Yu, H., Duan, Y., Zhang, J., Xu, N., & Zhao, Y. (2025). ASIP, AHCY and ITCH Genes Are Associated with the Coat Color of Local Goats (Capra hircus) of Southwestern China. Animals, 15(13), 1849. https://doi.org/10.3390/ani15131849







