Two Cases of Chromosome 27 Trisomy in Horses Detected Using Illumina BeadChip Genotyping
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Genotype Data
2.2. Genotype Intensity Data
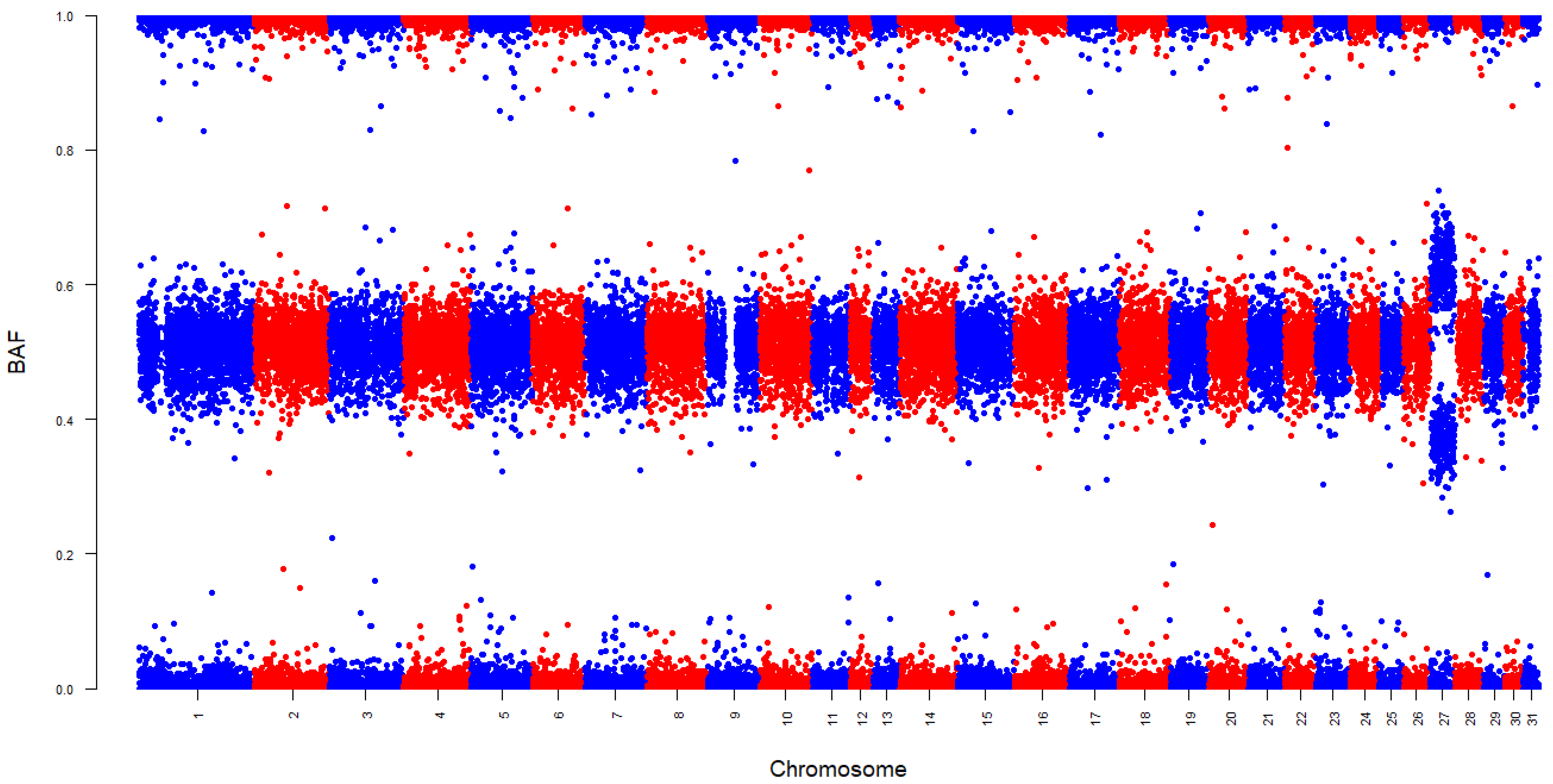
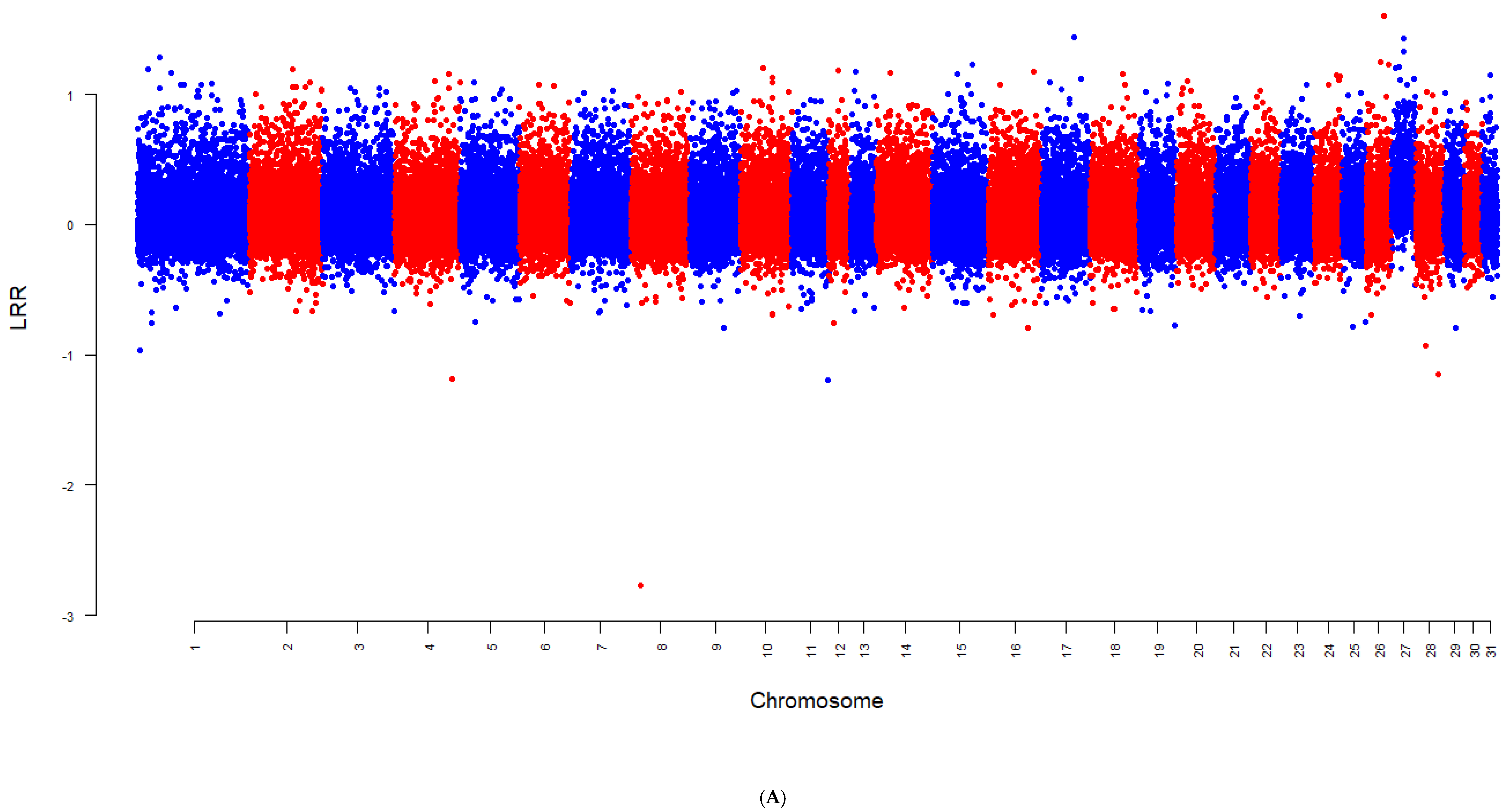
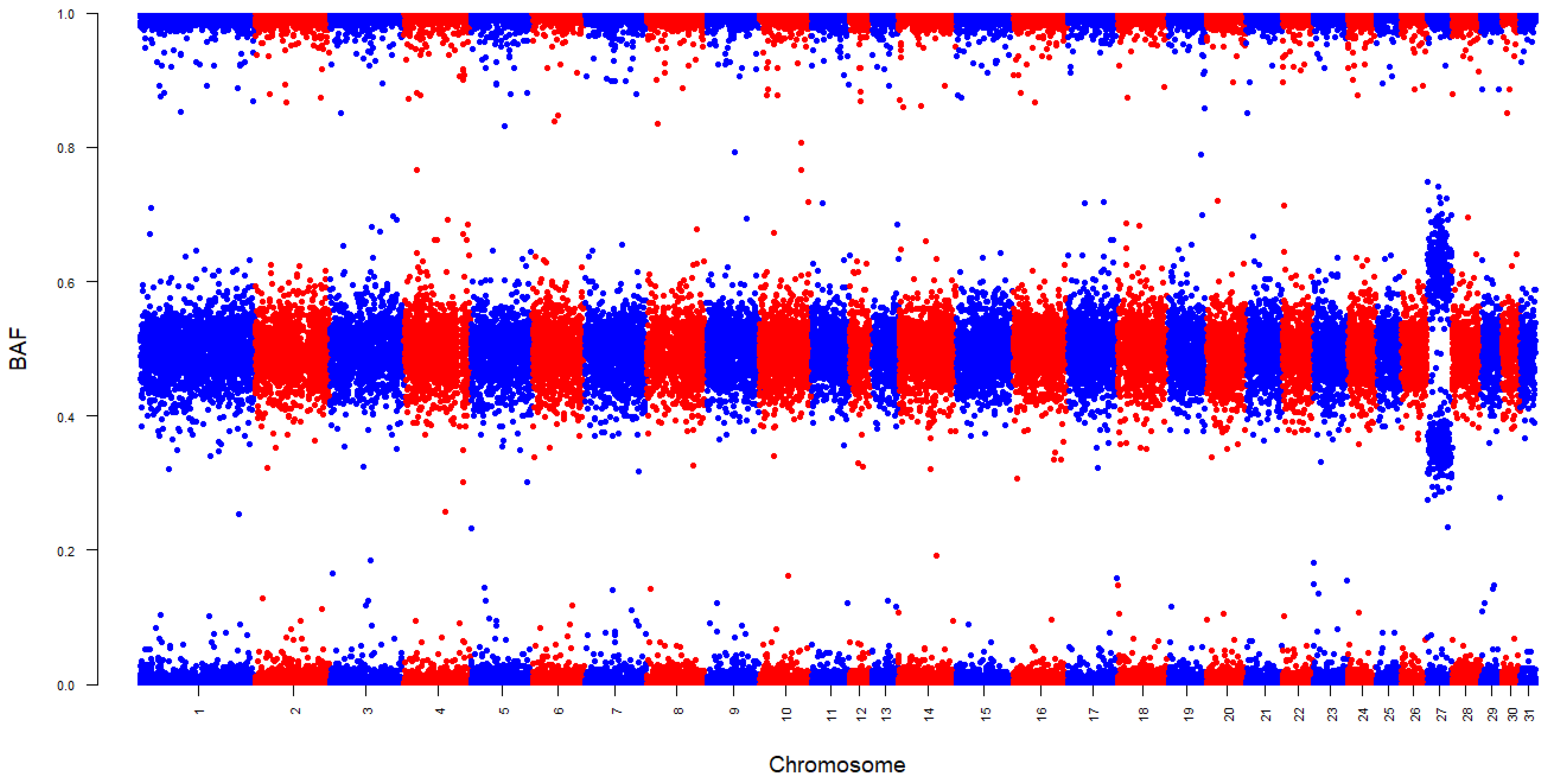
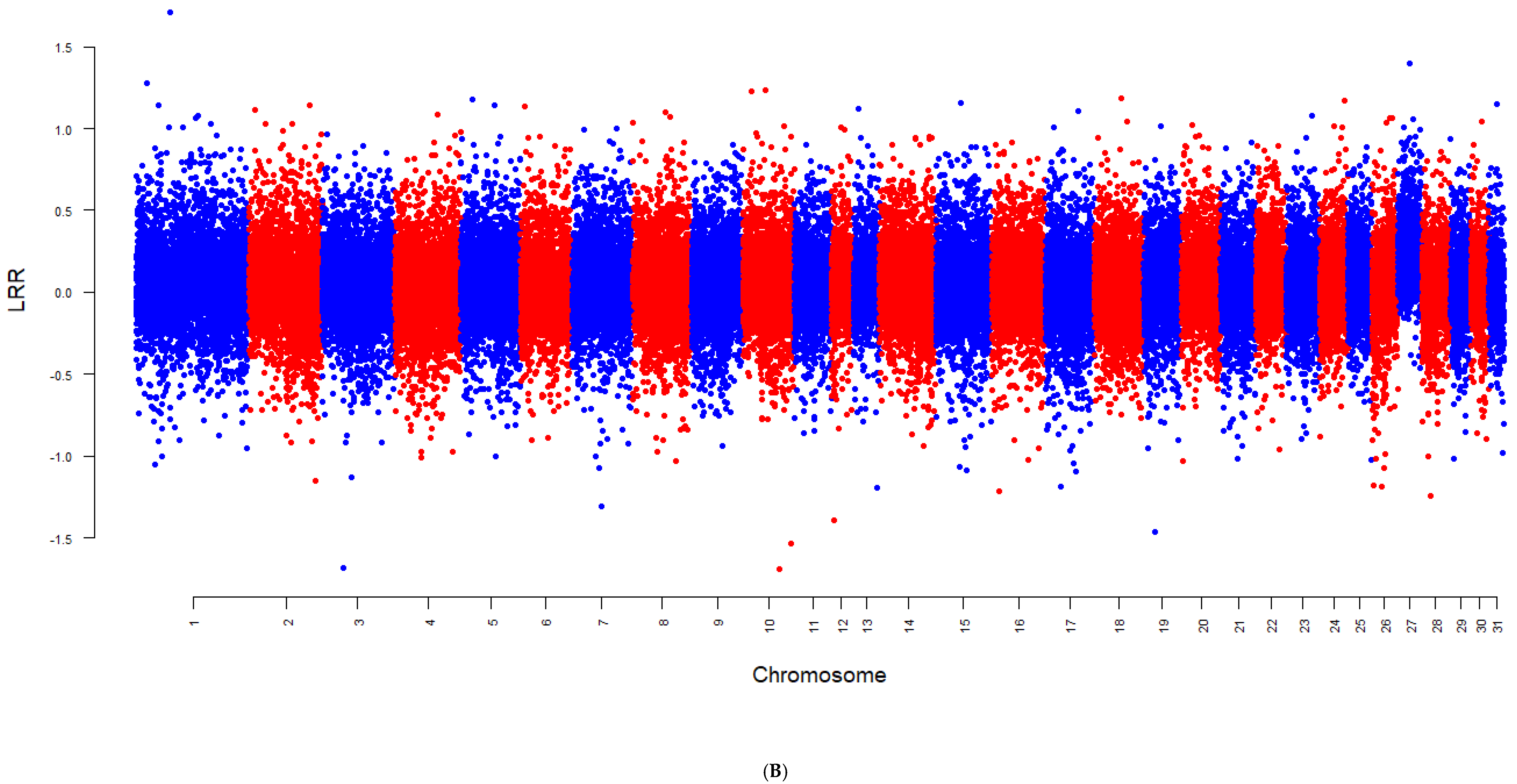
2.3. Detecting Aneuploidy
2.4. Karyotyping
2.5. Fluorescence In Situ Hybridization (FISH)
3. Results
3.1. Parental Origin
3.2. Phenotypic Assessment
4. Discussion
4.1. Prevalence of Aneuploidy
4.2. Parental Origin of Additional Chromosome
4.3. Chromosome-Specific Aneuploidy
4.4. Phenotypic Symptoms
4.5. Reduced Life Expectancy
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| SNP | Single Nucleotide Polymorphism |
| ISH | Irish Sport Horse |
| chr27 | Chromosome 27 |
| LRR | Log R Ratio |
| EDTA | Ethylenediaminetetraacetic Acid |
| BAC | Bacterial Artificial Chromosome |
| ISCNH | International System for Cytogenetic Nomenclature of the Horse |
References
- Bugno-Poniewierska, M.; Raudsepp, T. Horse Clinical Cytogenetics: Recurrent Themes and Novel Findings. Animals 2021, 11, 831. [Google Scholar] [CrossRef] [PubMed]
- Lawson, J.M.; Salem, S.E.; Miller, D.; Kahler, A.; van den Boer, W.J.; Shilton, C.A.; Sever, T.; Mouncey, R.R.; Ward, J.; Hampshire, D.J.; et al. Naturally occurring horse model of miscarriage reveals temporal relationship between chromosomal aberration type and point of lethality. Proc. Natl. Acad. Sci. USA 2024, 121, e2405636121. [Google Scholar] [CrossRef] [PubMed]
- Hassold, T.; Hunt, P. To err (meiotically) is human: The genesis of human aneuploidy. Nat. Rev. Genet. 2001, 2, 280–291. [Google Scholar] [CrossRef] [PubMed]
- Torres, E.M.; Williams, B.R.; Amon, A. Aneuploidy: Cells losing their balance. Genetics 2008, 179, 737–746. [Google Scholar] [CrossRef] [PubMed]
- Pai, G.S.; Lewandowski, R.C.; Borgaonkar, D.S. Handbook of Chromosomal Syndromes; Wiley-Liss: Hoboken, NJ, USA, 2003. [Google Scholar]
- Ryan, C.; Purfield, D.; Matthews, D.; Rathje, C.; Valldecabres, A.; Berry, D. Prevalence of Autosomal Monosomy and Trisomy Estimated Using Single Nucleotide Polymorphism Genotype Intensity Chip Information in a Large Population of Juvenile Dairy and Beef Cattle. J. Anim. Breed. Genet. 2024, 142, 277–286. [Google Scholar] [CrossRef]
- Klunder, L.R.; McFeely, R.A.; Beech, J.; McClune, W. Autosomal trisomy in a Standardbred colt. Equine Vet. J. 1989, 21, 69–70. [Google Scholar] [CrossRef]
- Bowling, A.T.; Millon, L.V. Two autosomal trisomies in the horse: 64,XX,−26,+t(26q26q) and 65,XX,+30. Genome 1990, 33, 679–682. [Google Scholar] [CrossRef]
- Brito, L.F.; Sertich, P.L.; Durkin, K.; Chowdhary, B.P.; Turner, R.M.; Greene, L.M.; McDonnell, S. Autosomic 27 trisomy in a standardbred colt. J. Equine Vet. Sci. 2008, 28, 431–436. [Google Scholar] [CrossRef]
- Power, M.M. Equine half sibs with an unbalanced X;15 translocation or trisomy 28. Cytogenet. Cell Genet. 1987, 45, 163–168. [Google Scholar] [CrossRef]
- Johannisson, R.; Gropp, A.; Winking, H.; Coerdt, W.; Rehder, H.; Schwinger, E. Down’s syndrome in the male. Reproductive pathology and meiotic studies. Hum. Genet. 1983, 63, 132–138. [Google Scholar] [CrossRef]
- Bugno, M.; Słota, E.; Kościelny, M. Karyotype evaluation among young horse populations in Poland. Schweiz. Arch. Tierheilkd. 2007, 149, 227–232. [Google Scholar] [CrossRef]
- Ryan, C.A.; Purfield, D.C.; Matthews, D.; Canedo-Ribeiro, C.; Valldecabres, A.; Berry, D.P. Prevalence of sex-chromosome aneuploidy estimated using SNP genotype intensity information in a large population of juvenile dairy and beef cattle. J. Anim. Breed. Genet. 2024, 141, 571–585. [Google Scholar] [CrossRef] [PubMed]
- Berry, D.P.; Wolfe, A.; O’Donovan, J.; Byrne, N.; Sayers, R.G.; Dodds, K.G.; McEwan, J.C.; O’Connor, R.E.; McClure, M.; Purfield, D.C. Characterization of an X-chromosomal non-mosaic monosomy (59, X0) dairy heifer detected using routinely available single nucleotide polymorphism genotype data. J. Anim. Sci. 2017, 95, 1042–1049. [Google Scholar] [CrossRef] [PubMed]
- Berry, D.P.; O’Brien, A.; O’Donovan, J.; McHugh, N.; Wall, E.; Randles, S.; McDermott, K.; O’Connor, R.E.; Patil, M.A.; Ho, J.; et al. Aneuploidy in dizygotic twin sheep detected using genome-wide single nucleotide polymorphism data from two commonly used commercial vendors. Animal 2018, 12, 2462–2469. [Google Scholar] [CrossRef]
- Treff, N.R.; Su, J.; Tao, X.; Levy, B.; Scott Jr, R.T. Accurate single cell 24 chromosome aneuploidy screening using whole genome amplification and single nucleotide polymorphism microarrays. Fertil. Steril. 2010, 94, 2017–2021. [Google Scholar] [CrossRef] [PubMed]
- Tuke, M.A.; Ruth, K.S.; Wood, A.R.; Beaumont, R.N.; Tyrrell, J.; Jones, S.E.; Yaghootkar, H.; Turner, C.L.; Donohoe, M.E.; Brooke, A.M. Phenotypes associated with female X chromosome aneuploidy in UK Biobank: An unselected, adult, population-based cohort. bioRxiv 2017. [Google Scholar] [CrossRef]
- Xiong, B.; Tan, K.; Tan, Y.Q.; Gong, F.; Zhang, S.P.; Lu, C.F.; Luo, K.L.; Lu, G.X.; Lin, G. Using SNP array to identify aneuploidy and segmental imbalance in translocation carriers. Genom. Data 2014, 2, 92–95. [Google Scholar] [CrossRef]
- Bouwman, A.C.; Hulsegge, I.; Hawken, R.J.; Henshall, J.M.; Veerkamp, R.F.; Schokker, D.; Kamphuis, C. Classifying aneuploidy in genotype intensity data using deep learning. J. Anim. Breed. Genet. 2023, 140, 304–315. [Google Scholar] [CrossRef]
- Peiffer, D.A.; Le, J.M.; Steemers, F.J.; Chang, W.; Jenniges, T.; Garcia, F.; Haden, K.; Li, J.; Shaw, C.A.; Belmont, J. High-resolution genomic profiling of chromosomal aberrations using Infinium whole-genome genotyping. Genome Res. 2006, 16, 1136–1148. [Google Scholar] [CrossRef]
- Staaf, J.; Vallon-Christersson, J.; Lindgren, D.; Juliusson, G.; Rosenquist, R.; Höglund, M.; Borg, Å.; Ringnér, M. Normalization of Illumina Infinium whole-genome SNP data improves copy number estimates and allelic intensity ratios. BMC Bioinform. 2008, 9, 409. [Google Scholar] [CrossRef]
- Hashem, E.M.; Mabrouk, M.S.; Eldeib, A.M. Novel altered region for biomarker discovery in hepatocellular carcinoma (HCC) using whole genome SNP array. Int. J. Adv. Comput. Sci. Appl. 2016, 7. [Google Scholar] [CrossRef]
- Wang, H.; Federoff, S. Trypsin technique to reveal G-bands. In Tissue Culture Methods and Applications; Academic Press: New York, NY, USA, 1974; pp. 782–787. [Google Scholar]
- Cribiu, E.P.; Di Berardino, D.; Di Meo, G.P.; Eggen, A.; Gallagher, D.S.; Gustavsson, I.; Hayes, H.; Iannuzzi, L.; Popescu, C.P.; Rubes, J.; et al. International System for Chromosome Nomenclature of Domestic Bovids (ISCNDB 2000). Cytogenet. Cell Genet. 2001, 92, 283–299. [Google Scholar] [CrossRef] [PubMed]
- Bugno-Poniewierska, M.; Jankowska, M.; Raudsepp, T.; Kowalska, K.; Pawlina-Tyszko, K.; Szmatola, T. Molecular cytogenetic screening of sex chromosome abnormalities in young horse populations. Equine Vet. J. 2024, 56, 786–795. [Google Scholar] [CrossRef] [PubMed]
- Raudsepp, T.; Chowdhary, B.P. FISH for mapping single copy genes. Phylogenomics 2008, 422, 31–49. [Google Scholar]
- Hook, E.B. Exclusion of chromosomal mosaicism: Tables of 90%, 95% and 99% confidence limits and comments on use. Am. J. Hum. Genet. 1977, 29, 94. [Google Scholar]
- Weatherbys. Weatherbys Fact Book; Weatherbys GSB: Wellingborough, UK, 2023. [Google Scholar]
- Berry, D.; Spangler, M.L. Animal board invited review: Practical applications of genomic information in livestock. Animal 2023, 17, 100996. [Google Scholar] [CrossRef]
- Descartes, M.; Korf, B.R.; Mikhail, F.M. 35-Chromosomes and Chromosomal Abnormalities. In Swaiman’s Pediatric Neurology, 6th ed.; Swaiman, K.F., Ashwal, S., Ferriero, D.M., Schor, N.F., Finkel, R.S., Gropman, A.L., Pearl, P.L., Shevell, M.I., Eds.; Elsevier: Amsterdam, The Netherlands, 2017; pp. 268–276. [Google Scholar]
- Shilton, C.A.; Kahler, A.; Davis, B.W.; Crabtree, J.R.; Crowhurst, J.; McGladdery, A.J.; Wathes, D.C.; Raudsepp, T.; de Mestre, A.M. Whole genome analysis reveals aneuploidies in early pregnancy loss in the horse. Sci. Rep. 2020, 10, 13314. [Google Scholar] [CrossRef]
- Silvestri, G.; Canedo-Ribeiro, C.; Serrano-Albal, M.; Labrecque, R.; Blondin, P.; Larmer, S.G.; Marras, G.; Tutt, D.A.R.; Handyside, A.H.; Farré, M.; et al. Preimplantation Genetic Testing for Aneuploidy Improves Live Birth Rates with In Vitro Produced Bovine Embryos: A Blind Retrospective Study. Cells 2021, 10, 2284. [Google Scholar] [CrossRef]
- Hassold, T.; Hall, H.; Hunt, P. The origin of human aneuploidy: Where we have been, where we are going. Hum. Mol. Genet. 2007, 16, R203–R208. [Google Scholar] [CrossRef]
- Switonski, M.; Szczerbal, I.; Nowacka-Woszuk, J. From cytogenetics to cytogenomics: A new era in the diagnosis of chromosomal abnormalities in domestic animals. J. Appl. Genet. 2025, 54, 1–13. [Google Scholar] [CrossRef]
- Ghosh, S.; Kjöllerström, J.; Metcalfe, L.; Reed, S.; Juras, R.; Raudsepp, T. The Second Case of Non-Mosaic Trisomy of Chromosome 26 with Homologous Fusion 26q;26q in the Horse. Animals 2022, 12, 803. [Google Scholar] [CrossRef] [PubMed]
- Lear, T.L.; Cox, J.H.; Kennedy, G.A. Autosomal trisomy in a Thoroughbred colt: 65,XY,+31. Equine Vet. J. 1999, 31, 85–88. [Google Scholar] [CrossRef] [PubMed]
- Buoen, L.C.; Zhang, T.Q.; Weber, A.F.; Turner, T.; Bellamy, J.; Ruth, G.R. Arthrogryposis in the foal and its possible relation to autosomal trisomy. Equine Vet. J. 1997, 29, 60–62. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Bellamy, J.; Buwn, L.; Weber, A.; Ruth, G. Autosomal trisomy in a foal with contracted tendon syndrome. In Proceedings of the 10th European Colloqium on Cytogenetics of Domestic Animals, Utrecht, The Netherlands, 18–21 August 1992. [Google Scholar]
- Rizzo, M.; du Preez, N.; Ducheyne, K.D.; Deelen, C.; Beitsma, M.M.; Stout, T.A.E.; de Ruijter-Villani, M. The horse as a natural model to study reproductive aging-induced aneuploidy and weakened centromeric cohesion in oocytes. Aging 2020, 12, 22220–22232. [Google Scholar] [CrossRef]
- Holečková, B.; Schwarzbacherová, V.; Galdíková, M.; Koleničová, S.; Halušková, J.; Staničová, J.; Verebová, V.; Jutková, A. Chromosomal Aberrations in Cattle. Genes 2021, 12, 1330. [Google Scholar] [CrossRef]
- Gersen, S.L.; Keagle, M.B. The Principles of Clinical Cytogenetics; Springer Nature: Dordrecht, The Netherlands, 2013. [Google Scholar] [CrossRef]
- Damas, J.; Corbo, M.; Kim, J.; Turner-Maier, J.; Farré, M.; Larkin, D.M.; Ryder, O.A.; Steiner, C.; Houck, M.L.; Hall, S.; et al. Evolution of the ancestral mammalian karyotype and syntenic regions. Proc. Natl. Acad. Sci. USA 2022, 119, e2209139119. [Google Scholar] [CrossRef]
- Ducos, A.; Seguela, A.; Pinton, A.; Berland, H.; Brun-Baronnat, C.; Darre, R.; Manesse, M.; Darre, A. Trisomy 26 mosaicism in a sterile Holstein-Friesian heifer. Vet. Rec. 2000, 146, 163–164. [Google Scholar] [CrossRef]
- Agerholm, J.; Christensen, K. Trisomy 22 in a calf. J. Vet. Med. Ser. A 1993, 40, 576–581. [Google Scholar] [CrossRef]
- Mäkinen, A.; Alitalo, I.; Alanko, M. Autosomal trisomy in a heifer. Acta Vet. Scand. 1987, 28, 1–8. [Google Scholar] [CrossRef]
- Holl, H.M.; Lear, T.L.; Nolen-Walston, R.D.; Slack, J.; Brooks, S.A. Detection of two equine trisomies using SNP-CGH. Mamm. Genome 2013, 24, 252–256. [Google Scholar] [CrossRef]
- Petersen, M.B.; Mikkelsen, M. Nondisjunction in trisomy 21: Origin and mechanisms. Cytogenet. Cell Genet. 2000, 91, 199–203. [Google Scholar] [CrossRef] [PubMed]
- Raudsepp, T.; Finno, C.J.; Bellone, R.R.; Petersen, J.L. Ten years of the horse reference genome: Insights into equine biology, domestication and population dynamics in the post-genome era. Anim. Genet. 2019, 50, 569–597. [Google Scholar] [CrossRef] [PubMed]




| Colt ID | Number of SNPs with Opposing Parental Homozygotes | % of Alleles Dam | % of Alleles from Sire | Inferred Parent of Origin of Extra Chromosome |
|---|---|---|---|---|
| ISH1 | 87 | 66.3% | 33.3% | Extra chromosome inherited from the dam |
| ISH2 | 102 | 33.7% | 66.0% | Extra chromosome inherited from the sire |
| Colt ID | Number of AB SNPs in Parent of Origin | Heterozygous Genotypes in the Colt | Homozygous Genotypes in the Colt | Interpretation |
|---|---|---|---|---|
| ISH1 | 240 | 240 | 0 | Heterodisomy |
| ISH2 | 262 | 242 | 20 | Heterodisomy |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ryan, C.A.; Berry, D.P.; Bugno-Poniewierska, M.; Burke, M.-K.; Raudsepp, T.; Egan, S.; Doyle, J.L. Two Cases of Chromosome 27 Trisomy in Horses Detected Using Illumina BeadChip Genotyping. Animals 2025, 15, 1842. https://doi.org/10.3390/ani15131842
Ryan CA, Berry DP, Bugno-Poniewierska M, Burke M-K, Raudsepp T, Egan S, Doyle JL. Two Cases of Chromosome 27 Trisomy in Horses Detected Using Illumina BeadChip Genotyping. Animals. 2025; 15(13):1842. https://doi.org/10.3390/ani15131842
Chicago/Turabian StyleRyan, Cliona A., Donagh P. Berry, Monika Bugno-Poniewierska, Mary-Kate Burke, Terje Raudsepp, Sonja Egan, and Jennifer L. Doyle. 2025. "Two Cases of Chromosome 27 Trisomy in Horses Detected Using Illumina BeadChip Genotyping" Animals 15, no. 13: 1842. https://doi.org/10.3390/ani15131842
APA StyleRyan, C. A., Berry, D. P., Bugno-Poniewierska, M., Burke, M.-K., Raudsepp, T., Egan, S., & Doyle, J. L. (2025). Two Cases of Chromosome 27 Trisomy in Horses Detected Using Illumina BeadChip Genotyping. Animals, 15(13), 1842. https://doi.org/10.3390/ani15131842






