Whole-Genome Resequencing Analysis of Copy Number Variations Associated with Athletic Performance in Grassland-Thoroughbred
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Library Construction and Sequencing
2.3. Data Filtering and Sequence Alignment
2.4. Phylogenetic Analysis
2.5. CNV and CNVR Detection
2.6. Genomic Selection Signals Based on CNVRs
2.7. RNA-Seq Library Construction
2.8. Quality Control and Alignment of RNA Sequencing Data
2.9. Functional Enrichment Analysis and QTL Association Analysis
2.10. PCR Validation
3. Results
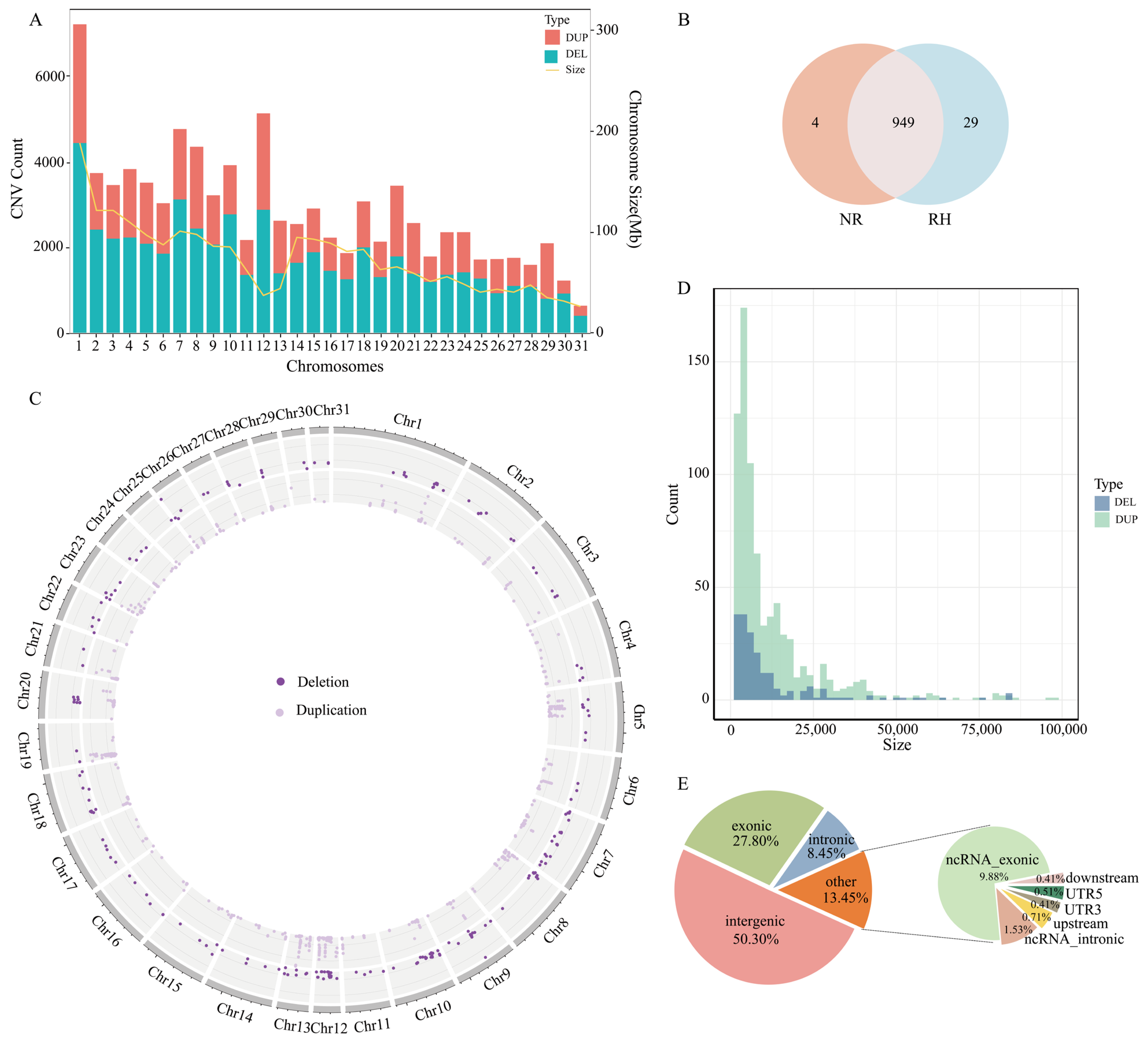
3.1. CNV Detection
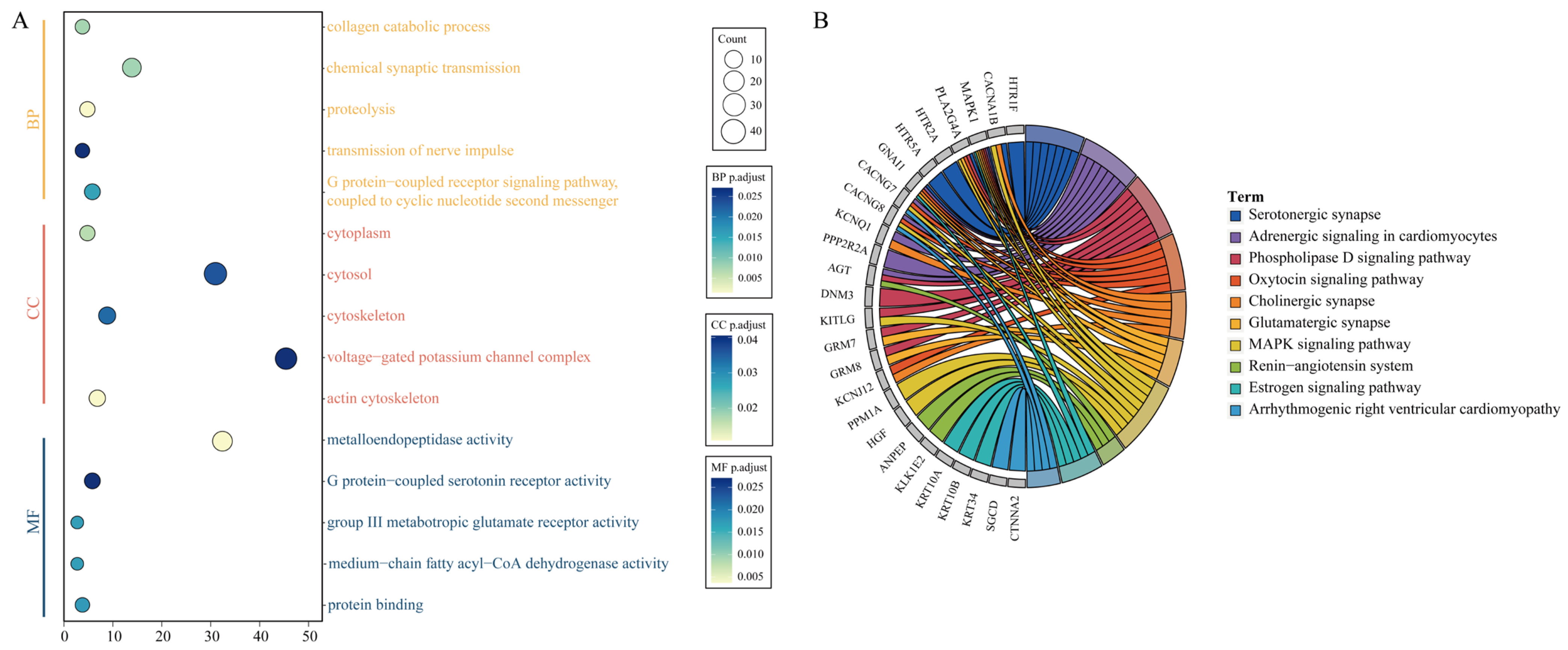
3.2. CNVR Functional Enrichment
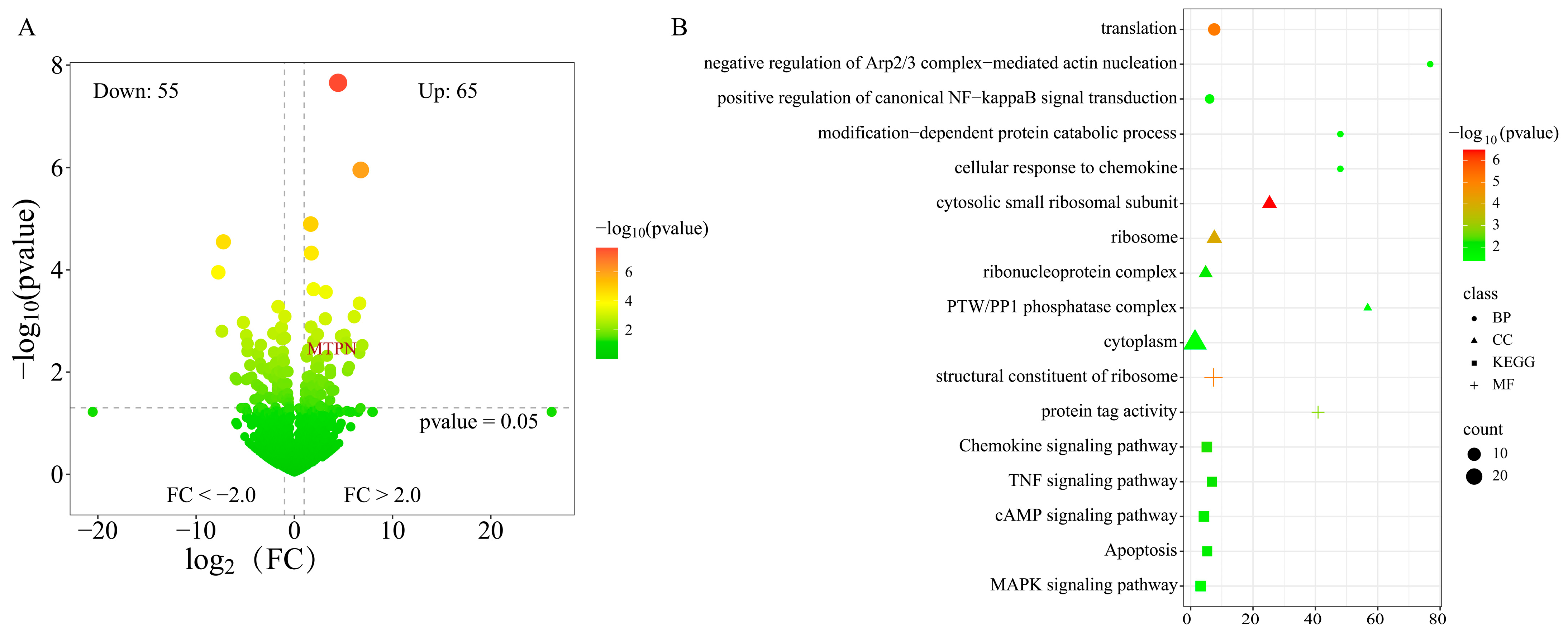
3.3. The Differential Expression Analysis of mRNA
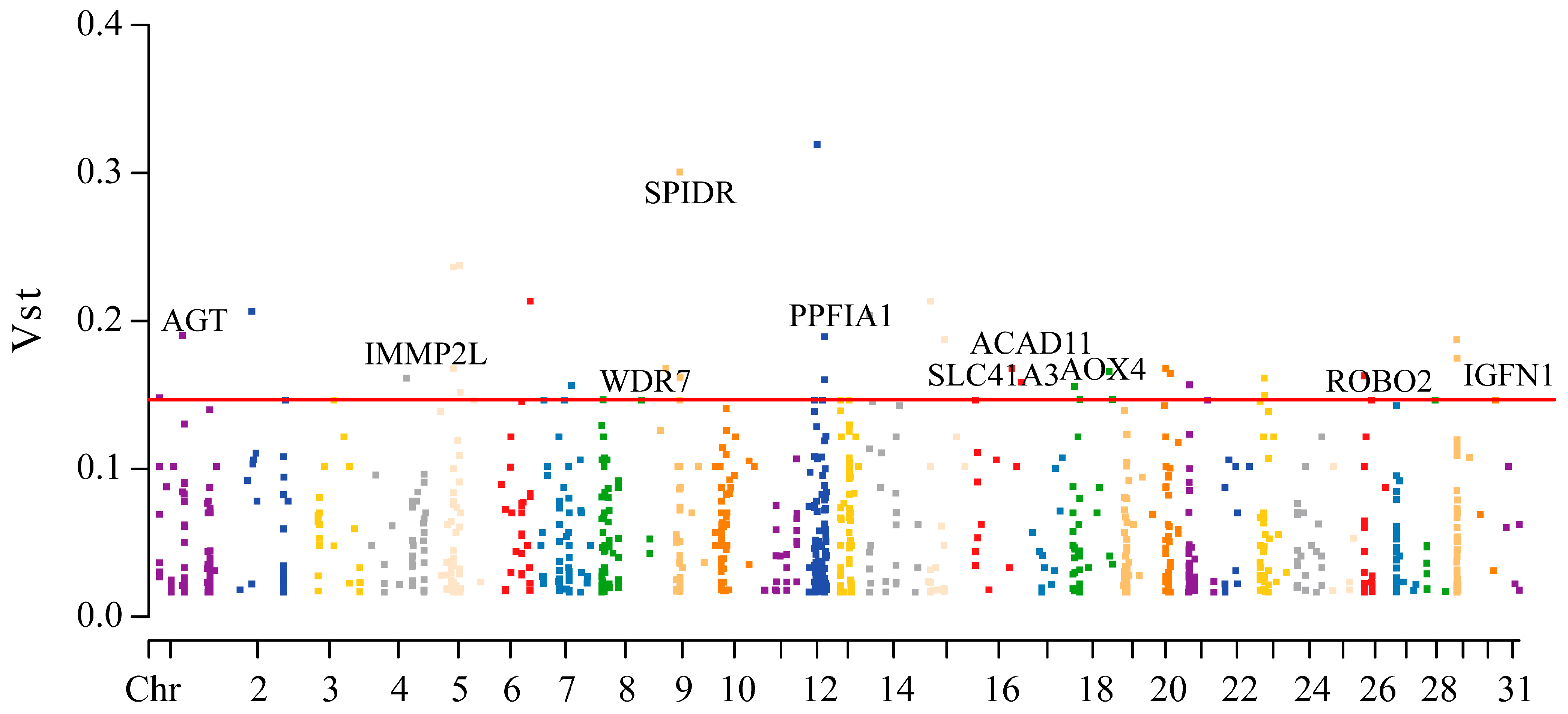
3.4. CNVR-Based Population Differentiation
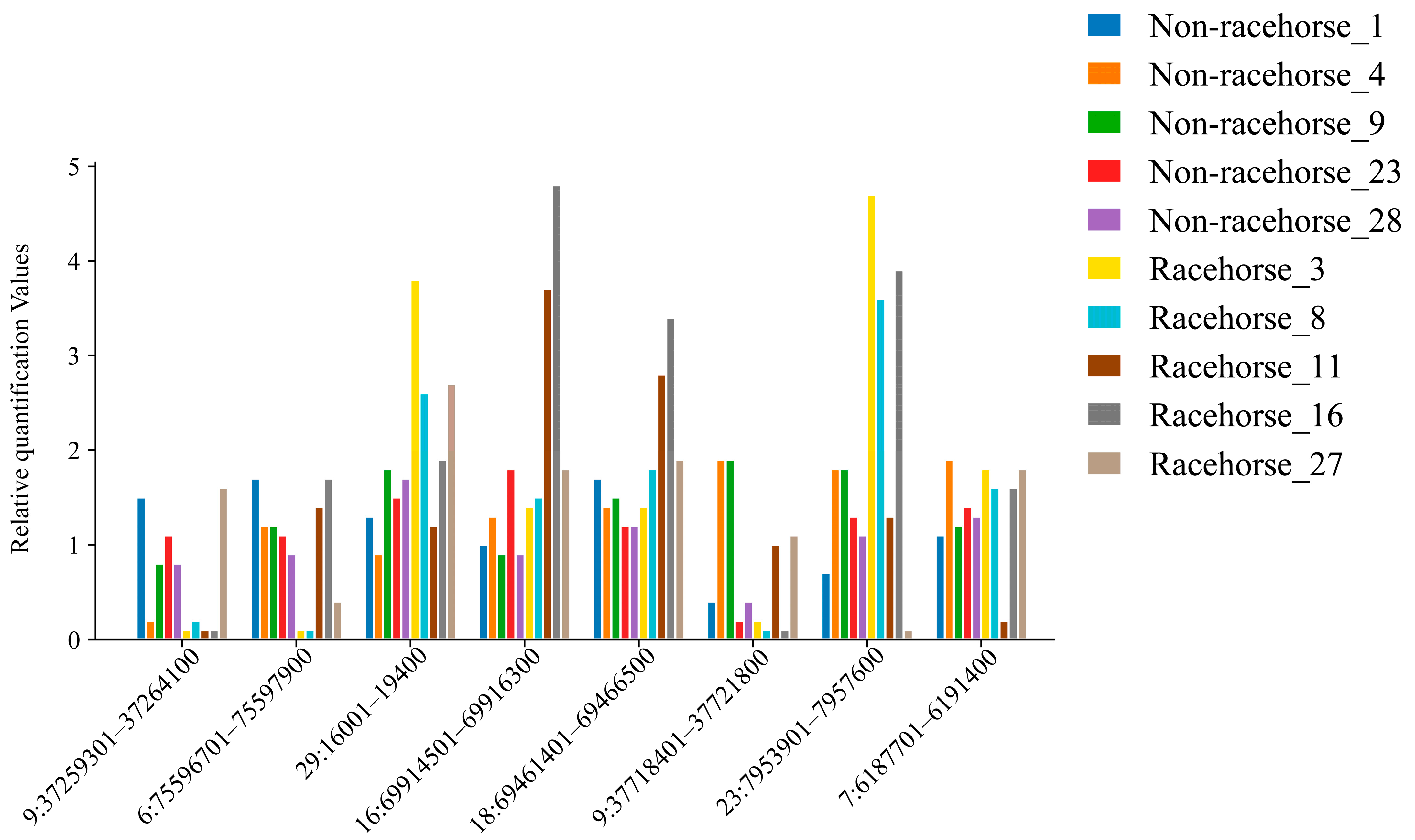
3.5. qPCR Validation of CNVRs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chien, P.M. Tourism and Hospitality in the Contemporary World: Trends, Changes & Complexity Proceedings of the 24th Annual Conference CAUTHE 2014. In Proceedings of the CAUTHE Conference 2014, Brisbane, Australia, 10–13 February 2014. [Google Scholar]
- Worthington, A.C. National exuberance: A note on the Melbourne Cup effect in Australian stock returns. Econ. Pap. A J. Appl. Econ. Policy 2007, 26, 170–179. [Google Scholar] [CrossRef]
- Narayan, P.K.; Smyth, R. The race that stops a nation: The demand for the Melbourne Cup. Econ. Rec. 2004, 80, 193–207. [Google Scholar] [CrossRef]
- Mostafavi, A.; Fozi, M.A.; Koshkooieh, A.E.; Mohammadabadi, M.; Babenko, O.I.; Klopenko, N.I. Effect of LCORL gene polymorphism on body size traits in horse populations. Acta Scientiarum. Anim. Sci. 2019, 42, e47483. [Google Scholar] [CrossRef]
- Davis, M. When Things Get Dark: A Mongolian Winter’s Tale; Macmillan: Tuggerah, Australia, 2010. [Google Scholar]
- Qi, B. Xilin Gol League Animal Husbandry Chronicle; Inner Mongolia People’s Publishing House: Hohhot, China, 2002. [Google Scholar]
- Zhang, F.; Gu, W.; Hurles, M.E.; Lupski, J.R. Copy number variation in human health, disease, and evolution. Annu. Rev. Genom. Hum. Genet. 2009, 10, 451–481. [Google Scholar] [CrossRef]
- Stranger, B.E.; Forrest, M.S.; Dunning, M.; Ingle, C.E.; Beazley, C.; Thorne, N.; Redon, R.; Bird, C.P.; De Grassi, A.; Lee, C. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science 2007, 315, 848–853. [Google Scholar] [CrossRef]
- Conrad, D.F.; Pinto, D.; Redon, R.; Feuk, L.; Gokcumen, O.; Zhang, Y.; Aerts, J.; Andrews, T.D.; Barnes, C.; Campbell, P. Origins and functional impact of copy number variation in the human genome. Nature 2010, 464, 704–712. [Google Scholar] [CrossRef]
- Scherer, S.W.; Lee, C.; Birney, E.; Altshuler, D.M.; Eichler, E.E.; Carter, N.P.; Hurles, M.E.; Feuk, L. Challenges and standards in integrating surveys of structural variation. Nat. Genet. 2007, 39, S7–S15. [Google Scholar] [CrossRef]
- Freeman, J.L.; Perry, G.H.; Feuk, L.; Redon, R.; McCarroll, S.A.; Altshuler, D.M.; Aburatani, H.; Jones, K.W.; Tyler-Smith, C.; Hurles, M.E.; et al. Copy number variation: New insights in genome diversity. Genome Res. 2006, 16, 949–961. [Google Scholar] [CrossRef]
- Sebat, J.; Lakshmi, B.; Troge, J.; Alexander, J.; Young, J.; Lundin, P.; Manér, S.; Massa, H.; Walker, M.; Chi, M. Large-scale copy number polymorphism in the human genome. Science 2004, 305, 525–528. [Google Scholar] [CrossRef]
- Clop, A.; Vidal, O.; Amills, M. Copy number variation in the genomes of domestic animals. Anim. Genet. 2012, 43, 503–517. [Google Scholar] [CrossRef]
- Conrad, D.F.; Bird, C.; Blackburne, B.; Lindsay, S.; Mamanova, L.; Lee, C.; Turner, D.J.; Hurles, M.E. Mutation spectrum revealed by breakpoint sequencing of human germline CNVs. Nat. Genet. 2010, 42, 385–391. [Google Scholar] [CrossRef] [PubMed]
- Stankiewicz, P.; Lupski, J.R. Structural variation in the human genome and its role in disease. Annu. Rev. Med. 2010, 61, 437–455. [Google Scholar] [CrossRef] [PubMed]
- Mazuet, C.; Legeay, C.; Sautereau, J.; Bouchier, C.; Criscuolo, A.; Bouvet, P.; Trehard, H.; Jourdan Da Silva, N.; Popoff, M. Characterization of Clostridium Baratii Type F Strains Responsible for an Outbreak of Botulism Linked to Beef Meat Consumption in France. PLoS Curr. 2017, 9. [Google Scholar]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Wingett, S.W.; Andrews, S. FastQ Screen: A tool for multi-genome mapping and quality control. F1000Res 2018, 7, 1338. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Subgroup, G.P.D.P. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Bathke, J.; Lühken, G. OVarFlow: A resource optimized GATK 4 based Open source Variant calling workFlow. BMC Bioinform. 2021, 22, 402. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.; Daly, M.J. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Abyzov, A.; Urban, A.E.; Snyder, M.; Gerstein, M. CNVnator: An approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 2011, 21, 974–984. [Google Scholar] [CrossRef]
- Pierce, M.D.; Dzama, K.; Muchadeyi, F.C. Genetic diversity of seven cattle breeds inferred using copy number variations. Front. Genet. 2018, 9, 163. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Kovaka, S.; Zimin, A.V.; Pertea, G.M.; Razaghi, R.; Salzberg, S.L.; Pertea, M. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 2019, 20, 278. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef]
- Aleman, M.; Nieto, J.E. Gene expression of proteolytic systems and growth regulators of skeletal muscle in horses with myopathy associated with pituitary pars intermedia dysfunction. Am. J. Vet. Res. 2010, 71, 664–670. [Google Scholar] [CrossRef]
- Doan, R.; Cohen, N.; Harrington, J.; Veazy, K.; Juras, R.; Cothran, G.; McCue, M.E.; Skow, L.; Dindot, S.V. Identification of copy number variants in horses. Genome Res. 2012, 22, 899–907. [Google Scholar] [CrossRef][Green Version]
- Ghosh, S.; Das, P.; McQueen, C.; Gerber, V.; Swiderski, C.; Lavoie, J.P.; Chowdhary, B.; Raudsepp, T. Analysis of genomic copy number variation in equine recurrent airway obstruction (heaves). Anim. Genet. 2016, 47, 334–344. [Google Scholar] [CrossRef]
- Wang, W.; Wang, S.; Hou, C.; Xing, Y.; Cao, J.; Wu, K.; Liu, C.; Zhang, D.; Zhang, L.; Zhang, Y. Genome-wide detection of copy number variations among diverse horse breeds by array CGH. PLoS ONE 2014, 9, e86860. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Solé, M.; Ablondi, M.; Binzer-Panchal, A.; Velie, B.D.; Hollfelder, N.; Buys, N.; Ducro, B.J.; François, L.; Janssens, S.; Schurink, A. Inter-and intra-breed genome-wide copy number diversity in a large cohort of European equine breeds. BMC Genom. 2019, 20, 1–12. [Google Scholar] [CrossRef]
- Tang, X.; Zhu, B.; Ren, R.; Chen, B.; Li, S.; Gu, J. Genome-wide copy number variation detection in a large cohort of diverse horse breeds by whole-genome sequencing. Front. Vet. Sci. 2023, 10, 1296213. [Google Scholar] [CrossRef]
- Schurink, A.; da Silva, V.H.; Velie, B.D.; Dibbits, B.W.; Crooijmans, R.P.; Franҫois, L.; Janssens, S.; Stinckens, A.; Blott, S.; Buys, N. Copy number variations in Friesian horses and genetic risk factors for insect bite hypersensitivity. BMC Genet. 2018, 19, 49. [Google Scholar] [CrossRef]
- Dupuis, M.-C.; Zhang, Z.; Durkin, K.; Charlier, C.; Lekeux, P.; Georges, M. Detection of copy number variants in the horse genome and examination of their association with recurrent laryngeal neuropathy. Anim. Genet. 2013, 44, 206–208. [Google Scholar] [CrossRef] [PubMed]
- Pawlina-Tyszko, K.; Gurgul, A.; Szmatoła, T.; Ropka-Molik, K.; Semik-Gurgul, E.; Klukowska-Rötzler, J.; Koch, C.; Mählmann, K.; Bugno-Poniewierska, M. Genomic landscape of copy number variation and copy neutral loss of heterozygosity events in equine sarcoids reveals increased instability of the sarcoid genome. Biochimie 2017, 140, 122–132. [Google Scholar] [CrossRef]
- Kader, A.; Liu, X.; Dong, K.; Song, S.; Pan, J.; Yang, M.; Chen, X.; He, X.; Jiang, L.; Ma, Y. Identification of copy number variations in three Chinese horse breeds using 70K single nucleotide polymorphism BeadChip array. Anim. Genet. 2016, 47, 560–569. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Qu, Z.; Das, P.J.; Fang, E.; Juras, R.; Cothran, E.G.; McDonell, S.; Kenney, D.G.; Lear, T.L.; Adelson, D.L. Copy number variation in the horse genome. PLoS Genet. 2014, 10, e1004712. [Google Scholar] [CrossRef]
- Wang, M.; Liu, Y.; Bi, X.; Ma, H.; Zeng, G.; Guo, J.; Guo, M.; Ling, Y.; Zhao, C. Genome-wide detection of copy number variants in Chinese indigenous horse breeds and verification of CNV-overlapped genes related to heat adaptation of the Jinjiang horse. Genes. 2022, 13, 603. [Google Scholar] [CrossRef]
- Metzger, J.; Philipp, U.; Lopes, M.S.; da Camara Machado, A.; Felicetti, M.; Silvestrelli, M.; Distl, O. Analysis of copy number variants by three detection algorithms and their association with body size in horses. BMC Genom. 2013, 14, 487. [Google Scholar] [CrossRef]
- Shiraishi, S.; Nakamura, Y.-N.; Iwamoto, H.; Haruno, A.; Sato, Y.; Mori, S.; Ikeuchi, Y.; Chikushi, J.; Hayashi, T.; Sato, M. S-myotrophin promotes the hypertrophy of skeletal muscle of mice in vivo. Int. J. Biochem. Cell Biol. 2006, 38, 1114–1122. [Google Scholar] [CrossRef] [PubMed]
- Anderson, K.M.; Berrebi-Bertrand, I.; Kirkpatrick, R.B.; McQueney, M.S.; Underwood, D.C.; Rouanet, S.; Chabot-Fletcher, M. cDNA sequence and characterization of the gene that encodes human myotrophin/V-1 protein, a mediator of cardiac hypertrophy. J. Mol. Cell. Cardiol. 1999, 31, 705–719. [Google Scholar] [CrossRef] [PubMed]
- Bordbar, F.; Jensen, J.; Du, M.; Abied, A.; Guo, W.; Xu, L.; Gao, H.; Zhang, L.; Li, J. Identification and validation of a novel candidate gene regulating net meat weight in Simmental beef cattle based on imputed next-generation sequencing. Cell Prolif. 2020, 53, e12870. [Google Scholar] [CrossRef] [PubMed]
- De Mello Costa, M.; Anderson, G.; Davies, H.; El-Hage, C.; Slocombe, R. Circulating angiotensin converting enzyme in endurance horses: Effect of exercise on blood levels and its value in predicting performance. Equine Vet. J. 2010, 42, 152–154. [Google Scholar] [CrossRef] [PubMed]
- Nader, G.A.; Esser, K.A. Intracellular signaling specificity in skeletal muscle in response to different modes of exercise. J. Appl. Physiol. 2001, 90, 1936–1942. [Google Scholar] [CrossRef]
- Widegren, U.; Jiang, X.J.; Krook, A.; Chibalin, A.V.; Björnholm, M.; Tally, M.; Roth, R.A.; Henriksson, J.; Wallberg-Henriksson, H.; Zierath, J.R. Divergent effects of exercise on metabolic and mitogenic signaling pathways in human skeletal muscle. FASEB J. 1998, 12, 1379–1389. [Google Scholar] [CrossRef]
- Yu, M.; Blomstrand, E.; Chibalin, A.V.; Krook, A.; Zierath, J.R. Marathon running increases ERK1/2 and p38 MAP kinase signalling to downstream targets in human skeletal muscle. J. Physiol. 2001, 536, 273–282. [Google Scholar] [CrossRef]
- Kramer, H.F.; Goodyear, L.J. Exercise, MAPK, and NF-κB signaling in skeletal muscle. J. Appl. Physiol. 2007, 103, 388–395. [Google Scholar] [CrossRef]
- Rauramaa, R.; Kuhanen, R.; Lakka, T.A.; Väisänen, S.B.; Halonen, P.; Alén, M.; Rankinen, T.; Bouchard, C. Physical exercise and blood pressure with reference to the angiotensinogen M235T polymorphism. Physiol. Genom. 2002, 10, 71–77. [Google Scholar] [CrossRef]
- Corvol, P.; Jeunemaitre, X. Molecular genetics of human hypertension: Role of angiotensinogen. Endocr. Rev. 1997, 18, 662–677. [Google Scholar] [CrossRef]
- Jones, A.; Woods, D.R. Skeletal muscle RAS and exercise performance. Int. J. Biochem. Cell Biol. 2003, 35, 855–866. [Google Scholar] [CrossRef] [PubMed]
- Ben-Zaken, S.; Eliakim, A.; Nemet, D.; Meckel, Y. Genetic Variability Among Power Athletes: The Stronger vs. the Faster. J. Strength. Cond. Res. 2019, 33, 1505–1511. [Google Scholar] [CrossRef] [PubMed]
- Aleksandra, Z.; Zbigniew, J.; Waldemar, M.; Agata, L.-D.; Mariusz, K.; Marek, S.; Agnieszka, M.-S.; Piotr, Ż.; Krzysztof, F.; Grzegorz, T. The AGT gene M235T polymorphism and response of power-related variables to aerobic training. J. Sports Sci. Med. 2016, 15, 616. [Google Scholar] [PubMed]
- Zarebska, A.; Sawczyn, S.; Kaczmarczyk, M.; Ficek, K.; Maciejewska-Karlowska, A.; Sawczuk, M.; LeoNska-Duniec, A.; Eider, J.; Grenda, A.; Cieszczyk, P. Association of rs699 (M235T) polymorphism in the AGT gene with power but not endurance athlete status. J. Strength. Cond. Res. 2013, 27, 2898–2903. [Google Scholar] [CrossRef]
- Perez, K.; Ciotlos, S.; McGirr, J.; Limbad, C.; Doi, R.; Nederveen, J.P.; Nilsson, M.I.; Winer, D.A.; Evans, W.; Tarnopolsky, M.; et al. Single nuclei profiling identifies cell specific markers of skeletal muscle aging, frailty, and senescence. Aging 2022, 14, 9393–9422. [Google Scholar] [CrossRef]
- Riedl, I.; Yoshioka, M.; Nishida, Y.; Tobina, T.; Paradis, R.; Shono, N.; Tanaka, H.; St-Amand, J. Regulation of skeletal muscle transcriptome in elderly men after 6 weeks of endurance training at lactate threshold intensity. Exp. Gerontol. 2010, 45, 896–903. [Google Scholar] [CrossRef]
- Li, X.; Baker, J.; Cracknell, T.; Haynes, A.R.; Blanco, G. IGFN1_v1 is required for myoblast fusion and differentiation. PLoS ONE 2017, 12, e0180217. [Google Scholar] [CrossRef]
- Kilpinen, S.; Ojala, K.; Kallioniemi, O. Analysis of kinase gene expression patterns across 5681 human tissue samples reveals functional genomic taxonomy of the kinome. PLoS ONE 2010, 5, e15068. [Google Scholar] [CrossRef]
- Lu, B.; Poirier, C.; Gaspar, T.; Gratzke, C.; Harrison, W.; Busija, D.; Matzuk, M.M.; Andersson, K.-E.; Overbeek, P.A.; Bishop, C.E. A mutation in the inner mitochondrial membrane peptidase 2-like gene (Immp2l) affects mitochondrial function and impairs fertility in mice. Biol. Reprod. 2008, 78, 601–610. [Google Scholar] [CrossRef]
- Sako, H.; Yada, K.; Suzuki, K. Genome-Wide Analysis of Acute Endurance Exercise-Induced Translational Regulation in Mouse Skeletal Muscle. PLoS ONE 2016, 11, e0148311. [Google Scholar] [CrossRef]
- Anunciado-Koza, R.P.; Zhang, J.; Ukropec, J.; Bajpeyi, S.; Koza, R.A.; Rogers, R.C.; Cefalu, W.T.; Mynatt, R.L.; Kozak, L.P. Inactivation of the mitochondrial carrier SLC25A25 (ATP-Mg2+/Pi transporter) reduces physical endurance and metabolic efficiency in mice. J. Biol. Chem. 2011, 286, 11659–11671. [Google Scholar] [CrossRef] [PubMed]
- Volpe, S.L. Magnesium in disease prevention and overall health. Adv. Nutr. 2013, 4, 378S–383S. [Google Scholar] [CrossRef] [PubMed]
- Mastrototaro, L.; Smorodchenko, A.; Aschenbach, J.R.; Kolisek, M.; Sponder, G. Solute carrier 41A3 encodes for a mitochondrial Mg(2+) efflux system. Sci. Rep. 2016, 6, 27999. [Google Scholar] [CrossRef] [PubMed]
- Williams, G.S.; Boyman, L.; Lederer, W.J. Mitochondrial calcium and the regulation of metabolism in the heart. J. Mol. Cell. Cardiol. 2015, 78, 35–45. [Google Scholar] [CrossRef]
- Fleig, A.; Schweigel-Röntgen, M.; Kolisek, M. Solute carrier family SLC41: What do we really know about it? Wiley Interdiscip. Rev. Membr. Transp. Signal. 2013, 2, 227–239. [Google Scholar] [CrossRef]
- Chiang, Y.-F.; Chen, H.-Y.; Lee, I.-T.; Chien, L.-S.; Huang, J.-H.; Kolisek, M.; Cheng, F.-C.; Tsai, S.-W. Magnesium-responsive genes are downregulated in diabetic patients after a three-month exercise program on a bicycle ergometer. J. Chin. Med. Assoc. 2019, 82, 495–499. [Google Scholar] [CrossRef]
- Terao, M.; Barzago, M.M.; Kurosaki, M.; Fratelli, M.; Bolis, M.; Borsotti, A.; Bigini, P.; Micotti, E.; Carli, M.; Invernizzi, R.W. Mouse aldehyde-oxidase-4 controls diurnal rhythms, fat deposition and locomotor activity. Sci. Rep. 2016, 6, 30343. [Google Scholar] [CrossRef]
- Slocum, N.; Durrant, J.R.; Bailey, D.; Yoon, L.; Jordan, H.; Barton, J.; Brown, R.H.; Clifton, L.; Milliken, T.; Harrington, W. Responses of brown adipose tissue to diet-induced obesity, exercise, dietary restriction and ephedrine treatment. Exp. Toxicol. Pathol. 2013, 65, 549–557. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, W.; Gong, W.; Bou, T.; Shi, L.; Lin, Y.; Shi, X.; Li, Z.; Wu, H.; Dugarjaviin, M.; Bai, D. Whole-Genome Resequencing Analysis of Copy Number Variations Associated with Athletic Performance in Grassland-Thoroughbred. Animals 2025, 15, 1458. https://doi.org/10.3390/ani15101458
Ding W, Gong W, Bou T, Shi L, Lin Y, Shi X, Li Z, Wu H, Dugarjaviin M, Bai D. Whole-Genome Resequencing Analysis of Copy Number Variations Associated with Athletic Performance in Grassland-Thoroughbred. Animals. 2025; 15(10):1458. https://doi.org/10.3390/ani15101458
Chicago/Turabian StyleDing, Wenqi, Wendian Gong, Tugeqin Bou, Lin Shi, Yanan Lin, Xiaoyuan Shi, Zheng Li, Huize Wu, Manglai Dugarjaviin, and Dongyi Bai. 2025. "Whole-Genome Resequencing Analysis of Copy Number Variations Associated with Athletic Performance in Grassland-Thoroughbred" Animals 15, no. 10: 1458. https://doi.org/10.3390/ani15101458
APA StyleDing, W., Gong, W., Bou, T., Shi, L., Lin, Y., Shi, X., Li, Z., Wu, H., Dugarjaviin, M., & Bai, D. (2025). Whole-Genome Resequencing Analysis of Copy Number Variations Associated with Athletic Performance in Grassland-Thoroughbred. Animals, 15(10), 1458. https://doi.org/10.3390/ani15101458




