Genome-Wide Microsatellite Characterization and Molecular Marker Development of Himalayan Griffon (Gyps himalayensis)
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Simple Sequence Repeats Mining in the Genome of Himalayan Griffon
2.2. Feather Sampling and DNA Extraction
2.3. SSR Primer Design
2.4. Sample Selection and PCR Amplification
2.5. Capillary Electrophoresis and Data Analysis
3. Results
3.1. Characteristic of Microsatellite Loci in the Genome of G. himalayensis
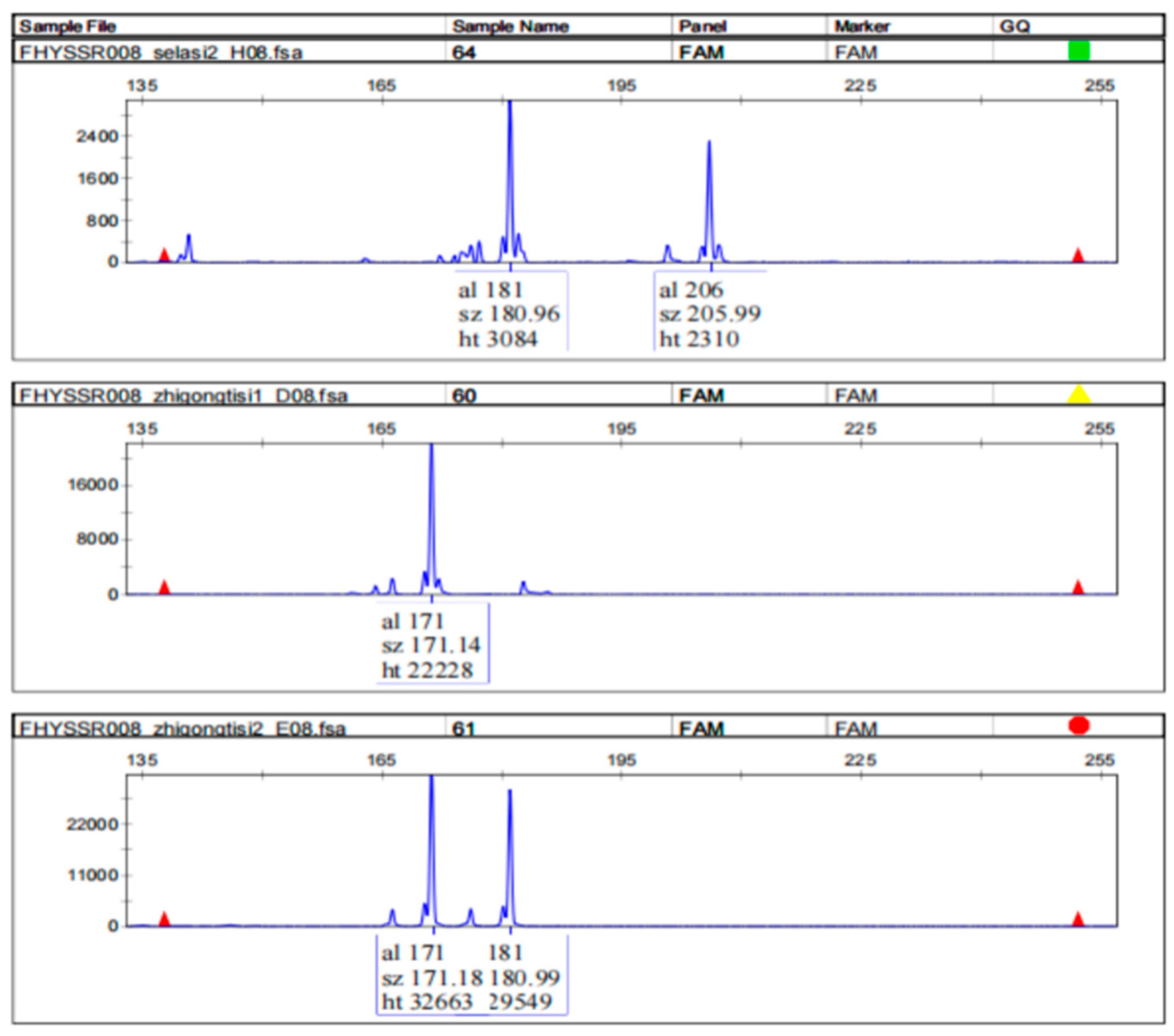
3.2. Design and Evaluation of SSR Primers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lu, X.; Ke, D.H.; Zeng, X.H.; Gong, G.H.; Ci, R. Status, ecology, and conservation of the Himalayan Griffon, G. himalayensis (Aves, Accipitridae) in the Tibetan Plateau. Ambio 2009, 38, 166–173. [Google Scholar] [CrossRef] [PubMed]
- Moleón, M.; Sánchez-Zapata, J.A.; Margalida, A.; Carrete, M.; Owen-Smith, N.; Donázar, J.A. Humans and scavengers: The evolution of interactions and ecosystem services. Bioscience 2014, 64, 394–403. [Google Scholar] [CrossRef]
- Siddique, M.; Khan, A.A. Spatio-temporal decline of Himalayan griffon (G. himalayensis Hume, 1869) in Azad Jammu and Kashmir, Pakistan. Pak. J. Zool. 2016, 48, 961–968. [Google Scholar]
- Frankham, R. Genetics and extinction. Biol. Conserv. 2005, 126, 131–140. [Google Scholar] [CrossRef]
- Bouzat, J.L. Conservation genetics of population bottlenecks: The role of chance, selection, and history. Conserv. Genet. 2010, 11, 463–478. [Google Scholar] [CrossRef]
- Schmidt, C.; Domaratzki, M.; Kinnunen, R.P.; Bowman, J.; Garroway, C.J. Continent-wide effects of urbanization on bird and mammal genetic diversity. Proc. R. Soc. B Biol. Sci. 2020, 287, 20192497. [Google Scholar] [CrossRef]
- Vieira, M.L.C.; Santini, L.; Diniz, A.L.; Munhoz, C.D. Microsatellite markers: What they mean and why they are so useful. Genet. Mol. Biol. 2016, 39, 312–328. [Google Scholar] [CrossRef]
- Kashi, Y.; King, D.G. Simple sequence repeats as advantageous mutators in evolution. Trends Genet. 2006, 22, 253–259. [Google Scholar] [CrossRef]
- Varela, M.A.; Amos, W. Heterogeneous distribution of SNPs in the human genome: Microsatellites as predictors of nucleotide diversity and divergence. Genomics 2010, 95, 151–159. [Google Scholar] [CrossRef]
- Feng, X.; Wang, Y.; Gong, X. Genetic diversity, genetic structure and demographic history of Cycas simplicipinna (Cycadaceae) assessed by DNA sequences and SSR markers. BMC Plant Biol. 2014, 14, 187. [Google Scholar] [CrossRef]
- Liu, Y.; Geng, Y.; Song, M.; Zhang, P.; Hou, J.; Wang, W. Genetic Structure and Diversity of Glycyrrhiza Populations Based on Transcriptome SSR Markers. Plant Mol. Biol. Rep. 2019, 37, 401–412. [Google Scholar] [CrossRef]
- Amos, W.; Rubinsztein, D.C. Microsatellites are subject to directional evolution. Nat. Genet. 1996, 12, 13–14. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Yang, T.; Yan, X.; Zheng, F.; Xu, X.; Zhou, C. Comparison of microsatellite distribution patterns in twenty-nine beetle genomes. Gene 2020, 757, 144919. [Google Scholar] [CrossRef]
- Feng, K.; Zhou, C.; Wang, L.; Zhang, C.; Yang, Z.; Hu, Z.; Yue, B.; Wu, Y. Comprehensive Comparative Analysis Sheds Light on the Patterns of Microsatellite Distribution across Birds Based on the Chromosome-Level Genomes. Animals 2023, 13, 655. [Google Scholar] [CrossRef]
- Fisher, P.J.; Gardner, R.C.; Richardson, T.E. Single locus microsatellites isolated using 5′ anchored PCR. Nucleic Acids Res. 1996, 24, 4369–4371. [Google Scholar] [CrossRef]
- Zane, L.; Bargelloni, L.; Patarnello, T. Strategies for microsatellite isolation: A review. Mol. Ecol. 2002, 11, 1–16. [Google Scholar] [CrossRef]
- Zhang, Z.; Deng, Y.; Tan, J.; Hu, S.; Yu, J.; Xue, Q. A genome-wide microsatellite polymorphism database for the Indica and Japonica rice. DNA Res. 2007, 14, 37–45. [Google Scholar] [CrossRef]
- Abdelkrim, J.; Robertson, B.C.; Stanton, J.A.L.; Gemmell, N.J. Fast, cost-effective development of species-specific microsatellite markers by genomic sequencing. Biotechniques 2009, 46, 185–192. [Google Scholar] [CrossRef]
- Song, Q.; Jia, G.; Zhu, Y.; Grant, D.; Nelson, R.T.; Hwang, E.-Y.; Hyten, D.L.; Cregan, P.B. Abundance of SSR motifs and development of candidate polymorphic SSR markers (BARCSOYSSR_1.0) in soybean. Crop Sci. 2010, 50, 1950–1960. [Google Scholar] [CrossRef]
- Xu, J.; Liu, L.; Xu, Y.; Chen, C.; Rong, T.; Ali, F.; Zhou, S.; Wu, F.; Liu, Y.; Wang, J.; et al. Development and characterization of simple sequence repeat markers providing genome-wide coverage and high resolution in maize. DNA Res. 2013, 20, 497–509. [Google Scholar] [CrossRef]
- Pandey, G.; Misra, G.; Kumari, K.; Gupta, S.; Parida, S.K.; Chattopadhyay, D.; Prasad, M. Genome-wide development and use of microsatellite markers for large-scale genotyping applications in foxtail millet Setaria italica (L.). DNA Res. 2013, 20, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Huang, S.; Zhan, J.; Yu, J.; Wang, X.; Hua, W.; Liu, S.; Liu, G.; Wang, H. Genomewide microsatellite characterization and marker development in the sequenced Brassica crop species. DNA Res. 2014, 21, 53–68. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, Z.K.; Chen, J.B.; Liu, C.Y.; Zhu, W.L.; Wang, L.Y.; Meng, L.H. De Novo transcriptome assembly and development of novel microsatellite markers for the traditional chinese medicinal herb, Veratrilla baillonii Franch (Gentianaceae). Evol. Bioinform. 2015, 11, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Qin, M.Z.; Yang, L.; Song, Z.Z.; Luo, L.; Bao, H.Y.; Ma, Z.G.; Zhou, Z.Y.; Xu, J.S. A genome-wide analysis of simple sequence repeats in Apis cerana, and its development as polymorphism markers. Gene 2016, 599, 53–59. [Google Scholar] [CrossRef]
- Kofler, R.; Schlotterer, C.; Lelley, T. SciRoKo: A new tool for whole genome microsatellite search and investigation. Bioinformatics 2007, 23, 1683–1685. [Google Scholar] [CrossRef]
- Jurka, J.; Pethiyagoda, C. Simple repetitive DNA sequences from primates: Compilation and analysis. J. Mol. Evol. 1995, 40, 120–126. [Google Scholar] [CrossRef]
- Li, C.Y.; Liu, L.; Yang, J.; Li, J.B.; Su, Y.; Zhang, Y.; Wang, Y.Y.; Zhu, Y.Y. Genome-wide analysis of microsatellite sequence in seven filamentous fungi. Interdiscip. Sci. Comput. Life Sci. 2009, 1, 141–150. [Google Scholar] [CrossRef]
- Huang, J.; Li, W.; Jian, Z.Y.; Yue, B.S.; Yan, Y.F. Genome-wide distribution and organization of microsatellites in six species of birds. Biochem. Syst. Ecol. 2016, 67, 95–102. [Google Scholar] [CrossRef]
- Subramanian, S.; Mishra, R.K.; Singh, L. Genome-wide analysis of microsatellite repeats in humans: Their abundance and density in specific genomic regions. Genome Biol. 2003, 4, R13. [Google Scholar] [CrossRef]
- Xu, Y.; Hu, Z.; Wang, C.; Zhang, X.; Li, J.; Yue, B. Characterization of perfect microsatellite based on genome-wide and chromosome level in rhesus monkey (Macaca mulatta). Gene 2016, 592, 269–275. [Google Scholar] [CrossRef]
- Liu, S.; Hou, W.; Sun, T.; Xu, Y.; Li, P.; Yue, B.; Fan, Z.; Li, J. Genome-wide mining and comparative analysis of microsatellites in three macaque species. Mol. Genet. Genom. 2017, 292, 537–550. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Li, W.; Hu, Z.; Zeng, T.; Shen, Y.; Liu, S.; Zhang, X.; Li, J.; Yue, B. Genome-wide mining of perfect microsatellites and tetranucleotide orthologous microsatellites estimates in six primate species. Gene 2018, 643, 124–132. [Google Scholar] [CrossRef] [PubMed]
- Qi, W.; Jiang, X.; Xiao, G.; Huang, X.; Du, L. Seeking and bioinformatics analysis of microsatellite sequence in the genomes of cow and sheep. Acta Vet. Zootech. Sin. 2013, 44, 1724–1733. [Google Scholar]
- Tong, X.; Dai, F.; Li, B.; Yu, Q.; Xia, Q.; Lu, C. Microsatellite repeats in mouse: Abundance, distribution and density. Acta Zool. Sin. 2006, 52, 138–152. [Google Scholar]
- Warren, W.C.; Hillier, L.W.; Graves, J.A.M.; Birney, E.; Ponting, C.P.; Grutzner, F.; Belov, K.; Miller, W.; Clarke, L.; Chinwalla, A.T.; et al. Genome analysis of the platypus reveals unique signatures of evolution. Nature 2008, 453, 175–183. [Google Scholar] [CrossRef][Green Version]
- Primmer, C.R.; Raudsepp, T.; Chowdhary, B.P.; Møller, A.P.; Ellegren, H. Low frequency of microsatellites in the avian genome. Genome Res. 1997, 7, 471–482. [Google Scholar] [CrossRef]
- Hughes, A.L.; Hughes, M.K. Small genomes for better flyers. Nature 1995, 377, 391. [Google Scholar] [CrossRef]
- Wright, N.A.; Gregory, T.R.; Witt, C.C. Metabolic ‘engines’ of flight drive genome size reduction in birds. Proc. Biol. Sci. 2014, 281, 20132780. [Google Scholar] [CrossRef]
- Garner, T.W.J. Genome size and microsatellites: The effect of nuclear size on amplification potential. Genome 2002, 45, 212–215. [Google Scholar] [CrossRef]
- Hughes, A.L.; Piontkivska, H. DNA repeat arrays in chicken and human genomes and the adaptive evolution of avian genome size. BMC Ecol. Evol. 2005, 5, 12. [Google Scholar] [CrossRef]
- Adams, R.; Blackmon, H.; Reyes Velasco, J.; Schield, D.; Card, D.; Andrew, A.; Waynewood, N.; Castoe, T. Microsatellite landscape evolutionary dynamics across 450 million years of vertebrate genome evolution. Genome 2016, 59, 295–310. [Google Scholar] [CrossRef] [PubMed]
- Tóth, G.; Gaspari, Z.; Jurka, J. Microsatellites in different eukaryotic genomes: Survey and analysis. Genome Res. 2000, 10, 967–981. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Li, C.; Li, Q.; Li, B.; Larkin, D.M.; Lee, C.; Storz, J.F.; Antunes, A.; Greenwold, M.J.; Meredith, R.W.; et al. Comparative genomics reveals insights into avian genome evolution and adaptation. Science 2014, 346, 1311–1320. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.; Avvaru, A.K.; Sowpati, D.T.; Mishra, R.K. Patterns of microsatellite distribution across eukaryotic genomes. BMC Genom. 2019, 20, 153. [Google Scholar] [CrossRef]
- Sharma, P.C.; Grover, A.; Kahl, G. Mining microsatellites in eukaryotic genomes. Trends Biotechnol. 2007, 25, 490–498. [Google Scholar] [CrossRef]
- Zhou, C.; Li, F.; Wen, Q.; Price, M.; Yang, N.; Yue, B. Characterization of microsatellites in the endangered snow leopard based on the chromosome-level genome. Mammal Res. 2021, 66, 385–398. [Google Scholar] [CrossRef]
- Harvey, S.C. Slipped structures in DNA triplet repeat sequences: Entropic contributions to genetic instabilities. Biochemistry 1997, 36, 3047–3049. [Google Scholar] [CrossRef]
- Chakraborty, R.; Kimmel, M.; Strivers, D.N.; Davison, L.J.; Deka, R. Relative mutation rates at di-, tri-, and tetra-nucleotide microsatellite loci. Proc. Natl. Acad. Sci. USA 1997, 94, 1041–1046. [Google Scholar] [CrossRef]
- Boyer, J.C.; Yamada, N.A.; Roques, C.N.; Hatch, S.B.; Riess, K.; Farber, R.A. Sequence dependent instability of mononucleotide microsatellites in cultured mismatch repair proficient and deficient mammalian cells. Hum. Mol. Genet. 2002, 11, 707–713. [Google Scholar] [CrossRef]
- Kelkar, Y.D.; Tyekucheva, S.; Chiaromonte, F.; Makova, K.D. The genome-wide determinants of human and chimpanzee microsatellite evolution. Genome Res. 2008, 18, 30–38. [Google Scholar] [CrossRef]
- Yu, Q.Y.; Li, B.; Li, G.R.; Fang, S.M.; Yan, H.; Tong, X.L.; Qian, J.F.; Xia, Q.Y.; Lu, C. Abundance and distribution of microsatellites in the entire mosquito genome. Prog. Biochem. Biophys. 2005, 32, 435–441. [Google Scholar]
- Wei, Z.M.; Kong, G.Y.; Lian, Z.M.; Liu, H.; Fan, Y.W.; Zhang, H. Abundance and distribution of microsatellites in the entire Apis mellifera genome. Chin. Bull. Entomol. 2007, 44, 501–504. [Google Scholar]
- Schlötterer, C.; Tautz, D. Slippage synthesis of simple sequence DNA. Nucleic Acids Res. 1992, 20, 211–215. [Google Scholar] [CrossRef]
- Kruglyak, S.; Durrett, R.T.; Schug, M.D.; Aquadro, C.F. Equilibrium distributions of microsatellite repeat length resulting from a balance between slippage events and point mutations. Proc. Natl. Acad. Sci. USA 1998, 95, 10774–10778. [Google Scholar] [CrossRef]
- Schlötterer, C. Genome evolution: Are microsatellites really simple sequences? Curr. Biol. 1998, 8, R132–R134. [Google Scholar] [CrossRef]
- Katti, M.V.; Ranjekar, P.K.; Gupta, V.S. Differential distribution of simple sequence repeats in eukaryotic genome sequences. Mol. Biol. Evol. 2001, 18, 1161–1167. [Google Scholar] [CrossRef]
- Qi, W.; Lu, T.; Zheng, C.; Jiang, X.; Jie, H.; Zhang, X.; Yue, B.; Zhao, G. Distribution patterns of microsatellites and development of its marker in different genomic regions of forest musk deer genome based on high throughput sequencing. Aging 2020, 12, 4445–4462. [Google Scholar] [CrossRef]
- Song, X.; Yang, T.; Zhang, X.; Yuan, Y.; Yan, X.; Wei, Y.; Zhang, J.; Zhou, C. Comparison of the Microsatellite Distribution Patterns in the Genomes of Euarchontoglires at the Taxonomic Level. Front. Genet. 2021, 12, 622724. [Google Scholar] [CrossRef]
- Chambers, G.K.; MacAvoy, E.S. Microsatellites: Consensus and controversy. Comp. Biochem. Physiol. B-Biochem. Mol. Biol. 2000, 126, 455–476. [Google Scholar] [CrossRef]
- Buschiazzo, E.; Gemmell, N.J. The rise, fall and renaissance of microsatellites in eukaryotic genomes. BioEssays 2006, 28, 1040–1050. [Google Scholar] [CrossRef]
- Neff, B.D.; Gross, M.R. Microsatellite evolution in vertebrates: Inference from AC dinucleotide repeats. Evolution 2001, 55, 1717–1733. [Google Scholar] [PubMed]
- Amos, W.; Clarke, A. Body temperature predicts maximum microsatellite length in mammals. Biol. Lett. 2008, 4, 399–401. [Google Scholar] [CrossRef] [PubMed]
- Frankham, R.; Ballou, J.D.; Briscoe, D.A. Introduction to Conservation Genetics; Cambridge University Press: Cambridge, UK, 2010. [Google Scholar]
- Sodhi, N.S.; Brook, B.W.; Bradshaw, C.J. Causes and consequences of species extinctions. In The Princeton Guide to Ecology; Princeton University Press: Princeton, NJ, USA, 2009; pp. 514–520. [Google Scholar]
- Lončar, V.; Kralj, J.; Stronen, A.V.; Grgurević, M.; Pavlinec, Ž.; Jurinović, L.; Svetličić, I.; Buzan, E.; Piro, S.; Herrmann, C.; et al. High genetic diversity yet weak population genetic structure in European common terns. Sci. Rep. 2024, 14, 29173. [Google Scholar] [CrossRef] [PubMed]
- Avise, J.C. Molecular Markers, Natural History and Evolution; Sinauer Associates: Sunderland, MA, USA, 2004. [Google Scholar]
- Jones, A.G.; Small, C.M.; Piertney, S.B.; Coltman, D.W. Molecular phylogenetics and the evolution of raptors. Mol. Phylogenetics Evol. 2012, 64, 550–562. [Google Scholar]

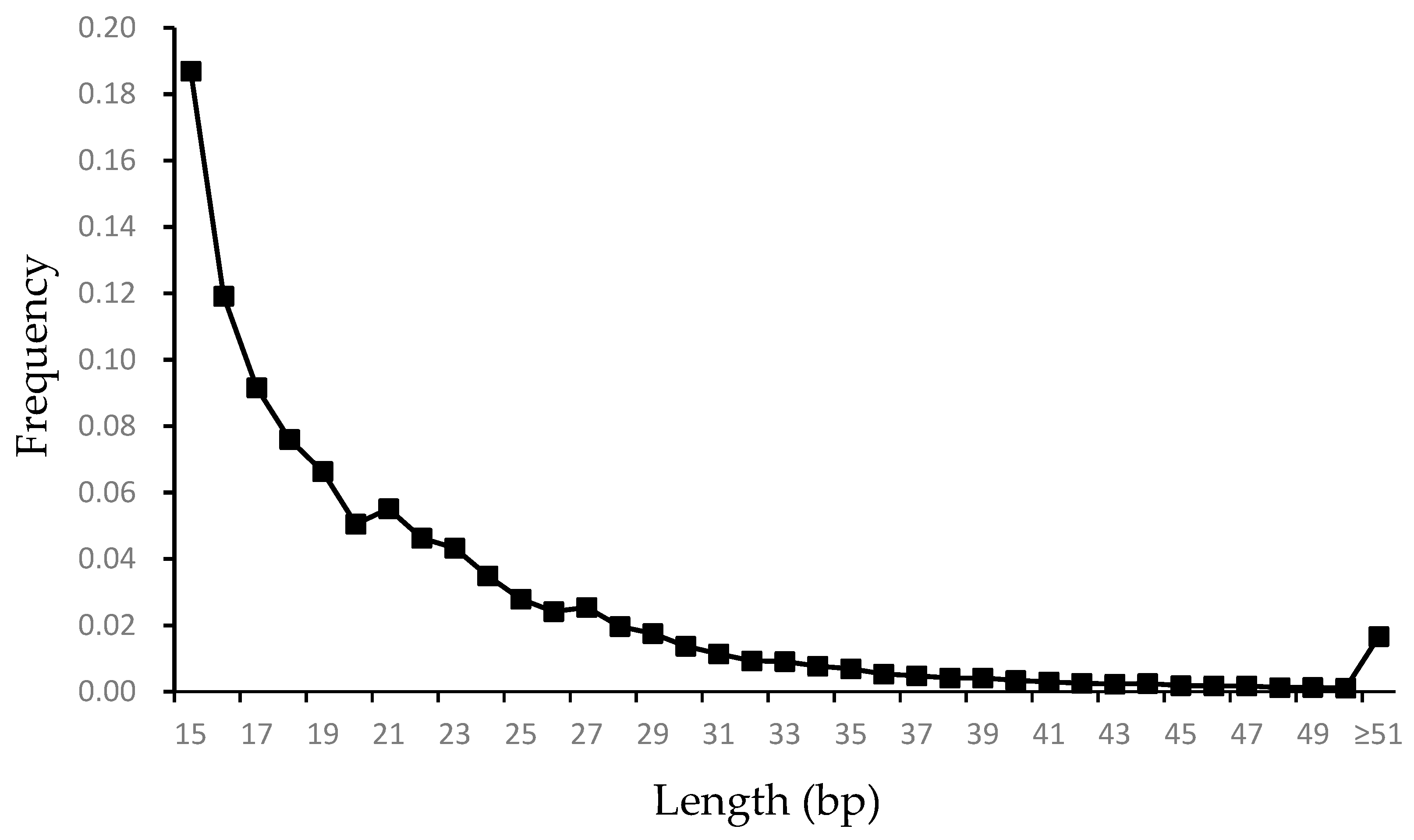
| Motif | Number of SSRs | Proportion (%) | Average Length (bp) | Average Mismatches | Counts/Mbp |
|---|---|---|---|---|---|
| Mononucleotide | 128,024 | 53.18 | 20.98 | 0.21 | 107.52 |
| Dinucleotide | 16,534 | 6.87 | 22.91 | 0.38 | 13.89 |
| Trinucleotide | 25,186 | 10.46 | 23.67 | 0.56 | 21.15 |
| Tetranucleotide | 31,514 | 13.09 | 19.93 | 0.28 | 26.47 |
| Pentanucleotide | 32,804 | 13.63 | 23.97 | 0.31 | 27.55 |
| Hexanucleotide | 6679 | 2.77 | 29.85 | 0.69 | 5.61 |
| Marker ID | Repeat | * Primer Sequences (5′ → 3′) | Alleles | Size (bp) | PIC # | Heterozygosity | |
|---|---|---|---|---|---|---|---|
| Expected (He) | Observed (Ho) | ||||||
| FHYSSR008 | (aggag)13 | F -tatttccaggtgcagggacc R -tccctctctcatgtcccagg | 8 | 161–206 | 0.817 | 0.836 | 0.5 |
| FHYSSR009 | (ccggc)4 | F -ctcgtttgctgatgcctgc R -cccaccacgttacctgagc | 5 | 168–188 | 0.717 | 0.758 | 0.75 |
| FHYSSR012 | (cctct)5 | F -gacagctctggatcagctcc R -ctgtcaacaaaacccgtccc | 3 | 174–184 | 0.427 | 0.477 | 0.625 |
| FHYSSR016 | (ggctc)4 | F -ggtcagttgtccgcgatcc R -gaggaaccgacccaaccc | 4 | 143–174 | 0.53 | 0.609 | 0.75 |
| FHYSSR017 | (gccga)8 | F -ggtctgtgcaccctgagg R -atgcaaggagaggggaggg | 4 | 230–250 | 0.458 | 0.492 | 0.375 |
| FHYSSR024 | (gtttg)13 | F -atccaggtttgtcttgggcg R -tgctggctgcttatttcaagc | 4 | 205–222 | 0.644 | 0.695 | 0.625 |
| FHYSSR037 | (tg)8 | F -gttaaggactctgatgctgatgc R -acattagacgtgcagcgacc | 3 | 147–152 | 0.294 | 0.32 | 0.375 |
| FHYSSR043 | (ca)10 | F -ggtgaccacgtcttactcgg R -gctgtgcaccttgtctttcc | 3 | 177–183 | 0.294 | 0.32 | 0.375 |
| FHYSSR044 | (at)11 | F -acaatttcaaggctgcaatgc R -gtctccttcagcatccctcc | 3 | 206–212 | 0.428 | 0.508 | 0.125 |
| FHYSSR045 | (tg)12 | F -tcaggatccaacacagtgagc R -ctccctagtgcatcacgtgg | 4 | 187–193 | 0.458 | 0.492 | 0.375 |
| FHYSSR046 | (tc)7 | F -cgcctaccctaccccacc R -tgtgcaagtgattaagggattgg | 2 | 168–170 | 0.258 | 0.305 | 0.125 |
| FHYSSR048 | (tat)5 | F -cgtttcctgttcacacaagcc R -tgcttaaaacactggacacgc | 2 | 179–188 | 0.375 | 0.5 | 0.25 |
| FHYSSR059 | (gct)5 | F -gttcctcagaccgtctctgg R -atgtcagaagctcacctgcg | 3 | 180–189 | 0.521 | 0.586 | 0.5 |
| FHYSSR063 | (gag)8 | F -gaaaggttccccggaccg R -ctgcccaagtcctccagc | 2 | 178–181 | 0.384 | 0.43 | 0.375 |
| FHYSSR066 | (ttc)9 | F -cagttgttgctgatcgctgg R -gtcctcttctgcagggtgc | 2 | 181–184 | 0.371 | 0.492 | 0.625 |
| FHYSSR074 | (act)6 | F -ccccttcttaccttagtgacttcc R -cgactcatagctcttctagtggg | 3 | 183–189 | 0.503 | 0.594 | 0.75 |
| FHYSSR095 | (tttg)5 | F -tctggcattgtataccgggc R -tgtcccaaaggtcctttcagg | 2 | 149–153 | 0.337 | 0.43 | 0.375 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, W.; Ke, D.; Wang, C.; Fan, H. Genome-Wide Microsatellite Characterization and Molecular Marker Development of Himalayan Griffon (Gyps himalayensis). Animals 2025, 15, 1438. https://doi.org/10.3390/ani15101438
Guo W, Ke D, Wang C, Fan H. Genome-Wide Microsatellite Characterization and Molecular Marker Development of Himalayan Griffon (Gyps himalayensis). Animals. 2025; 15(10):1438. https://doi.org/10.3390/ani15101438
Chicago/Turabian StyleGuo, Weibin, Dianhua Ke, Changcao Wang, and Haiying Fan. 2025. "Genome-Wide Microsatellite Characterization and Molecular Marker Development of Himalayan Griffon (Gyps himalayensis)" Animals 15, no. 10: 1438. https://doi.org/10.3390/ani15101438
APA StyleGuo, W., Ke, D., Wang, C., & Fan, H. (2025). Genome-Wide Microsatellite Characterization and Molecular Marker Development of Himalayan Griffon (Gyps himalayensis). Animals, 15(10), 1438. https://doi.org/10.3390/ani15101438





