Association of Novel Mutations in the Vasoactive Intestinal Peptide Receptor-1 Gene with Egg Shell Thickness in Three Strains of Laying-Type Quail
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Treatment
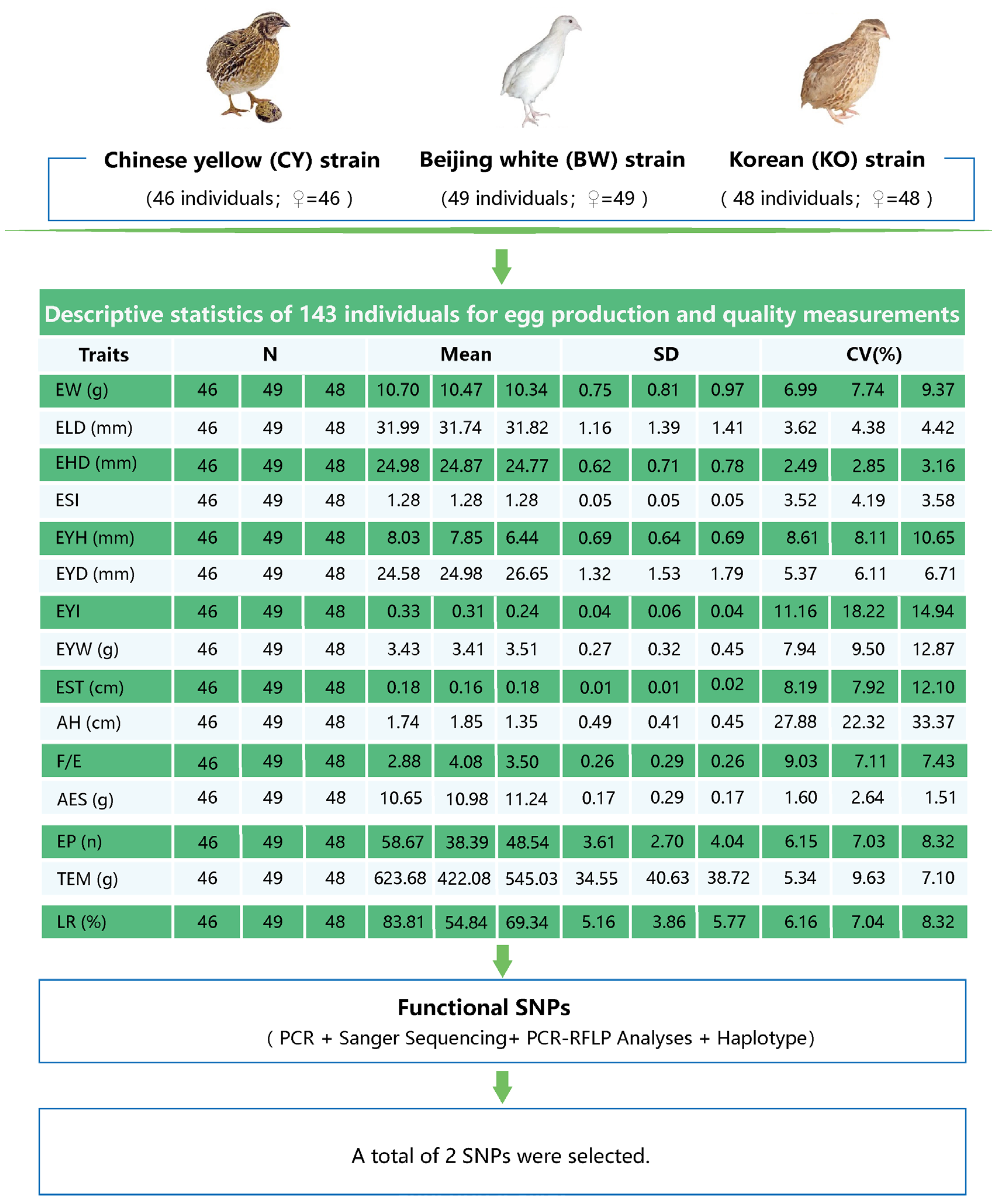
2.2. Experimental Populations and Phenotypic Data Collection
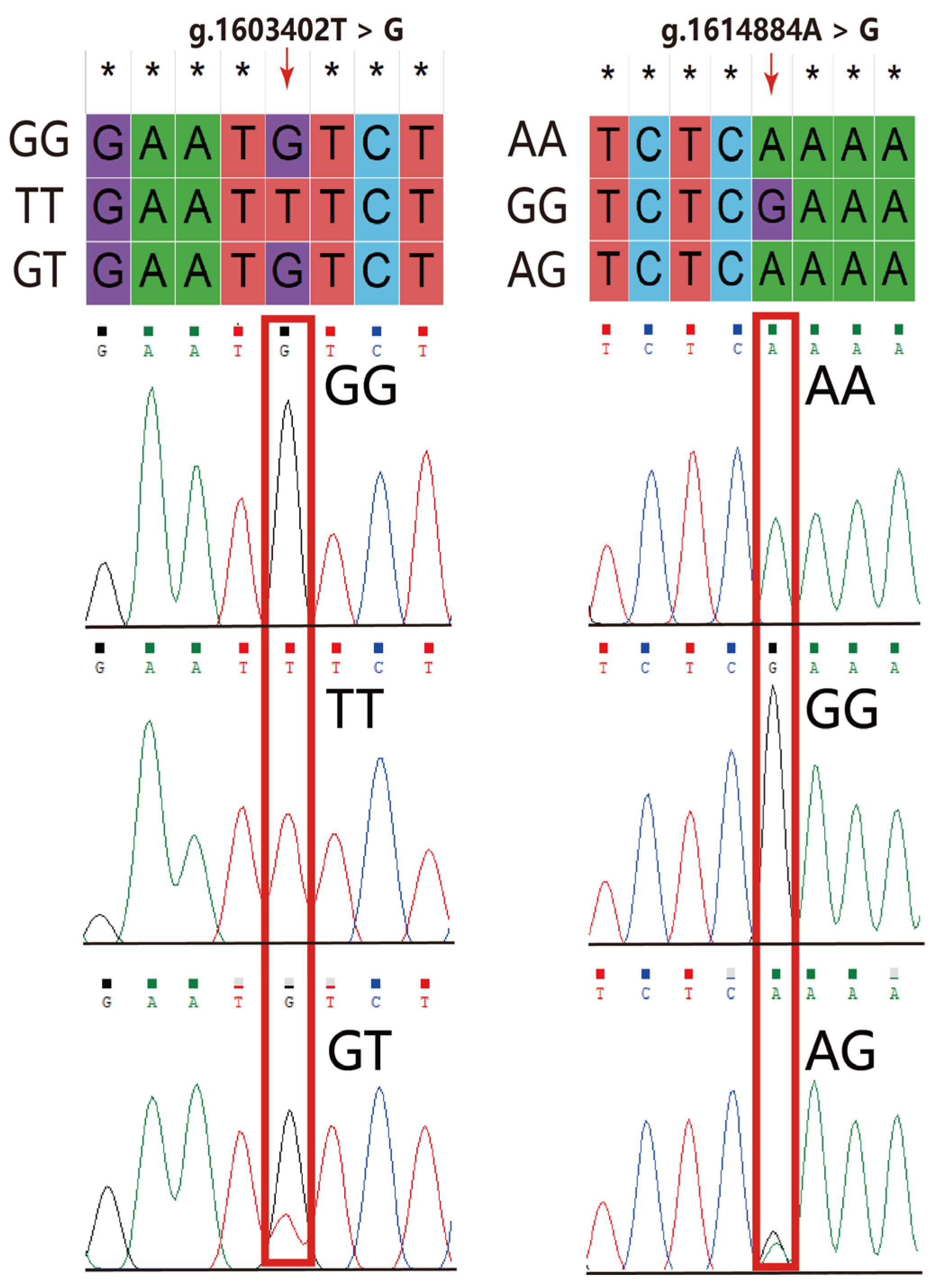
2.3. DNA Sample Collection, Primers, and Polymerase Chain Reaction
2.4. Sequencing and PCR-RFLP Analysis
2.5. Statistical Analysis
3. Results
3.1. Descriptive Statistics
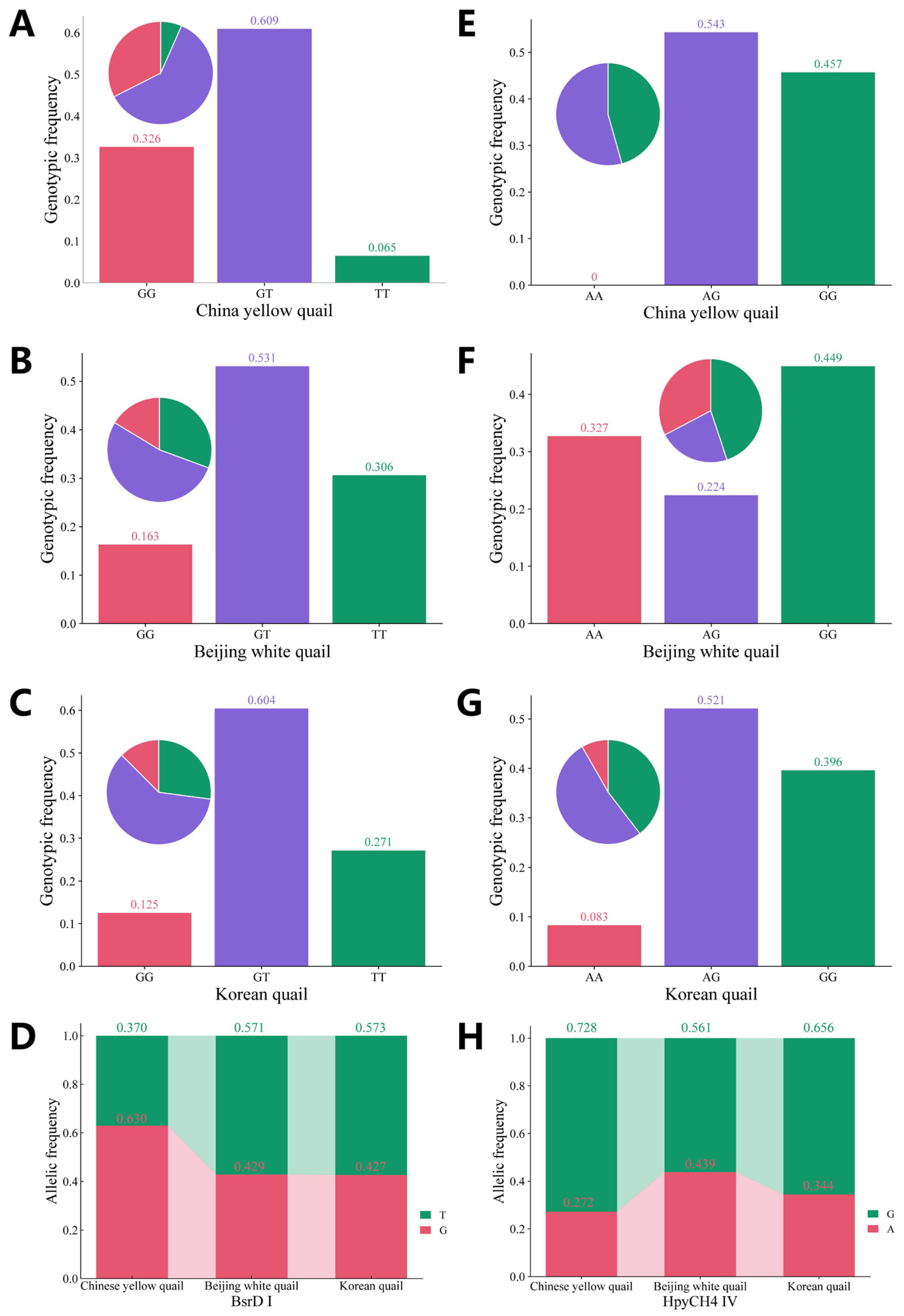
3.2. Polymorphism of the VIPR-1 Gene in Quail
3.3. Association Analysis of SNPs and Haplotype Combinations with Egg Quality in Quail
3.4. Association Analysis of SNPs and Haplotype Combinations with Laying Performance in Quail
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Thiele, I.; Fleming, R.M.T. Whole-body metabolic modelling predicts isoleucine dependency of SARS-CoV-2 replication. Comput. Struct. Biotechnol. J. 2022, 20, 4098–4109. [Google Scholar] [CrossRef] [PubMed]
- Quaresma, M.A.G.; Antunes, I.C.; Ferreira, B.G.; Parada, A.; Elias, A.; Barros, M.; Santos, C.; Partidário, A.; Mourato, M.; Roseiro, L.C. The composition of the lipid, protein and mineral fractions of quail breast meat obtained from wild and farmed specimens of Common quail (Coturnix coturnix) and farmed Japanese quail (Coturnix japonica domestica). Poult. Sci. 2022, 101, 101505. [Google Scholar] [CrossRef] [PubMed]
- Arunrao, K.V.; Kannan, D.; Amutha, R.; Thiruvenkadan, A.K.; Yakubu, A. Production performance of four lines of Japanese quail reared under tropical climatic conditions of Tamil Nadu, India. Front. Genet. 2023, 14, 1128944. [Google Scholar] [CrossRef]
- Shen, Y.Y.; Dai, K.; Cao, X.; Murphy, R.W.; Shen, X.J.; Zhang, Y.P. The updated phylogenies of the phasianidae based on combined data of nuclear and mitochondrial DNA. PLoS ONE 2014, 9, e95786. [Google Scholar] [CrossRef]
- Nunome, M.; Nakano, M.; Tadano, R.; Kawahara-Miki, R.; Kono, T.; Takahashi, S.; Kawashima, T.; Fujiwara, A.; Nirasawa, K.; Mizutani, M.; et al. Genetic Divergence in Domestic Japanese Quail Inferred from Mitochondrial DNA D-Loop and Microsatellite Markers. PLoS ONE 2017, 12, e0169978. [Google Scholar] [CrossRef]
- Zhai, Z.; Zhao, W.; He, C.; Yang, K.; Tang, L.; Liu, S.; Zhang, Y.; Huang, Q.; Meng, H. SNP discovery and genotyping using restriction-site-associated DNA sequencing in chickens. Anim. Genet. 2015, 46, 216–219. [Google Scholar] [CrossRef]
- Geibel, J.; Praefke, N.P.; Weigend, S.; Simianer, H.; Reimer, C. Assessment of linkage disequilibrium patterns between structural variants and single nucleotide polymorphisms in three commercial chicken populations. BMC Genom. 2022, 23, 193. [Google Scholar] [CrossRef] [PubMed]
- Chaiseha, Y.; Youngren, O.M.; El Halawani, M.E. Expression of Vasoactive Intestinal Peptide Receptor Messenger RNA in the Hypothalamus and Pituitary Throughout the Turkey Reproductive Cycle1. Biol. Reprod. 2004, 70, 593–599. [Google Scholar] [CrossRef][Green Version]
- Kosonsiriluk, S.; Sartsoongnoen, N.; Chaiyachet, O.-a.; Prakobsaeng, N.; Songserm, T.; Rozenboim, I.; El Halawani, M.; Chaiseha, Y. Vasoactive intestinal peptide and its role in continuous and seasonal reproduction in birds. Gen. Comp. Endocrinol. 2008, 159, 88–97. [Google Scholar] [CrossRef]
- Kansaku, N.; Shimada, K.; Ohkubo, T.; Saito, N.; Suzuki, T.; Matsuda, Y.; Zadworny, D. Molecular cloning of chicken vasoactive intestinal polypeptide receptor complementary DNA, tissue distribution and chromosomal localization. Biol. Reprod. 2001, 64, 1575–1581. [Google Scholar] [CrossRef]
- Zhou, M.; Lei, M.; Rao, Y.; Nie, Q.; Zeng, H.; Xia, M.; Liang, F.; Zhang, D.; Zhang, X. Polymorphisms of vasoactive intestinal peptide receptor-1 gene and their genetic effects on broodiness in chickens. Poult. Sci. 2008, 87, 893–903. [Google Scholar] [CrossRef]
- Pu, Y.J.; Wu, Y.; Xu, X.J.; Du, J.P.; Gong, Y.Z. Association of VIPR-1 gene polymorphisms and haplotypes with egg production in laying quails. J. Zhejiang Univ. Sci. B 2016, 17, 591–596. [Google Scholar] [CrossRef]
- Bai, J.Y.; Pang, Y.Z.; Zhang, X.H.; Yun, Y.X.; Qi, Y.X. Microsatellite Analysis of Genetic Diversity in Quail Populations from China. Rev. Bras. Ciênc. Avíc. 2016, 18, 519–524. [Google Scholar] [CrossRef][Green Version]
- He, M.; Liang, X.; Pu, H.; Hu, Y.; Ye, G.; Li, X.; Liu, L. Immunohistochemical localization of vasoactive intestinal peptide in bursa of fabricius of Chinese yellow quail. Indian J. Anim. Res. 2016, 50, 101–104. [Google Scholar] [CrossRef]
- Zhao, S.; Cui, X.; Pang, Y.; Zhang, X.; You, X.; Yang, Y.; Lei, Y. Cloning, genome structure and expression analysis of MHC class I gene in Korean quail. Br. Poult. Sci. 2022, 63, 291–297. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Wang, X.; Li, J.; Chen, M.; Zeng, F.; Lu, X.; He, Y. Research Note: Association of VIPR-1 gene polymorphism with growth traits in meat type Japanese quail (Coturnix japonica). Poult. Sci. 2023, 102, 102781. [Google Scholar] [CrossRef]
- Liu, R.; Fang, X.; Lu, X.; Liu, Y.; Li, Y.; Bai, X.; Ding, X.; Yang, R. Polymorphisms of the SCD1 Gene and Its Association Analysis with Carcass, Meat Quality, Adipogenic Traits, Fatty Acid Composition, and Milk Production Traits in Cattle. Animals 2024, 14, 1759. [Google Scholar] [CrossRef]
- Alzohairy, A. BioEdit: An important software for molecular biology. GERF Bull. Biosci. 2011, 2, 60–61. [Google Scholar]
- Excoffier, L.; Lischer, H.E. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Cuaresma, B.; Ermube, H.; Fabros, R.; Lantano, R.; Berdos, J.; Viloria, M.; Briones, R. Laying performance of quail (Coturnix coturnix) fed diets formulated based on crude protein restriction. Philipp. J. Vet. Med. 2021, 47, 51–58. [Google Scholar]
- Cullere, M.; Woods, M.J.; van Emmenes, L.; Pieterse, E.; Hoffman, L.C.; Dalle Zotte, A. Hermetia illucens Larvae Reared on Different Substrates in Broiler Quail Diets: Effect on Physicochemical and Sensory Quality of the Quail Meat. Animals 2019, 9, 525. [Google Scholar] [CrossRef]
- Han, X.; Song, Z.; Wang, W.; Tang, H. Polymorphism in the 5′ regulatory region of CTNNB1 gene and association with age at first lay and egg production. Br. Poult. Sci. 2022, 63, 510–518. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.; Vu, C.; Nhan, N.; Xuan, N.; An, N.; Dung, T. Effects of genetic polymorphisms on egg production in indigenous Noi chicken. J. Exp. Biol. Agric. Sci. 2015, 3, 487–493. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, Y.; Bai, J. Research Note: Association of LEPR gene polymorphism with growth and carcass traits in Savimalt and French Giant meat-type quails. Poult. Sci. 2023, 102, 103047. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.P.; Zeng, H.; Zhang, D.X.; Jia, X.L.; Luo, C.L.; Fang, M.X.; Nie, Q.H.; Zhang, X.Q. Polymorphisms associated with egg number at 300 days of age in chickens. Genet. Mol. Res. GMR 2011, 10, 2279–2289. [Google Scholar] [CrossRef]
- Hosseinpoor, L.; Nikbin, S.; Hedayat-Evrigh, N.; Elyasi, G. Association of polymorphisms of vasoactive intestinal peptide and its receptor with reproductive traits of turkey hens. S. Afr. J. Anim. Sci. 2021, 50, 345–352. [Google Scholar] [CrossRef]

| SNP | S | HWE | Ho | He | PIC | Ne | |
|---|---|---|---|---|---|---|---|
| χ2 | p | ||||||
| g.1603402T>G | CY (46) | 4.315 | 0.038 | 0.609 | 0.466 | 0.357 | 1.873 |
| BW (49) | 0.340 | 0.560 | 0.531 | 0.490 | 0.370 | 1.960 | |
| KO (48) | 2.642 | 0.104 | 0.604 | 0.489 | 0.370 | 1.958 | |
| g.1614884A>G | CY (46) | 6.405 | 0.011 | 0.543 | 0.396 | 0.317 | 1.655 |
| BW (49) | 14.511 | 0.000 | 0.224 | 0.493 | 0.371 | 1.970 | |
| KO (48) | 1.144 | 0.285 | 0.521 | 0.451 | 0.349 | 1.822 | |
| S | EQ | BsrD I (Mean ± SE) | |||
|---|---|---|---|---|---|
| GG | GT | TT | ANOVA/Kruskal–Wallis p-Value | ||
| BW (49) | EW (g) | 10.50 ± 0.17 | 10.41 ± 0.18 | 10.55 ± 0.22 | 0.877 |
| ELD (mm) | 31.8 ± 0.46 | 31.7 ± 0.29 | 31.6 ± 0.35 | 0.960 | |
| EHD (mm) | 24.8 ± 0.12 | 24.8 ± 0.16 | 24.9 ± 0.16 | 0.742 | |
| ESI | 1.28 ± 0.02 | 1.28 ± 0.01 | 1.26 ± 0.01 | 0.833 | |
| EYH (mm) | 7.80 ± 0.17 | 7.76 ± 0.11 | 8.01 ± 0.20 | 0.500 | |
| EYD (mm) | 25.33 ± 0.27 | 24.992 ± 0.29 | 25.78 ± 0.49 | 0.575 | |
| EYI | 0.26 ± 0.039 | 0.31 ± 0.006 | 0.32 ± 0.011 | 0.223 | |
| EYW (g) | 3.36 ± 0.06 | 3.40 ± 0.06 | 3.43 ± 0.10 | 0.888 | |
| EST (cm) | 0.15 ± 0.002 ab | 0.16 ± 0.003 a | 0.15 ± 0.003 b | 0.043 | |
| AH (cm) | 1.88 ± 0.10 | 1.82 ± 0.08 | 1.86 ± 0.11 | 0.922 | |
| KO (48) | EW (g) | 10.40 ± 0.51 | 10.25 ± 0.18 | 10.47 ± 0.24 | 0.795 |
| ELD (mm) | 31.8 ± 0.6 | 31.68 ± 0.2 | 32.09 ± 0.3 | 0.697 | |
| EHD (mm) | 24.8 ± 0.4 | 24.72 ± 0.1 | 24.82 ± 0.2 | 0.896 | |
| ESI | 1.28 ± 0.01 | 1.28 ± 0.01 | 1.293 ± 0.01 | 0.773 | |
| EYH (mm) | 6.63 ± 0.13 | 6.38 ± 0.15 | 6.47 ± 0.11 | 0.776 | |
| EYD (mm) | 25.95 ± 0.70 | 26.65 ± 0.36 | 26.96 ± 0.43 | 0.533 | |
| EYI | 0.25 ± 0.01 | 0.24 ± 0.01 | 0.24 ± 0.01 | 0.316 | |
| EYW (g) | 3.45 ± 0.20 | 3.47 ± 0.08 | 3.63 ± 0.13 | 0.568 | |
| EST (cm) | 0.17 ± 0.01 | 0.17 ± 0.01 | 0.17 ± 0.00 | 0.825 | |
| AH (cm) | 1.31 ± 0.18 | 1.32 ± 0.07 | 1.41 ± 0.15 | 0.939 | |
| S | EQ | HpyCH4 IV (Mean ± SE) | |||
|---|---|---|---|---|---|
| AA | AG | GG | ANOVA/Kruskal–Wallis p-Value | ||
| KO (48) | EW (g) | 10.00 ± 0.33 | 10.44 ± 0.20 | 10.26 ± 0.23 | 0.665 |
| ELD (mm) | 30.6 ± 0.5 | 32.1 ± 0.3 | 31.7 ± 0.2 | 0.154 | |
| EHD (mm) | 24.8 ± 0.3 | 24.7 ± 0.1 | 24.7 ± 0.2 | 0.975 | |
| ESI | 1.23 ± 0.02 | 1.29 ± 0.01 | 1.28 ± 0.01 | 0.056 | |
| EYH (mm) | 6.52 ± 0.35 | 6.56 ± 0.12 | 6.26 ± 0.17 | 0.367 | |
| EYD (mm) | 26.72 ± 1.07 | 26.71 ± 0.29 | 26.55 ± 0.50 | 0.956 | |
| EYI | 0.24 ± 0.02 | 0.24 ± 0.01 | 0.23 ± 0.01 | 0.735 | |
| EYW (g) | 3.67 ± 0.25 | 3.60 ± 0.07 | 3.36 ± 0.11 | 0.151 | |
| EST (cm) | 0.17 ± 0.01 | 0.17 ± 0.00 | 0.18 ± 0.01 | 0.155 | |
| AH (cm) | 1.49 ± 0.27 | 1.31 ± 0.08 | 1.36 ± 0.11 | 0.568 | |
| D | Traits (Mean ± SE) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| EW (g) | ELD (mm) | EHD (mm) | ESI | EYH (mm) | EYD (mm) | EYI | EYW (g) | EST (cm) | AH (cm) | |
| GGAG (4.2%) | 11.40 ± 1.10 | 33.5 ± 1.2 | 25.5 ± 0.8 | 1.31 ± 0.01 | 6.70 ± 0.50 | 27.67 ± 0.02 | 0.24 ± 0.02 | 4.00 ± 0.20 | 0.17 ± 0.01 | 0.80 ± 0.10 |
| GGGG (8.3%) | 9.90 ± 0.46 | 31.1 ± 0.1 | 24.5 ± 0.5 | 1.26 ± 0.02 | 6.60 ± 0.07 | 25.09 ± 0.70 | 0.26 ± 0.01 | 3.17 ± 0.14 | 0.18 ± 0.01 | 1.56 ± 0.14 |
| GTAA (4.2%) | 10.20 ± 0.60 | 30.9 ± 0.1 | 25.0 ± 0.7 | 1.23 ± 0.03 | 6.30 ± 0.80 | 26.16 ± 2.29 | 0.24 ± 0.05 | 3.45 ± 0.45 | 0.17 ± 0.00 | 1.68 ± 0.05 |
| GTAG (29.2%) | 10.31 ± 0.27 | 31.7 ± 0.4 | 24.7 ± 0.1 | 1.28 ± 0.01 | 6.63 ± 0.19 | 26.64 ± 0.39 | 0.25 ± 0.01 | 3.62 ± 0.08 | 0.17 ± 0.01 | 1.35 ± 0.12 |
| GTGG (27.1%) | 10.20 ± 0.28 | 31.6 ± 0.3 | 24.6 ± 0.2 | 1.28 ± 0.01 | 6.12 ± 0.25 | 26.73 ± 0.64 | 0.23 ± 0.01 | 3.32 ± 0.14 | 0.18 ± 0.01 | 1.24 ± 0.10 |
| TTAA (4.2%) | 9.80 ± 0.50 | 30.3 ± 1.3 | 24.6 ± 0.2 | 1.23 ± 0.04 | 6.75 ± 0.05 | 27.28 ± 0.98 | 0.24 ± 0.01 | 3.90 ± 0.30 | 0.17 ± 0.02 | 1.30 ± 0.62 |
| TTAG (18.8%) | 10.42 ± 0.29 | 32.2 ± 0.4 | 24.7 ± 0.2 | 1.30 ± 0.02 | 6.41 ± 0.15 | 26.60 ± 0.55 | 0.24 ± 0.01 | 3.47 ± 0.16 | 0.17 ± 0.01 | 1.35 ± 0.10 |
| TTGG (4.2%) | 11.40 ± 0.10 | 33.0 ± 0.2 | 25.5 ± 0.0 | 1.20 ± 0.01 | 6.50 ± 0.30 | 28.27 ± 0.16 | 0.23 ± 0.01 | 4.05 ± 0.15 | 0.18 ± 0.01 | 1.80 ± 0.90 |
| ANOVA/Kruskal–Wallis p-value | 0.537 | 0.247 | 0.710 | 0.449 | 0.838 | 0.605 | 0.879 | 0.093 | 0.655 | 0.276 |
| S | LP | BsrD I (Mean ± SE) | |||
|---|---|---|---|---|---|
| GG | GT | TT | Kruskal–Wallis p-Value | ||
| BW (49) | F/E | 4.20 ± 0.11 | 4.08 ± 0.04 | 4.01 ± 0.08 | 0.431 |
| EP (n) | 38.4 ± 0.8 | 38.4 ± 0.5 | 38.2 ± 0.8 | 0.985 | |
| TEM (g) | 421.21 ± 13.80 | 422.63 ± 7.88 | 421.57 ± 11.57 | 0.985 | |
| AES (g) | 10.93 ± 0.10 | 10.97 ± 0.05 | 11.00 ± 0.08 | 0.991 | |
| LR (%) | 54.9 ± 1.2 | 54.9 ± 0.7 | 54.6 ± 1.1 | 0.959 | |
| KO (48) | F/E | 3.36 ± 0.08 | 3.56 ± 0.05 | 3.41 ± 0.06 | 0.127 |
| EP (n) | 50.9 ± 1.3 | 47.4 ± 0.7 | 49.7 ± 1.0 | 0.060 | |
| TEM (g) | 568.08 ± 14.49 | 535.21 ± 7.11 | 556.29 ± 9.92 | 0.060 | |
| AES (g) | 11.15 ± 0.02 | 11.28 ± 0.03 | 11.187 ± 0.03 | 0.231 | |
| LR (%) | 72.78 ± 1.97 | 67.84 ± 1.07 | 71.08 ± 1.44 | 0.074 | |
| S | LP | HpyCH4 IV (Mean ± SE) | |||
|---|---|---|---|---|---|
| AA | AG | GG | Kruskal–Wallis p-Value | ||
| KO(48) | F/E | 3.31 ± 0.06 | 3.49 ± 0.05 | 3.53 ± 0.06 | 0.510 |
| EP (n) | 52.0 ± 1.3 | 48.5 ± 0.7 | 47.8 ± 1.0 | 0.089 | |
| TEM (g) | 580.05 ± 13.10 | 543.19 ± 7.47 | 540.07 ± 9.27 | 0.089 | |
| AES (g) | 11.15 ± 0.02 | 11.19 ± 0.02 | 11.31 ± 0.04 | 0.200 | |
| LR (%) | 74.28 ± 1.85 | 69.33 ± 1.06 | 68.30 ± 1.45 | 0.072 | |
| D | Traits (Mean ± SE) | ||||
|---|---|---|---|---|---|
| F/E | EP (n) | TEM (g) | AES (g) | LR (%) | |
| GGAG (4.2%) | 3.49 ± 0.23 | 49.3 ± 3.9 | 550.91 ± 42.24 | 11.16 ± 0.04 | 70.50 ± 5.64 |
| GGGG (8.3%) | 3.30 ± 0.06 | 51.7 ± 1.2 | 576.67 ± 12.39 | 11.14 ± 0.03 | 73.92 ± 1.77 |
| GTAA (4.2%) | 3.25 ± 0.00 | 53.3 ± 0.0 | 593.16 ± 0.00 | 11.16 ± 0.00 | 76.14 ± 0.00 |
| GTAG (29.2%) | 3.56 ± 0.07 | 47.4 ± 0.9 | 532.43 ± 9.61 | 11.23 ± 0.04 | 67.77 ± 1.40 |
| GTGG (27.1%) | 3.60 ± 0.07 | 46.6 ± 1.1 | 529.29 ± 10.45 | 11.36 ± 0.06 | 66.64 ± 1.65 |
| TTAA (4.2%) | 3.37 ± 0.12 | 50.7 ± 2.6 | 566.95 ± 26.21 | 11.18 ± 0.06 | 72.42 ± 3.71 |
| TTAG (18.8%) | 3.39 ± 0.07 | 50.0 ± 1.1 | 558.22 ± 11.39 | 11.15 ± 0.02 | 71.50 ± 1.57 |
| TTGG (4.2%) | 3.52 ± 0.34 | 47.5 ± 4.8 | 536.94 ± 42.70 | 11.32 ± 0.24 | 67.85 ± 6.85 |
| Kruskal–Wallis p-value | 0.414 | 0.067 | 0.067 | 0.316 | 0.083 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Chen, H.; Lei, Y.; Wang, Q.; Li, G.; Bai, J. Association of Novel Mutations in the Vasoactive Intestinal Peptide Receptor-1 Gene with Egg Shell Thickness in Three Strains of Laying-Type Quail. Animals 2025, 15, 1373. https://doi.org/10.3390/ani15101373
Wang X, Chen H, Lei Y, Wang Q, Li G, Bai J. Association of Novel Mutations in the Vasoactive Intestinal Peptide Receptor-1 Gene with Egg Shell Thickness in Three Strains of Laying-Type Quail. Animals. 2025; 15(10):1373. https://doi.org/10.3390/ani15101373
Chicago/Turabian StyleWang, Xinle, Huricha Chen, Ying Lei, Qiankun Wang, Gan Li, and Junyan Bai. 2025. "Association of Novel Mutations in the Vasoactive Intestinal Peptide Receptor-1 Gene with Egg Shell Thickness in Three Strains of Laying-Type Quail" Animals 15, no. 10: 1373. https://doi.org/10.3390/ani15101373
APA StyleWang, X., Chen, H., Lei, Y., Wang, Q., Li, G., & Bai, J. (2025). Association of Novel Mutations in the Vasoactive Intestinal Peptide Receptor-1 Gene with Egg Shell Thickness in Three Strains of Laying-Type Quail. Animals, 15(10), 1373. https://doi.org/10.3390/ani15101373





