Differential Mitochondrial Genome Expression of Three Sympatric Lizards in Response to Low-Temperature Stress
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
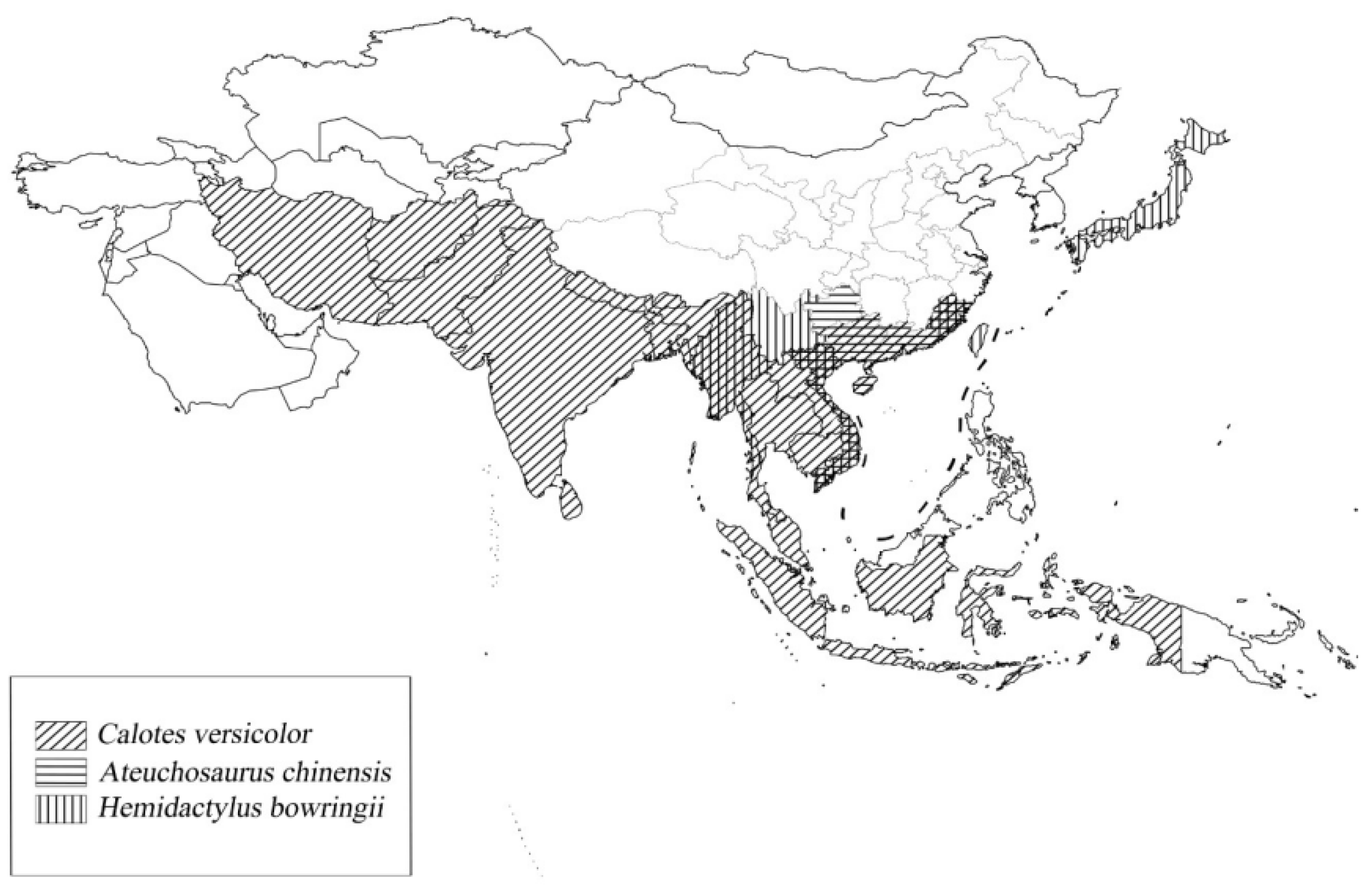
2.1. Sample Collection, Acclimatization, and Low-Temperature Stress
2.2. DNA Extraction, PCR Amplification, and Sequencing
2.3. mRNA Extraction and cDNA Synthesis
2.4. RT-qPCR Primer Design and Reaction
2.5. qPCR Data Analyses
3. Results
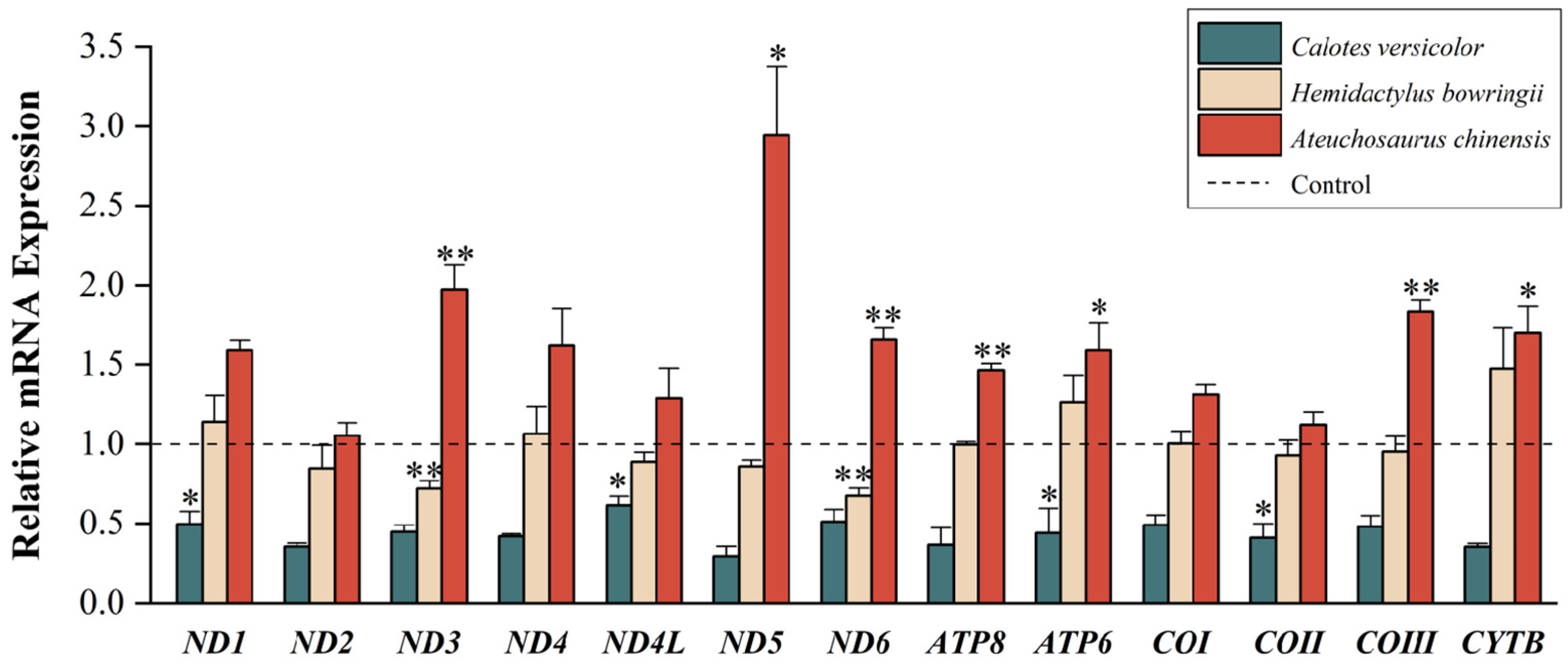
3.1. Quantitative Analyses of Mitochondrial PCGs
3.2. Comparison of Cold Tolerance Plasticity
4. Discussion
4.1. Low-Temperature Stress on Mitochondrial Gene Expression
4.2. Differential Gene Expression of Different Lizards in Regions of Sympatry
4.3. Conservation Strategy for Lizards
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Araújo, M.B.; Thuiller, W.; Pearson, R.G. Climate warming and the decline of amphibians and reptiles in Europe. J. Biogeogr. 2006, 33, 1712–1728. [Google Scholar] [CrossRef]
- Boucek, R.E.; Gaiser, E.E.; Liu, H.; Rehage, J.S. A review of subtropical community resistance and resilience to extreme cold spells. Ecosphere 2016, 7, e01455. [Google Scholar] [CrossRef]
- Osland, M.J.; Stevens, P.W.; Lamont, M.M.; Brusca, R.C.; Hart, K.M.; Waddle, J.H.; Langtimm, C.A.; Williams, C.M.; Keim, B.D.; Terando, A.J. Tropicalization of temperate ecosystems in North America: The northward range expansion of tropical organisms in response to warming winter temperatures. Glob. Chang. Biol. 2021, 27, 3009–3034. [Google Scholar] [CrossRef] [PubMed]
- Bickford, D.; Howard, S.D.; Ng, D.J.; Sheridan, J.A. Impacts of climate change on the amphibians and reptiles of Southeast Asia. Biodivers. Conserv. 2010, 19, 1043–1062. [Google Scholar] [CrossRef]
- Maxwell, S.L.; Butt, N.; Maron, M.; McAlpine, C.A.; Chapman, S.; Ullmann, A.; Segan, D.B.; Watson, J.E. Conservation implications of ecological responses to extreme weather and climate events. Divers. Distrib. 2019, 25, 613–625. [Google Scholar] [CrossRef]
- Zhang, Q.; Han, X.Z.; Burraco, P.; Hao, X.; Teng, L.W.; Liu, Z.S.; Zhang, F.S.; Du, W.G. Telomere length, oxidative stress and their links with growth and survival in a lizard facing climate warming. Sci. Total Environ. 2023, 891, 164424. [Google Scholar] [CrossRef] [PubMed]
- Mi, C.; Ma, L.; Wang, Y.; Wu, D.; Du, W.; Sun, B. Temperate and tropical lizards are vulnerable to climate warming due to increased water loss and heat stress. Proc. R. Soc. B 2022, 289, 20221074. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.P.; Tu, M.C. Cold tolerance and altitudinal distribution of Takydromus lizards in Taiwan. Zool. Stud. 2008, 47, 438–444. [Google Scholar]
- Capraro, A.; O’Meally, D.; Waters, S.A.; Patel, H.R.; Georges, A.; Waters, P.D. Waking the sleeping dragon: Gene expression profiling reveals adaptive strategies of the hibernating reptile Pogona vitticeps. BMC Genom. 2019, 20, 460. [Google Scholar] [CrossRef]
- Sun, B.J.; Li, T.; Gao, J.; Ma, L.; Du, W.G. High incubation temperatures enhance mitochondrial energy metabolism in reptile embryos. Sci. Rep. 2015, 5, 8861. [Google Scholar] [CrossRef]
- Campbell-Staton, S.C.; Cheviron, Z.A.; Rochette, N.; Catchen, J.; Losos, J.B.; Edwards, S.V. Winter storms drive rapid phenotypic, regulatory, and genomic shifts in the green anole lizard. Science 2017, 357, 495–498. [Google Scholar] [CrossRef] [PubMed]
- Yatsu, R.; Miyagawa, S.; Kohno, S.; Parrott, B.B.; Yamaguchi, K.; Ogino, Y.; Miyakawa, H.; Lowers, R.H.; Shigenobu, S.; Guillette, L.J. RNA-seq analysis of the gonadal transcriptome during Alligator mississippiensis temperature-dependent sex determination and differentiation. BMC Genom. 2016, 17, 77. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.; Pan, Y.; Liu, W.; Xie, Y.; Hao, W.; Xu, P.; Wang, Y. Acute temperature adaptation mechanisms in the native reptile species Eremias argus. Sci. Total Environ. 2022, 818, 151773. [Google Scholar] [CrossRef] [PubMed]
- Landman, M.; Schoeman, D.S.; Kerley, G.I. Shift in black rhinoceros diet in the presence of elephant: Evidence for competition? PLoS ONE 2013, 8, e69771. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, S.; Li, L.; Zhang, F.; Han, X.; Bi, J.; Sun, B. Thermal-physiological strategies underlying the sympatric occurrence of three desert lizard species. Asian Herpetol. Res. 2019, 10, 190–196B. [Google Scholar]
- Osellame, L.D.; Blacker, T.S.; Duchen, M.R. Cellular and molecular mechanisms of mitochondrial function. Best Pract. Res. Clin. Endocrinol. Metab. 2012, 26, 711–723. [Google Scholar] [CrossRef] [PubMed]
- Alvanou, M.V.; Apostolidis, A.P.; Lattos, A.; Michaelidis, B.; Giantsis, I.A. The coding mitogenome of the freshwater crayfish Pontastacus leptodactylus (Decapoda: Astacidea: Astacidae) from Lake Vegoritida, Greece and its taxonomic classification. Genes 2023, 14, 494. [Google Scholar] [CrossRef]
- Amer, S.A.; Kumazawa, Y. The mitochondrial genome of the lizard Calotes versicolor and a novel gene inversion in South Asian draconine agamids. Mol. Biol. Evol. 2007, 24, 1330–1339. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.J.; Wu, Q.Q.; Wang, Y.M.; Guo, K.; Ding, G.H.; Luo, S.T. The first complete mitochondrial DNA of the Chinese short-limbed skink (Ateuchosaurus chinensis Gray, 1845) determined by next-generation sequencing. Mitochondrial DNA B Resour. 2021, 6, 995–996. [Google Scholar] [CrossRef]
- Lajmi, A.; Giri, V.B.; Karanth, K.P. Molecular data in conjunction with morphology help resolve the Hemidactylus brookii complex (Squamata: Gekkonidae). Org. Diver. Evol. 2016, 16, 659–677. [Google Scholar] [CrossRef]
- Friedman, J.R.; Nunnari, J. Mitochondrial form and function. Nature 2014, 505, 335–343. [Google Scholar] [CrossRef]
- Storey, J.M.; Wu, S.; Storey, K.B. Mitochondria and the frozen frog. Antioxidants 2021, 10, 543. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Luu, B.E.; Yu, D.N.; Zhang, L.P.; Al-attar, R.; Storey, K.B. The complete mitochondrial genome of Dryophytes versicolor: Phylogenetic relationship among Hylidae and mitochondrial protein-coding gene expression in response to freezing and anoxia. Int. J. Biol. Macromol. 2019, 132, 461–469. [Google Scholar] [CrossRef]
- Healy, T.M.; Bryant, H.J.; Schulte, P.M. Mitochondrial genotype and phenotypic plasticity of gene expression in response to cold acclimation in killifish. Mol. Ecol. 2017, 26, 814–830. [Google Scholar] [CrossRef]
- Cai, L.N.; Zhang, L.H.; Lin, Y.J.; Wang, J.Y.; Storey, K.B.; Zhang, J.Y.; Yu, D.N. Two-fold ND5 genes, three-fold control regions, incRNA, and the “missing” ATP8 found in the mitogenomes of polypedates megacephalus (Rhacophridae: Polypedates). Animals 2023, 13, 2857. [Google Scholar] [CrossRef] [PubMed]
- Teplitsky, C.; Mills, J.A.; Alho, J.S.; Yarrall, J.W.; Merilä, J. Bergmann’s rule and climate change revisited: Disentangling environmental and genetic responses in a wild bird population. Proc. Natl. Acad. Sci. USA 2008, 105, 13492–13496. [Google Scholar] [CrossRef]
- Einum, S.; Burton, T. Divergence in rates of phenotypic plasticity among ectotherms. Ecol. Lett. 2023, 26, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Kirigin, A.J.; Naya, D.E. Latitudinal patterns in phenotypic plasticity: The case of seasonal flexibility in lizards’ fat body size. Oecologia 2013, 173, 745–752. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Pesquera, L.M.; Tejedo, M.; Olalla-Tárraga, M.; Duarte, H.; Nicieza, A.; Solé, M. Testing the climate variability hypothesis in thermal tolerance limits of tropical and temperate tadpoles. J. Biogeogr. 2016, 43, 1166–1178. [Google Scholar] [CrossRef]
- Pallotta, M.M.; Fogliano, C.; Carotenuto, R. Temperature incubation influences gonadal gene expression during Leopard gecko development. Animals 2022, 12, 3186. [Google Scholar] [CrossRef]
- Wohlgemuth, R.P.; Haro, D.; Liwanag, H.E. Osmotic and metabolic responses to cold acclimation and acute cold challenge in a freeze avoidant lizard, Podarcis siculus. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2023, 283, 111471. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Jiang, Z.; Wang, J.; Cui, L.; Zhang, M. The complete mitochondrial genome of the blue-crested lizard, Calotes mystaceus (Squamata, Agamidae) in China. Mitochondrial DNA B Resour. 2020, 5, 3512–3513. [Google Scholar] [CrossRef] [PubMed]
- Zizka, V.M.; Geiger, M.F.; Hörren, T.; Kirse, A.; Noll, N.W.; Schäffler, L.; Scherges, A.M.; Sorg, M. Repeated subsamples during DNA extraction reveal increased diversity estimates in DNA metabarcoding of Malaise traps. Ecol. Evol. 2022, 12, e9502. [Google Scholar] [CrossRef] [PubMed]
- Kumazawa, Y.; Endo, H. Mitochondrial genome of the Komodo dragon: Efficient sequencing method with reptile-oriented primers and novel gene rearrangements. DNA Res. 2004, 11, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Lalitha, S. Primer premier 5. Biotech Softw. Internet Rep. 2000, 1, 270–272. [Google Scholar] [CrossRef]
- Velo-Antón, G.; Henrique, M.; Liz, A.V.; Martínez-Freiría, F.; Pleguezuelos, J.M.; Geniez, P.; Crochet, P.-A.; Brito, J.C. DNA barcode reference library for the West Sahara-Sahel reptiles. Sci. Data 2022, 9, 459. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, S.; Basumatary, P.; Nath, D.; Paul, S.; Uddin, A. Compositional features and pattern of codon usage for mitochondrial CO genes among reptiles. Mitochondrion 2022, 62, 111–121. [Google Scholar] [CrossRef]
- Sakuma, T.; Takenaga, M.; Kawabe, Y.; Nakamura, T.; Kamihira, M.; Yamamoto, T. Homologous recombination-independent large gene cassette knock-in in CHO cells using TALEN and MMEJ-directed donor plasmids. Int. J. Mol. Sci. 2015, 16, 23849–23866. [Google Scholar] [CrossRef]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef] [PubMed]
- Nassief, A.M. Conventional data science techniques to bioinformatics and utilizing a grid computing approach to computational medicine. Preprints 2020, 2020010274. [Google Scholar] [CrossRef]
- Smith, L.B.; Anderson, C.V.; Withangage, M.H.H.; Koch, A.; Roberts, T.J.; Liebl, A.L. Relationship between gene expression networks and muscle contractile physiology differences in Anolis lizards. J. Comp. Physiol. B 2022, 192, 489–499. [Google Scholar] [CrossRef]
- Wittmeier, P.; Hummel, S. Agarose gel electrophoresis to assess PCR product yield: Comparison with spectrophotometry, fluorometry and qPCR. Biotechniques 2022, 72, 155–158. [Google Scholar] [CrossRef]
- Booze, M.L.; Eyster, K.M. Extraction of RNA and analysis of estrogen-responsive genes by RT-qPCR. In Estrogen Receptors: Methods and Protocols; Springer: New York, NY, USA, 2022; pp. 113–127. [Google Scholar]
- Ji, C.; Wang, M.; Wen, X. Cloning of mouse β-actin gene. Anat. Sci. 2018, 1, 298. [Google Scholar] [CrossRef]
- Chang, J.; Pan, Y.; Yang, L.; Xie, Y.; Xu, P.; Wang, H. Environmental relevant concentration of λ-cyhalothrin and 3-phenoxybenzoic acid caused endocrine-disrupting effects on male lizards (Eremias argus). Environ. Pollut. 2020, 265, 115077. [Google Scholar] [CrossRef]
- Wang, J.Y.; Zhang, L.H.; Hong, Y.H.; Cai, L.N.; Storey, K.B.; Zhang, J.Y.; Zhang, S.S.; Yu, D.N. How does mitochondrial protein-coding gene expression in Fejervarya kawamurai (Anura: Dicroglossidae) respond to extreme temperatures? Animals 2023, 13, 3015. [Google Scholar] [CrossRef]
- Moeller, A.H.; Ivey, K.; Cornwall, M.B.; Herr, K.; Rede, J.; Taylor, E.N.; Gunderson, A.R. The lizard gut microbiome changes with temperature and is associated with heat tolerance. Appl. Environ. Microbiol. 2020, 86, e01181-20. [Google Scholar] [CrossRef]
- May, R.A.; Stevenson, K.J. Software review of Origin 8. J. Am. Chem. Soc. 2009, 131, 872. [Google Scholar] [CrossRef]
- Münch, C. The different axes of the mammalian mitochondrial unfolded protein response. BMC Biol. 2018, 16, 81. [Google Scholar] [CrossRef]
- Montaña-Lozano, P.; Balaguera-Reina, S.A.; Prada-Quiroga, C.F. Comparative analysis of codon usage of mitochondrial genomes provides evolutionary insights into reptiles. Gene 2023, 851, 146999. [Google Scholar] [CrossRef]
- James, J.E.; Piganeau, G.; Eyre-Walker, A. The rate of adaptive evolution in animal mitochondria. Mol. Ecol. 2016, 25, 67–78. [Google Scholar] [CrossRef]
- Formosa, L.E.; Dibley, M.G.; Stroud, D.A.; Ryan, M.T. Building a complex complex: Assembly of mitochondrial respiratory chain complex I. Semin. Cell Dev. Biol. 2018, 76, 154–162. [Google Scholar] [CrossRef]
- Apweiler, R.; Bairoch, A.; Wu, C.H.; Barker, W.C.; Boeckmann, B.; Ferro, S.; Gasteiger, E.; Huang, H.; Lopez, R.; Magrane, M. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2004, 32, D115–D119. [Google Scholar] [CrossRef]
- Baranowska, E.; Niedzwiecka, K.; Panja, C.; Charles, C.; Dautant, A.; Poznanski, J.; di Rago, J.-P.; Tribouillard-Tanvier, D.; Kucharczyk, R. Probing the pathogenicity of patient-derived variants of MT-ATP6 in yeast. Dis. Models Mech. 2023, 16, dmm049783. [Google Scholar] [CrossRef]
- Okajima, Y.; Kumazawa, Y. Mitochondrial genomes of acrodont lizards: Timing of gene rearrangements and phylogenetic and biogeographic implications. BMC Evol. Biol. 2010, 10, 141. [Google Scholar] [CrossRef]
- Reinecke, F.; Smeitink, J.A.; van Der Westhuizen, F.H. OXPHOS gene expression and control in mitochondrial disorders. Biochim. Biophys. Acta. Mol. Basis. Dis. 2009, 1792, 1113–1121. [Google Scholar] [CrossRef]
- Jin, W.T.; Guan, J.Y.; Dai, X.Y.; Wu, G.J.; Zhang, L.P.; Storey, K.B.; Zhang, J.Y.; Zheng, R.Q.; Yu, D.N. Mitochondrial gene expression in different organs of Hoplobatrachus rugulosus from China and Thailand under low-temperature stress. BMC Zool. 2022, 7, 24. [Google Scholar] [CrossRef]
- Guppy, M.; Fuery, C.; Flanigan, J. Biochemical principles of metabolic depression. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 1994, 109, 175–189. [Google Scholar] [CrossRef]
- Mark, F.C.; Lucassen, M.; Strobel, A.; Barrera-Oro, E.; Koschnick, N.; Zane, L.; Patarnello, T.; Pörtner, H.O.; Papetti, C. Mitochondrial function in Antarctic nototheniids with ND6 translocation. PLoS ONE 2012, 7, e31860. [Google Scholar] [CrossRef][Green Version]
- Guderley, H.; Seebacher, F. Thermal acclimation, mitochondrial capacities and organ metabolic profiles in a reptile (Alligator mississippiensis). J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 2011, 181, 53–64. [Google Scholar] [CrossRef]
- Dos Santos, R.S.; Galina, A.; Da-Silva, W.S. Cold acclimation increases mitochondrial oxidative capacity without inducing mitochondrial uncoupling in goldfish white skeletal muscle. Biol. Open 2012, 2, 82–87. [Google Scholar] [CrossRef]
- O’Brien, K.M. Mitochondrial biogenesis in cold-bodied fishes. J. Exp. Biol. 2011, 214, 275–285. [Google Scholar] [CrossRef]
- Wodtke, E. Temperature adaptation of biological membranes. Compensation of the molar activity of cytochrome c oxidase in the mitochondrial energy-transducing membrane during thermal acclimation of the carp (Cyprinus carpio L.). Biochim. Biophys. Acta. Biomembr. 1981, 640, 710–720. [Google Scholar] [CrossRef]
- El-Banna, A.; Al-Johany, A. Effect of cold and hot temperature on behavioral and selected physiological measures of Uromastyx aegyptius (Agamidae). Sultan Qaboos Univ. J. Sci. 2003, 8, 1–10. [Google Scholar] [CrossRef]
- Ulmasov, K.A.; Shammakov, S.; Karaev, K.; Evgen’ev, M.B. Heat shock proteins and thermoresistance in lizards. Proc. Natl. Acad. Sci. USA 1992, 89, 1666–1670. [Google Scholar] [CrossRef] [PubMed]
- Feiner, N.; Rago, A.; While, G.M.; Uller, T. Developmental plasticity in reptiles: Insights from temperature-dependent gene expression in wall lizard embryos. J. Exp. Zool. Part A 2018, 329, 351–361. [Google Scholar] [CrossRef] [PubMed]
- Scudiero, R.; Verderame, M.; Motta, C.M.; Migliaccio, V.; Simoniello, P. HSP70 localization in Podarcis siculus embryos under natural thermal regime and following a non-lethal cold shock. Comptes Rendus Biol. 2019, 342, 299–308. [Google Scholar] [CrossRef] [PubMed]
- Brattstrom, B.H. Body temperatures of reptiles. Am. Midl. Nat. 1965, 73, 376–422. [Google Scholar] [CrossRef]
- Lin, T.E.; Chen, T.Y.; Wei, H.L.; Richard, R.; Huang, S.P. Low cold tolerance of the invasive lizard Eutropis multifasciata constrains its potential elevation distribution in Taiwan. J. Therm. Biol. 2019, 82, 115–122. [Google Scholar] [CrossRef]
- Overgaard, J.; Kristensen, T.N.; Mitchell, K.A.; Hoffmann, A.A. Thermal tolerance in widespread and tropical Drosophila species: Does phenotypic plasticity increase with latitude? Am. Nat. 2011, 178, S80–S96. [Google Scholar] [CrossRef]
- Cowles, R.B.; Bogert, C.M. Preliminary study of the thermal requirements of desert reptiles. Bull. Am. Mus. Nat. Hist. 2006, 13, 53–60. [Google Scholar]
- McMahan, C.D.; Zug, G.R. Burmese Hemidactylus (Reptilia, Squamata, Gekkonidae): Geographic variation in the morphology of Hemidactylus bowringii in Myanmar and Yunnan. Proc. Calif. Acad. Sci. 2007, 58, 485–509. [Google Scholar]
- Lazell, J.; Kolby, J.; Lin, Y.M.; Zhuang, D.H.; Lu, W. Reptiles and amphibians from Nan Ao island, China. Postilla 1999, 217, 3–18. [Google Scholar]
- Guo, C.; Zhong, M.; Wang, X.; Yang, S.; Tang, K.; Jia, L.; Zhang, C.; Hu, J. An updated species checklist of amphibians and reptiles in Fujian Province, China. Biodivers. Sci. 2022, 30, 22090. [Google Scholar] [CrossRef]
- Gawor, A.; Nguyen, T.Q.; Nguyen, T.T.; Schmitz, A.; Ziegler, T. The herpetofauna of the Bai Tu Long National Park, northeastern Vietnam. Salamandra 2016, 52, 23–41. [Google Scholar]
- Mazzotti, F.J.; Cherkiss, M.S.; Parry, M.; Beauchamp, J.; Rochford, M.; Smith, B.; Hart, K.; Brandt, L.A. Large reptiles and cold temperatures: Do extreme cold spells set distributional limits for tropical reptiles in Florida? Ecosphere 2016, 7, e01439. [Google Scholar] [CrossRef]
- Huang, S.-P.; Hsu, Y.; Tu, M.-C. Thermal tolerance and altitudinal distribution of two Sphenomorphus lizards in Taiwan. J. Therm. Biol. 2006, 31, 378–385. [Google Scholar] [CrossRef]
- Storey, K.B. Reptile freeze tolerance: Metabolism and gene expression. Cryobiology 2006, 52, 1–16. [Google Scholar] [CrossRef]
- Munoz, M.M.; Stimola, M.A.; Algar, A.C.; Conover, A.; Rodriguez, A.J.; Landestoy, M.A.; Bakken, G.S.; Losos, J.B. Evolutionary stasis and lability in thermal physiology in a group of tropical lizards. Proc. R. Soc. B 2014, 281, 20132433. [Google Scholar] [CrossRef] [PubMed]
- Schmid, M.; Dallo, R.; Guillaume, F. Species’ range dynamics affect the evolution of spatial variation in plasticity under environmental change. Am. Nat. 2019, 193, 798–813. [Google Scholar] [CrossRef]
- Wu, J. Can changes in the distribution of lizard species over the past 50 years be attributed to climate change? Theor. Appl. Climato. 2016, 125, 785–798. [Google Scholar] [CrossRef]
- Cosendey, B.N.; Rocha, C.F.D.; Menezes, V.A. Climate change, lizard populations, and species vulnerability/persistence: Trends in ecological and predictive climate studies. Environ. Dev. Sustain. 2023, 25, 8929–8950. [Google Scholar] [CrossRef]
- Caldwell, A.J.; While, G.M.; Beeton, N.J.; Wapstra, E. Potential for thermal tolerance to mediate climate change effects on three members of a cool temperate lizard genus, Niveoscincus. J. Therm. Biol. 2015, 52, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Williams, C.M.; Li, T.; Speakman, J.R.; Jin, Z.; Lu, H.; Luo, L.; Du, W. Higher metabolic plasticity in temperate compared to tropical lizards suggests increased resilience to climate change. Ecol. Monogr. 2022, 92, e1512. [Google Scholar] [CrossRef]
- Bonino, M.F.; Azócar, D.L.M.; Schulte II, J.A.; Abdala, C.S.; Cruz, F.B. Thermal sensitivity of cold climate lizards and the importance of distributional ranges. Zoology 2015, 118, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Vicenzi, N.; Corbalán, V.; Miles, D.; Sinervo, B.; Ibargüengoytía, N. Range increment or range detriment? Predicting potential changes in distribution caused by climate change for the endemic high-Andean lizard Phymaturus palluma. Biol. Conserv. 2017, 206, 151–160. [Google Scholar] [CrossRef]
- Laspiur, A.; Santos, J.; Medina, S.M.; Pizarro, J.E.; Sanabria, E.A.; Sinervo, B.; Ibargüengoytía, N. Vulnerability to climate change of a microendemic lizard species from the central Andes. Sci. Rep. 2021, 11, 11653. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Nolasco, F.J.; Arenas-Moreno, D.M.; Gandarilla-Aizpuro, F.J.; Bautista-del Moral, A.; Santos-Bibiano, R.; Miles, D.B.; Méndez-de la Cruz, F.R. Physiological ecology and vulnerability to climate change of a microendemic, habitat-specialist lizard in a tropical dry forest of Mexico. Clim. Chang. Ecol. 2023, 5, 100066. [Google Scholar] [CrossRef]
- Fordham, D.A.; Watts, M.J.; Delean, S.; Brook, B.W.; Heard, L.M.; Bull, C. Managed relocation as an adaptation strategy for mitigating climate change threats to the persistence of an endangered lizard. Glob. Chang. Biol. 2012, 18, 2743–2755. [Google Scholar] [CrossRef]
- Huang, C.; Yu, H.; Wu, Z.; Li, Y.; Wei, F.; Gong, M. Population and conservation strategies for the Chinese crocodile lizard (Shinisaurus crocodilurus) in China. Anim. Biodiv. Conserv. 2008, 31, 63–70. [Google Scholar] [CrossRef]
- Kacoliris, F.P.; Velasco, M.A.; Kass, C.; Kass, N.; Simoy, V.; Grilli, P.G.; Aguirre, T.M.; Di Pietro, D.O.; Williams, J.D.; Berkunsky, I. A management strategy for the long-term conservation of the Endangered sand-dune lizard Liolaemus multimaculatus in the Pampean coastal dunes of Argentina. Oryx 2019, 53, 561–569. [Google Scholar] [CrossRef]
- Brito, J.C.; Godinho, R.; Luís, C.; Paulo, O.S.; Crespo, E.G. Management strategies for conservation of the lizard Lacerta schreiberi in Portugal. Biol. Conserv. 1999, 89, 311–319. [Google Scholar] [CrossRef]
- Martín, J.; Lopez, P. The effect of Mediterranean dehesa management on lizard distribution and conservation. Biol. Conserv. 2002, 108, 213–219. [Google Scholar] [CrossRef]
| No. | Primer Name | Nucleotide Sequence (5′ to 3′) | Amplicon Size (bp) |
|---|---|---|---|
| 1 | xiyi-12S-J505 | ACAAACTAGGATTAGATACCC | 730 |
| xiyi-12S-N1225 | CANBTTTCCCTTGCGGTACT | ||
| 2 | 16SL | AACCCYYGTACCTYTTGCATCATG | 886 |
| 16SH | TCCACAGGGTCTTYTCGTC | ||
| 3 | ND1-ND2L | CGATTTCGCTATGACCAACT | 891 |
| ND1-ND2H | ATTGATGAGWAKGCTATRATTTTTCG | ||
| 4 | ND2-CO1L | GCCCCMYTMCACTTCTGA | 1142 |
| ND2-CO1H | GTAHAGGGTGCCRATRTCTTT | ||
| 5 | SX-ND5-ND6-J | GARCARGACCTYCGACTAATRGG | 1067 |
| SX-ND5-ND6-N | ATATTAGTAGTGTTTGTSTATAC | ||
| 6 | CO1-CO2L | TACTCAGACTACCCAGAYGC | 971 |
| CO1-CO2H | CCRCARATTTCTGAGCATTG | ||
| 7 | ND4-CUNL | CCMAAAGCCCAYGTAGAAGC | 909 |
| ND4-CUNH | CTTHTACTTGGADTTGCACC | ||
| 8 | SX-ND5-GLU-J | YTYATTAACGCCTGAGCCTT | 805 |
| SX-ND5-GLU-N | ATAACAACGAYGGTTTTTC | ||
| 9 | CYTB-ProL | TGAGGACAAATATCMTTCTGAGG | 861 |
| CYTB-ProH | TTAAAATKCTAGTTTTGG | ||
| 10 | Thr-CRL | YAAAGCMTTGRTCTTGTAA | 1635 |
| Thr-CRH | CTCGAKTTTWGGGGTTTGRCGA | ||
| 11 | CR-12SL | TCGYCAAACCCCWAAAMCGAG | 627 |
| CR-12SH | TRTAACCGCGGTKGCTGGCAC |
| Primer Name | Forward Primers (5′ to 3′) | Reverse Primers (5′ to 3′) | Amplicon Size (bp) |
|---|---|---|---|
| DLLX-β-ACTIN | GCTCTGCTATGTTGCCCTTG | ACCTGAACCGCTCATTACCA | 125 |
| DLLX-ND1 | TCCTTCTTAGTAGCCGTAGCA | TCCGTCTGCCATTGGTTGA | 123 |
| DLLX-ND2 | CCTCATGCCTGCTTCTCCTA | GGTTAGTAGTGTGCTGCCTTG | 182 |
| DLLX-ND3 | GCCCTACGAATGCGGATTT | ACAGTTGGTGTTGGTGCTAG | 156 |
| DLLX-ND4 | CTAACCAACCTGGCACTTCC | GGTGGCTGAGGCAATAATTGT | 105 |
| DLLX-ND4L | CTAACACTAAATACCCCCCAC | AGAAGAGGTGACGAGGAGTGT | 102 |
| DLLX-ND5 | GCCACAGCAGGAAGTCTTCT | GCTTGGAGTGCGGATGAGT | 119 |
| DLLX-ND6 | CGGTGGCGTGTTATTATTCG | ACCAGCACCAACAATTAGGAG | 173 |
| DLLX-ATP6 | CGCCTGACCGCTAACCTAA | TGGATGAGTGCTACGGCTATT | 155 |
| DLLX-ATP8 | GCAACTTAACCCAAACCCAT | TTGGATTTGTGGTTGTGGTG | 104 |
| DLLX-COI | CTTGTGAGCCTTCTTGTACGA | TTCCGAAGCCGCCGATTA | 145 |
| DLLX-COII | CACGACTACGCCATAACTACC | TCGGTTAGTACGGTGGTGAA | 101 |
| DLLX-COIII | GCCAATTCTAGCCGCCATATC | TGTGCCTTCACGAATGATGTC | 151 |
| DLLX-CYTB | TAGCCGCCTCAGTCCTAATC | CCGCTTGGTTGTCCTCCTA | 151 |
| DLGX-β-ACTIN | AAGGAGAAGCTGTGCTATGTG | AGGAAGGAAGGCTGGAAGAG | 164 |
| DLGX-ND1 | GACCATCCTCCTCTTCACCAA | AGTAGACCGCAGTGCTTGATA | 159 |
| DLGX-ND2 | CCGAGCAACAGAAGCAACAA | AAGTATCGTACAGGCGTAGGG | 145 |
| DLGX-ND3 | AACCCTCCCAGACACAGAAA | GCAAGAATAGGATGGCGACTA | 113 |
| DLGX-ND4 | CCTTCTCCGCCACAGACTT | TAGCCGTTCAGCTTGATTACC | 104 |
| DLGX-ND4L | GCTGCCTACCAACACAATGT | GTTCTGTATGTGGTCGGTTCC | 121 |
| DLGX-ND5 | CAAGACCGCCTTATCACTCTG | TGCTAGTTGTGGTTGGTTGAG | 166 |
| DLGX-ND6 | GGACCCGTATCCTGAGACT | GCACGAATCAACCCAAATC | 162 |
| DLGX-ATP6 | TACCAGAAGGCACTCCTACAC | GCTGTGAGGTTGGCAGTTAG | 112 |
| DLGX-ATP8 | ATGCCACAACTAAATCCCG | TTGATTTGGGTTGAGGCTG | 104 |
| DLGX-COI | GCTCCACGACACTTACTACG | GCCTGCGAATATTACTCCGAA | 163 |
| DLGX-COII | CAGACTACGAGGACCTGTTGT | ACGGCTCACGAGTGGAGAA | 163 |
| DLGX-COIII | GGCTTCGCTACGGAATAGTC | GTTAATGCCGCTTGGAGGTC | 134 |
| DLGX-CYTB | CCTTGTCATAGCCACAGCATT | CGCCTCAGATCCACTCTACTA | 137 |
| DLXH-β-ACTIN | GAGGGAGATTGTGCGGGATAT | AGGAAGGACGGCTGGAAGA | 186 |
| DLXH-ND1 | TCGCCGTAGCATTCCTGAC | GTTGTTGGTCGTGTTGGTTCT | 151 |
| DLXH-ND2 | CACCATACCACCAGCACTAAT | TAAGGCAACCAGGAGTCACC | 142 |
| DLXH-ND3 | TCCCATTCTCAATACGCTTCTT | TGTTGTAAGGTGTAGTGTTGTG | 131 |
| DLXH-ND4 | AGCCTGTATAGCCGCACTAC | GATGATTCCGTATCCGCCAAG | 142 |
| DLXH-ND4L | AACTATAAGCACCACCACAGC | TTAGGTTGTCCGAGGCGTAT | 125 |
| DLXH-ND5 | ATCCGCACCTACCACGATTC | CGGTTGAAGATTACGGCTTGAA | 196 |
| DLXH-ND6 | GGGATGCTTGTTGTGTTTGC | AACCACCGCCTCCATTACA | 175 |
| DLXH-ATP6 | ATCGCACAACAGCACTACAG | GGTGAAGGTATATGGCAGGAG | 161 |
| DLXH-ATP8 | TATAACTGCTGTCGTCACCTG | CTGATTGTGTTGATGGGTTTGG | 101 |
| DLXH-COI | CTCGCCGCTACTCTGACTAC | GCTGAGAAGTGTGGTTGATGTT | 158 |
| DLXH-COII | CCGCCTCACCAACCATAGA | GGGCAGCACAGTTCAAATAGT | 173 |
| DLXH-COIII | GCAAGCGATAGAGTACGGAGA | CCAGGCATACAGTGAGGAATG | 131 |
| DLXH-CYTB | ACCACCACATATTAAGCCAGAG | GCGACTGATATAAGGAGTGCTA | 105 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, J.; Zhan, L.; Meng, S.; Wang, Z.; Gao, L.; Wang, W.; Storey, K.B.; Zhang, Y.; Yu, D. Differential Mitochondrial Genome Expression of Three Sympatric Lizards in Response to Low-Temperature Stress. Animals 2024, 14, 1158. https://doi.org/10.3390/ani14081158
He J, Zhan L, Meng S, Wang Z, Gao L, Wang W, Storey KB, Zhang Y, Yu D. Differential Mitochondrial Genome Expression of Three Sympatric Lizards in Response to Low-Temperature Stress. Animals. 2024; 14(8):1158. https://doi.org/10.3390/ani14081158
Chicago/Turabian StyleHe, Jingyi, Lemei Zhan, Siqi Meng, Zhen Wang, Lulu Gao, Wenjing Wang, Kenneth B. Storey, Yongpu Zhang, and Danna Yu. 2024. "Differential Mitochondrial Genome Expression of Three Sympatric Lizards in Response to Low-Temperature Stress" Animals 14, no. 8: 1158. https://doi.org/10.3390/ani14081158
APA StyleHe, J., Zhan, L., Meng, S., Wang, Z., Gao, L., Wang, W., Storey, K. B., Zhang, Y., & Yu, D. (2024). Differential Mitochondrial Genome Expression of Three Sympatric Lizards in Response to Low-Temperature Stress. Animals, 14(8), 1158. https://doi.org/10.3390/ani14081158





