Identification of Growth-Related Gene BAMBI and Analysis of Gene Structure and Function in the Pacific White Shrimp Litopenaeus vannamei
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Animal
2.2. Screening for Growth Trait-Related Genes
2.3. Analysis of Gene Structure
2.4. Prediction of Transcription Factors
2.5. Gene Expression Pattern Analysis
2.6. In vivo RNA Interference
2.7. Real-Time Quantitative PCR
2.8. RNA-seq Analysis
2.9. Combined Metabolomic and Lipidomic Analyses
2.10. Statistical Analysis
3. Results
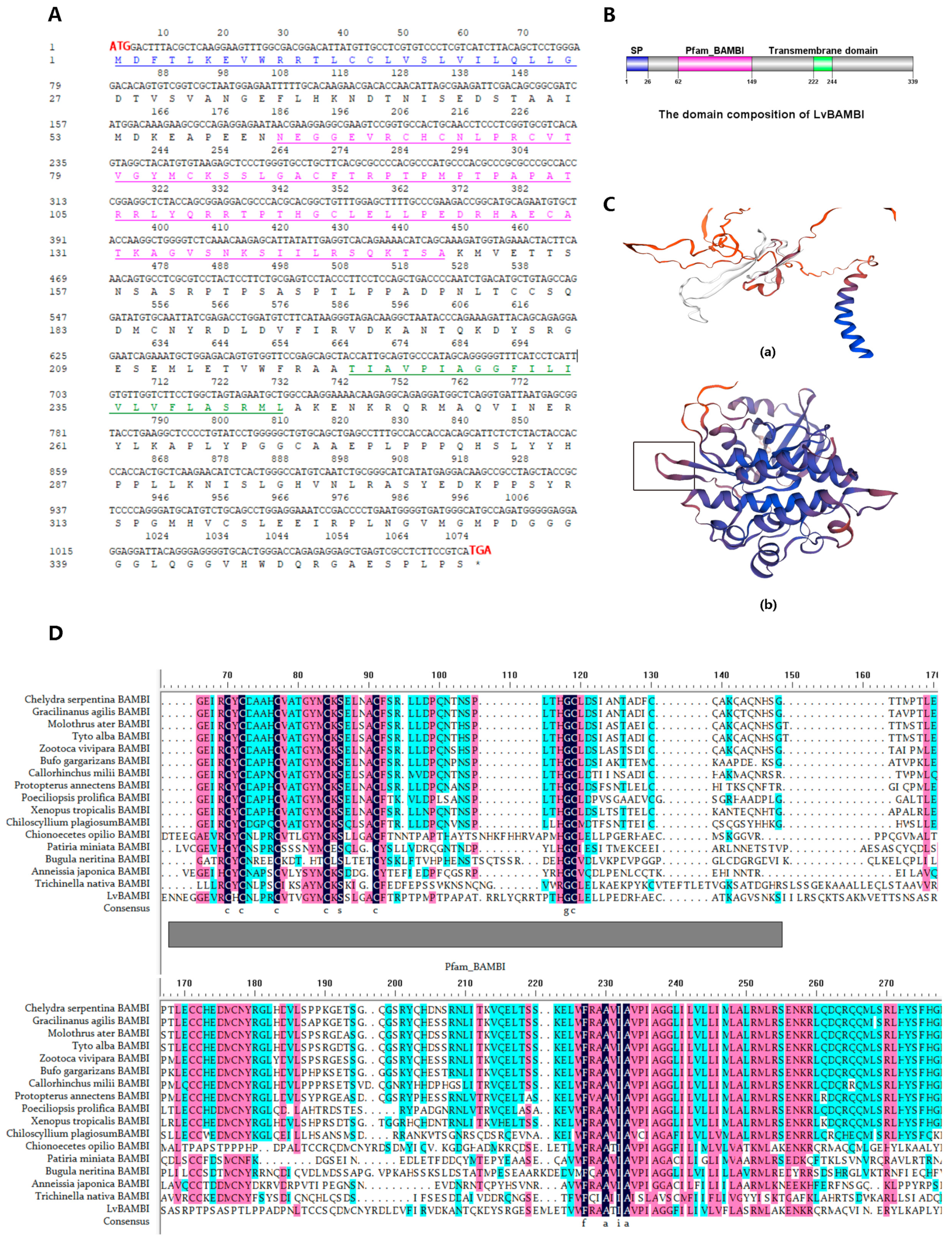
3.1. Identification of LvBAMBI
3.2. Structural Characterization of LvBAMBI
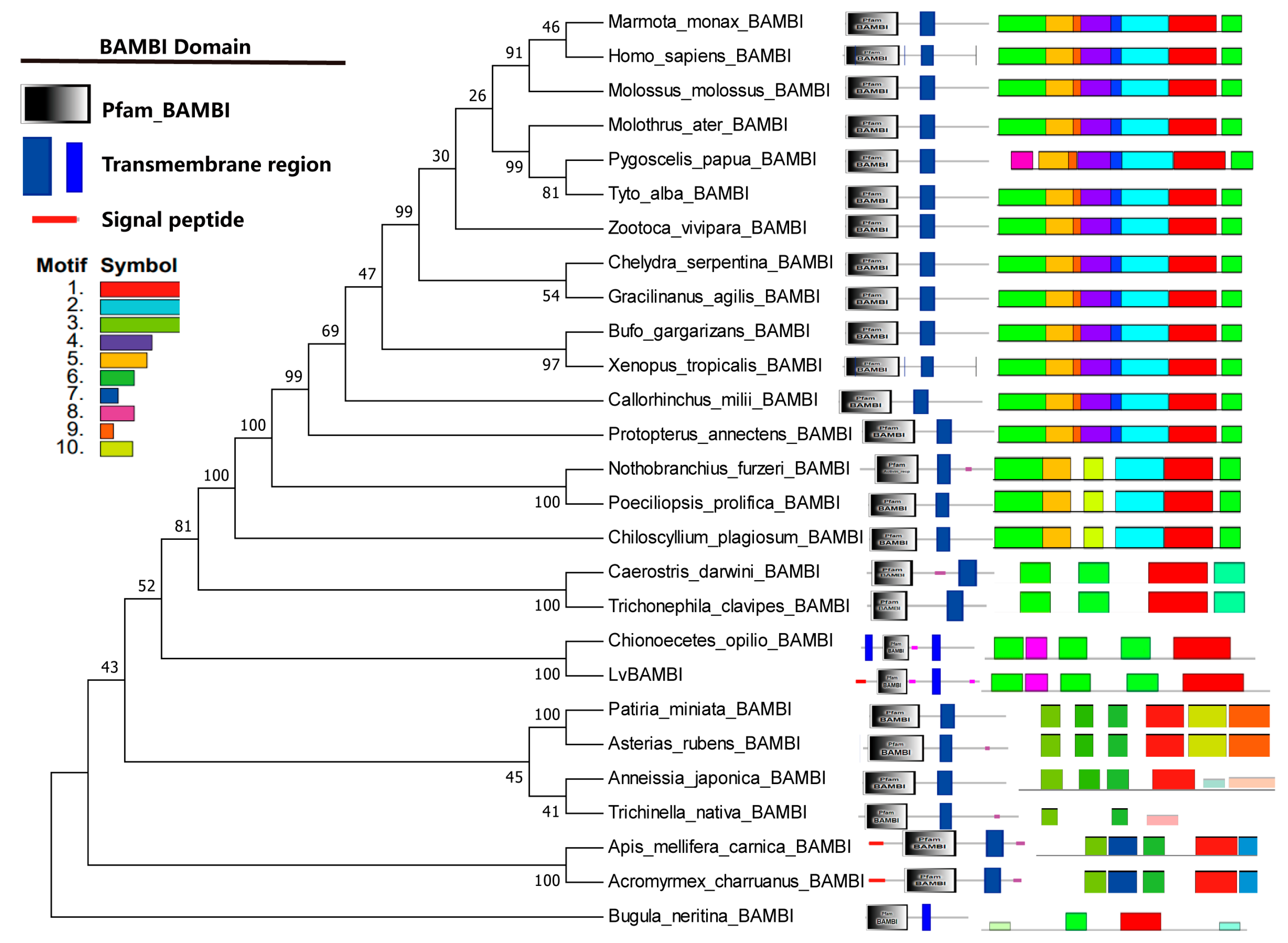
3.3. Phylogenetic Analysis of BAMBI Genes
3.4. Transcription Factor Prediction of LvBAMBI
3.5. Expression Pattern of LvBAMBI
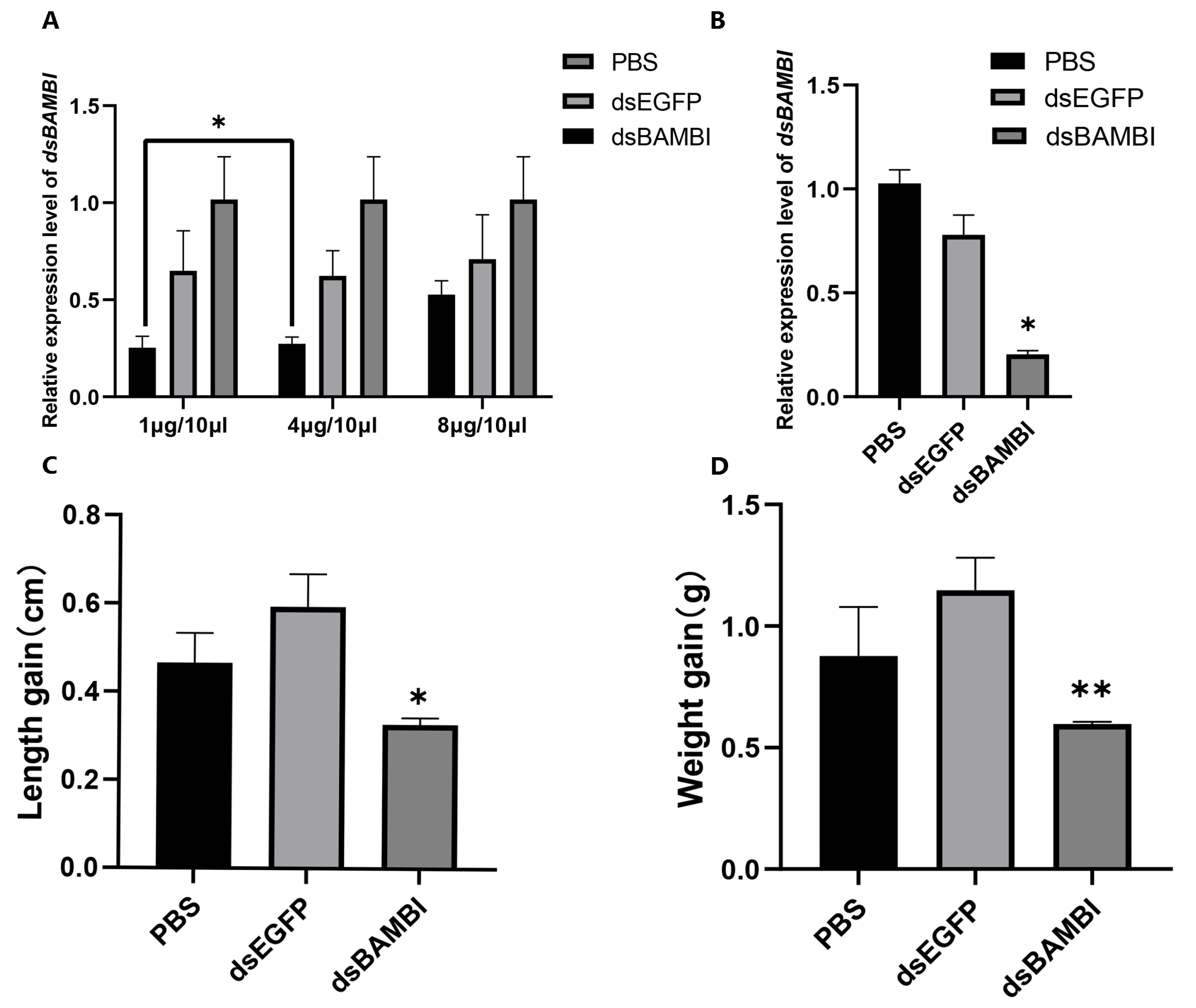
3.6. RNA Interference of LvBAMBI
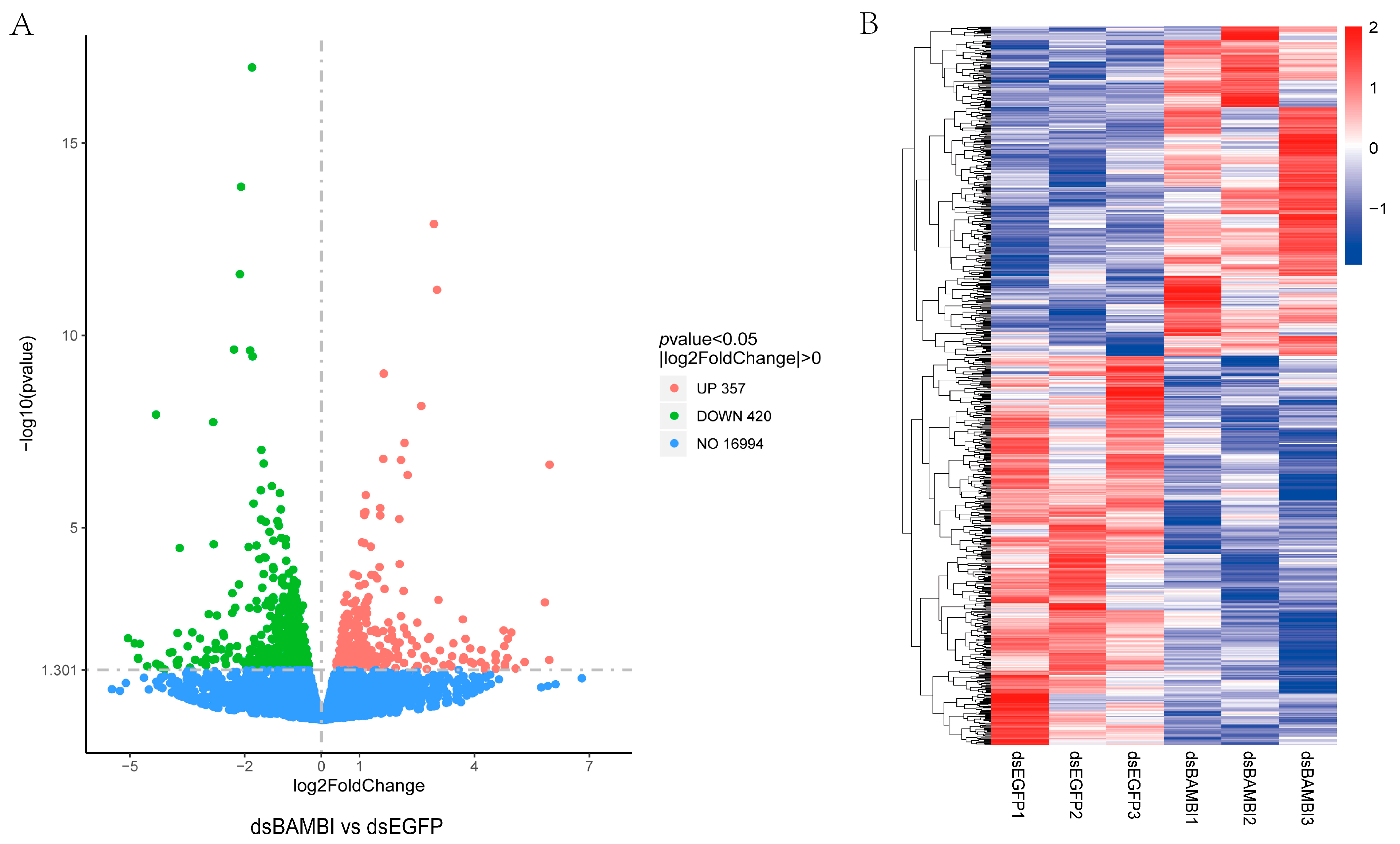
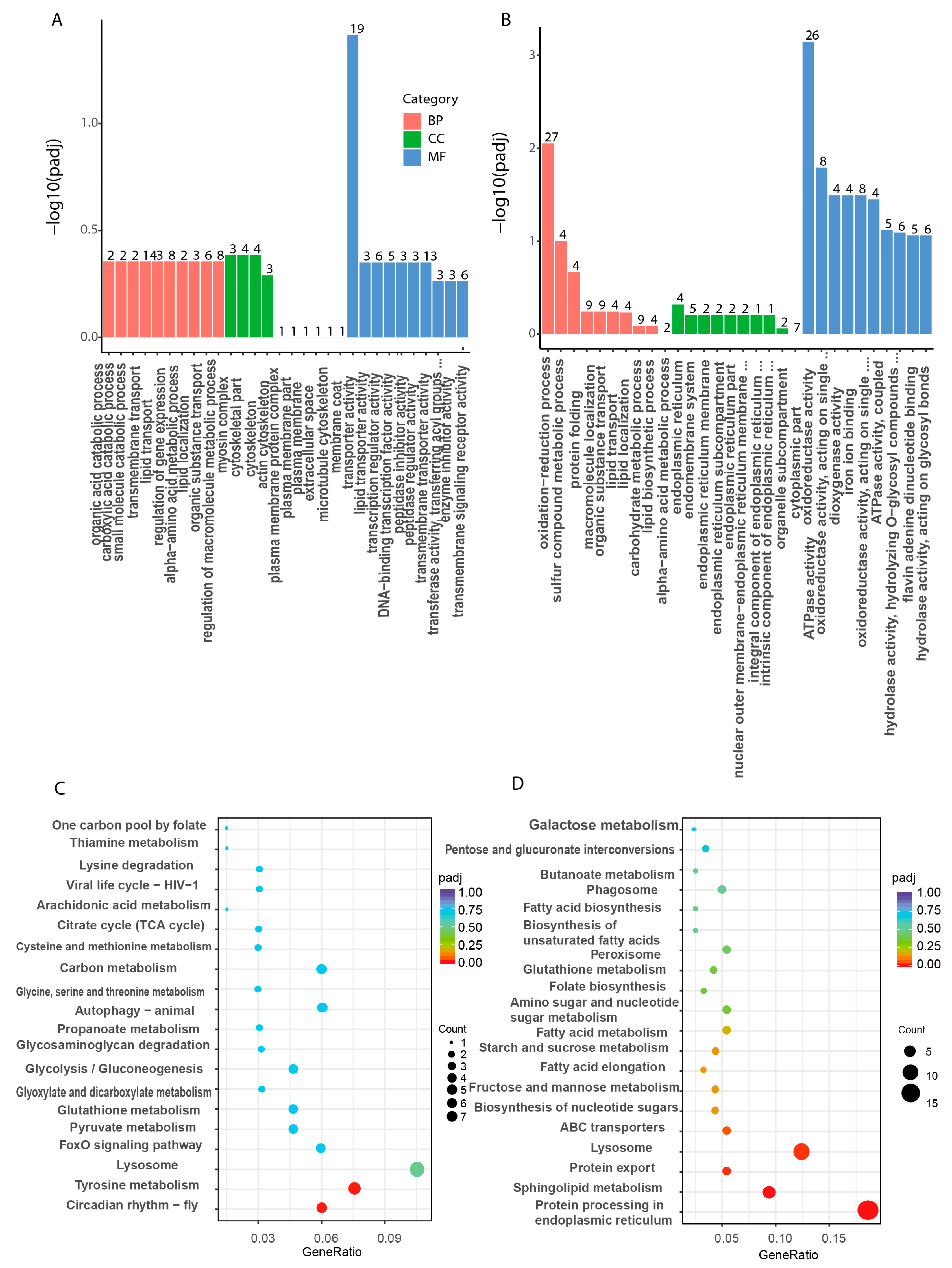
3.7. Gene Expression Changes after LvBAMBI Knockdown
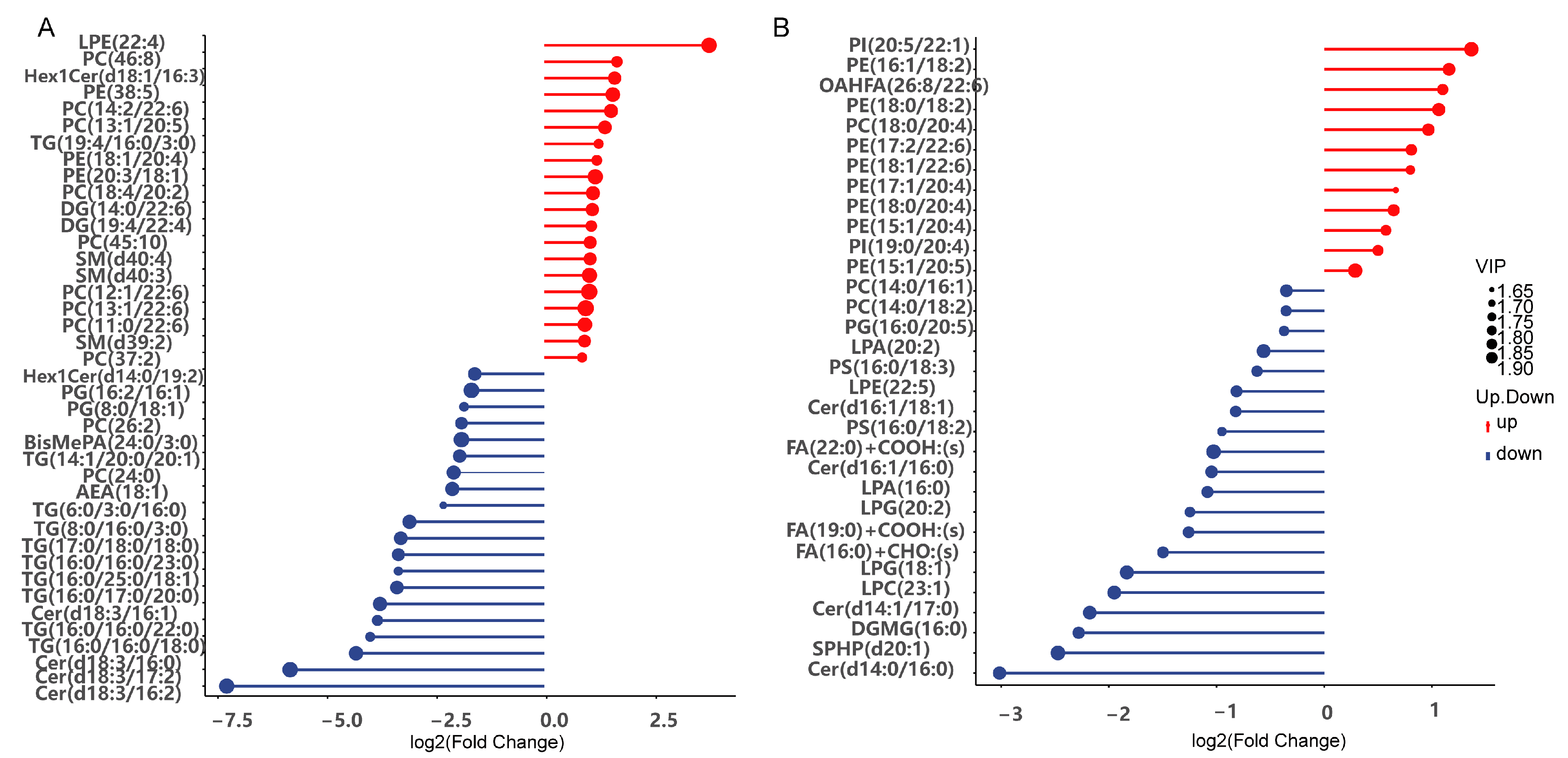
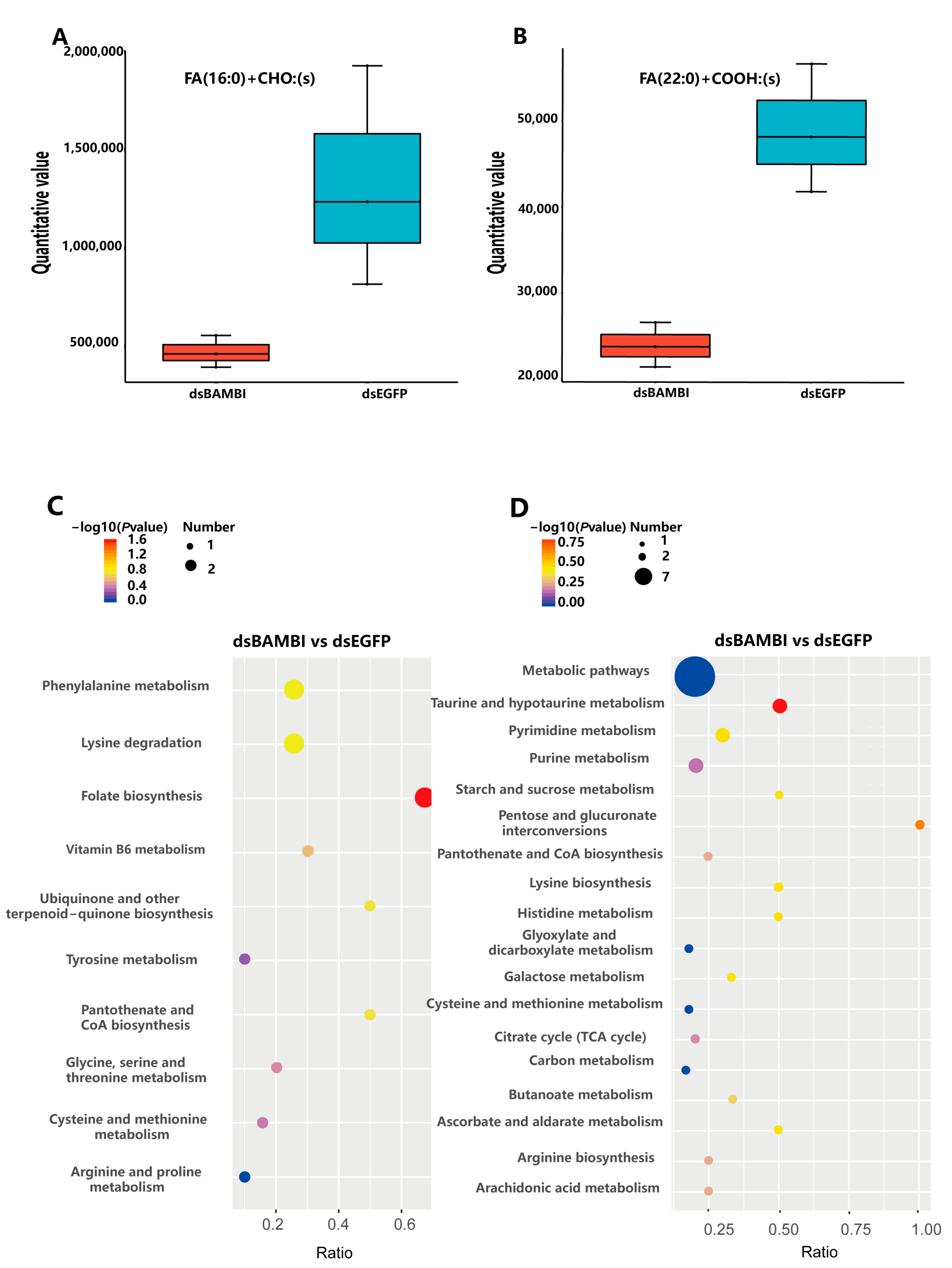
3.8. Effects of LvBAMBI Knockdown on Lipid Metabolism
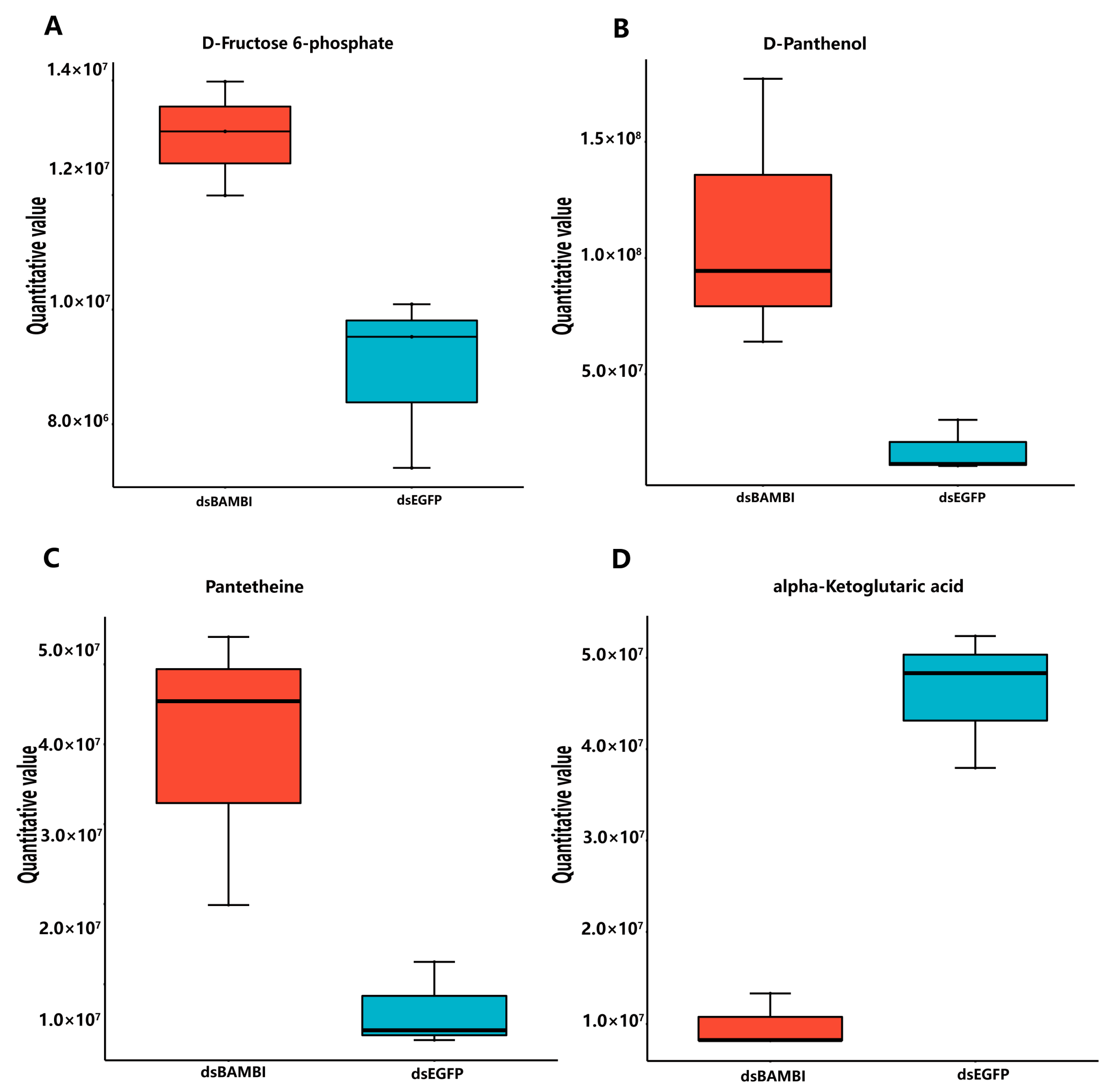
3.9. Effects of LvBAMBI Knockdown on Glucose Metabolism
4. Discussion
4.1. Conservation of BAMBI Gene Structure and Function
4.2. The Effect of LvBAMBI on Growth
4.3. Effect of LvBAMBI on Lipid Metabolism and Glucose Metabolism
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, H.; Teng, M.; Liu, P.; Zhao, M.; Wang, S.; Hu, J.; Bao, Z.; Zeng, Q. Selection Signatures of Pacific White Shrimp Litopenaeus vannamei Revealed by Whole-Genome Resequencing Analysis. Front. Mar. Sci. 2022, 9. [Google Scholar] [CrossRef]
- Argue, B.J.; Arce, S.M.; Lotz, J.M.; Moss, S.M. Selective breeding of Pacific white shrimp (Litopenaeus vannamei) for growth and resistance to Taura Syndrome Virus. Aquaculture 2002, 204, 447–460. [Google Scholar] [CrossRef]
- Huang, W.-J.; Leu, J.-H.; Tsau, M.-T.; Chen, J.-C.; Chen, L.-L. Differential expression of LvHSP60 in shrimp in response to environmental stress. Fish Shellfish. Immunol. 2011, 30, 576–582. [Google Scholar] [CrossRef]
- Andriantahina, F.; Liu, X.; Feng, T.; Xiang, J. Current Status of Genetics and Genomics of Reared Penaeid Shrimp: Information Relevant to Access and Benefit Sharing. Mar. Biotechnol. 2013, 15, 399–412. [Google Scholar] [CrossRef] [PubMed]
- Sui, J.; Luan, S.; Luo, K.; Meng, X.; Cao, B.; Liu, N.; Li, W.; Lu, X.; Chai, Z.; Kong, J. Genetic parameters and response to selection of harvest body weight of the Chinese shrimp Fenneropenaeus chinensis after five generations of multi-trait selection. Aquaculture 2016, 452, 134–141. [Google Scholar] [CrossRef]
- Nolasco-Alzaga, H.R.; Perez-Enriquez, R.; Enez, F.; Bestin, A.; Palacios-Mechetnov, E.; Haffray, P. Quantitative genetic parameters of growth and fatty acid content in the hemolymph of the Whiteleg shrimp Litopenaeus vannamei. Aquaculture 2018, 482, 17–23. [Google Scholar] [CrossRef]
- Andriantahina, F.; Liu, X.; Huang, H.; Xiang, J. Erratum to: Response to selection, heritability and genetic correlations between body weight and body size in Pacific white shrimp, Litopenaeus vannamei. Chin. J. Oceanol. Limnol. 2012, 30, 506. [Google Scholar] [CrossRef][Green Version]
- Yu, Y.; Wang, Q.; Zhang, Q.; Luo, Z.; Wang, Y.; Zhang, X.; Huang, H.; Xiang, J.; Li, F. Genome Scan for Genomic Regions and Genes Associated with Growth Trait in Pacific White Shrimp Litopeneaus vannamei. Mar. Biotechnol. 2019, 21, 374–383. [Google Scholar] [CrossRef] [PubMed]
- Glenn, K.L.; Grapes, L.; Suwanasopee, T.; Harris, D.L.; Li, Y.; Wilson, K.; Rothschild, M.F. SNP analysis of AMY2 and CTSL genes in Litopenaeus vannamei and Penaeus monodon shrimp. Anim. Genet. 2005, 36, 235–236. [Google Scholar] [CrossRef]
- Yuan, D.; Zhou, S.; Liu, S.; Li, K.; Zhao, H.; Long, S.; Liu, H.; Xie, Y.; Su, Y.; Yu, F.; et al. The AMPK-PP2A axis in insect fat body is activated by 20-hydroxyecdysone to antagonize insulin/IGF signaling and restrict growth rate. Proc. Natl. Acad. Sci. 2020, 117, 9292–9301. [Google Scholar] [CrossRef]
- Yang, Y.; Zhao, T.; Li, Z.; Qian, W.; Peng, J.; Wei, L.; Yuan, D.; Li, Y.; Xia, Q.; Cheng, D. Histone H3K27 methylation–mediated repression of Hairy regulates insect developmental transition by modulating ecdysone biosynthesis. Proc. Natl. Acad. Sci. 2021, 118, e2101442118. [Google Scholar] [CrossRef]
- Kim, M.; Sim, C.; Denlinger, D.L. RNA interference directed against ribosomal protein S3a suggests a link between this gene and arrested ovarian development during adult diapause in Culex pipiens. Insect Mol. Biol. 2010, 19, 27–33. [Google Scholar] [CrossRef]
- Katewa, S.D.; Kapahi, P. Role of TOR signaling in aging and related biological processes in Drosophila melanogaster. Exp. Gerontol. 2011, 46, 382–390. [Google Scholar] [CrossRef] [PubMed]
- Pang, Y.; Zhang, X.; Yuan, J.; Zhang, X.; Xiang, J.; Li, F. Characterization and Expression Analysis of Insulin Growth Factor Binding Proteins (IGFBPs) in Pacific White Shrimp Litopenaeus vannamei. Int. J. Mol. Sci. 2021, 22, 1056. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.-H.; Sarbassov, D.D.; Ali, S.M.; King, J.E.; Latek, R.R.; Erdjument-Bromage, H.; Tempst, P.; Sabatini, D.M. mTOR Interacts with Raptor to Form a Nutrient-Sensitive Complex that Signals to the Cell Growth Machinery. Cell 2002, 110, 163–175. [Google Scholar] [CrossRef]
- Lewis, K.A.; Gray, P.C.; Blount, A.L.; MacConell, L.A.; Wiater, E.; Bilezikjian, L.M.; Vale, W. Betaglycan binds inhibin and can mediate functional antagonism of activin signalling. Nature 2000, 404, 411–414. [Google Scholar] [CrossRef] [PubMed]
- Heldin, C.-H.; Moustakas, A. Signaling Receptors for TGF-β Family Members. Cold Spring Harb. Perspect. Biol. 2016, 8, a022053. [Google Scholar] [CrossRef]
- Hata, A.; Chen, Y.-G. TGF-β Signaling from Receptors to Smads. Cold Spring Harb. Perspect. Biol. 2016, 8, a022061. [Google Scholar] [CrossRef]
- Yan, Y.; Lu, X.; Kong, J.; Meng, X.; Luan, S.; Dai, P.; Chen, B.; Cao, B.; Luo, K. Molecular characterization of myostatin and its inhibitory function on myogenesis and muscle growth in Chinese Shrimp, Fenneropenaeus chinensis. Gene 2020, 758, 144986. [Google Scholar] [CrossRef]
- Chaikuad, A.; Bullock, A.N. Structural Basis of Intracellular TGF-β Signaling: Receptors and Smads. Cold Spring Harb. Perspect. Biol. 2016, 8, a022111. [Google Scholar] [CrossRef]
- Onichtchouk, D.; Chen, Y.-G.; Dosch, R.; Gawantka, V.; Delius, H.; Massague´, J.; Niehrs, C. Silencing of TGF-β signalling by the pseudoreceptor BAMBI. Nature 1999, 401, 480–485. [Google Scholar] [CrossRef] [PubMed]
- Eivers, E.; Demagny, H.; De Robertis, E. Integration of BMP and Wnt signaling via vertebrate Smad1/5/8 and Drosophila Mad. Cytokine Growth Factor Rev. 2009, 20, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Hutley, L.J.; Webster, J.A.; Kim, Y.-H.; Liu, D.-F.; Newell, F.S.; Widberg, C.H.; Bachmann, A.; Turner, N.; Schmitz-Peiffer, C.; et al. Identification of BMP and Activin Membrane-Bound Inhibitor (BAMBI) as a Potent Negative Regulator of Adipogenesis and Modulator of Autocrine/Paracrine Adipogenic Factors. Diabetes 2011, 61, 124–136. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Ning, Y.; Mei, C.; Zhang, W.; Sun, J.; Wang, S.; Zan, L. The role of BAMBI in regulating adipogenesis and myogenesis and the association between its polymorphisms and growth traits in cattle. Mol. Biol. Rep. 2020, 47, 5963–5974. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yuan, J.; Sun, Y.; Li, S.; Gao, Y.; Yu, Y.; Liu, C.; Wang, Q.; Lv, X.; Zhang, X.; et al. Penaeid shrimp genome provides insights into benthic adaptation and frequent molting. Nat. Commun. 2019, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.; Zhong, H.; Lin, Y.; Chen, B.; Han, M.; Ren, H.; Lu, H.; Luber, J.M.; Xia, M.; Li, W.; et al. Assessment of the cPAS-based BGISEQ-500 platform for metagenomic sequencing. GigaScience 2017, 7, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows—Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple hypothesis testing. J R Stat Soc Series B Stat Methodol. 1995, 57. [Google Scholar] [CrossRef]
- Dunn, W.B.; Broadhurst, D.; Begley, P.; Zelena, E.; Francis-McIntyre, S.; Anderson, N.; Brown, M.; Knowles, J.D.; Halsall, A.; Haselden, J.N.; et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2011, 6, 1060–1083. [Google Scholar] [CrossRef] [PubMed]
- Want, E.J.; Wilson, I.D.; Gika, H.; Theodoridis, G.; Plumb, R.S.; Shockcor, J.; Holmes, E.; Nicholson, J.K. Global metabolic profiling procedures for urine using UPLC–MS. Nat. Protoc. 2010, 5, 1005–1018. [Google Scholar] [CrossRef] [PubMed]
- Summers, S.A.; Chaurasia, B.; Holland, W.L. Metabolic Messengers: Ceramides. Nat. Metab. 2019, 1, 1051–1058. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Zheng, L.; Ye, J.; Zhang, C.; Zhou, J.; Huang, Q.; Guo, Y.; Wang, L.; Yu, P.; Liu, S.; et al. CREB1 contributes colorectal cancer cell plasticity by regulating lncRNA CCAT1 and NF-κB pathways. Sci. China Life Sci. 2022, 65, 1481–1497. [Google Scholar] [CrossRef] [PubMed]
- Lancho, O.; Herranz, D. The MYC Enhancer-ome: Long-Range Transcriptional Regulation of MYC in Cancer. Trends Cancer 2018, 4, 810–822. [Google Scholar] [CrossRef] [PubMed]
- Kunej, T. Integrative Map of HIF1A Regulatory Elements and Variations. Genes 2021, 12, 1526. [Google Scholar] [CrossRef] [PubMed]
- Makita, S.; Tobinai, K. Targeting EZH2 with tazemetostat. Lancet Oncol. 2018, 19, 586–587. [Google Scholar] [CrossRef]
- Dustin, D.; Gu, G.; Fuqua, S.A.W. ESR1 mutations in breast cancer. Cancer 2019, 125, 3714–3728. [Google Scholar] [CrossRef]
- Yan, X.; Lin, Z.; Chen, F.; Zhao, X.; Chen, H.; Ning, Y.; Chen, Y.-G. Human BAMBI Cooperates with Smad7 to Inhibit Transforming Growth Factor-β Signaling. J. Biol. Chem. 2009, 284, 30097–30104. [Google Scholar] [CrossRef]
- Pfeifer, C.G.; Karl, A.; Berner, A.; Zellner, J.; Schmitz, P.; Loibl, M.; Koch, M.; Angele, P.; Nerlich, M.; Mueller, M.B. Expression of BMP and Actin Membrane Bound Inhibitor Is Increased during Terminal Differentiation of MSCs. Stem Cells Int. 2016, 2016, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Chen, X.; Zhang, R. BAMBI promotes macrophage proliferation and differentiation in gliomas. Mol. Med. Rep. 2017, 17, 3960–3966. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Shi, X.-E.; Song, C.; Sun, S.; Yang, G.; Li, X. BAMBI Promotes C2C12 Myogenic Differentiation by Enhancing Wnt/β-Catenin Signaling. Int. J. Mol. Sci. 2015, 16, 17734–17745. [Google Scholar] [CrossRef] [PubMed]
- Bai, L.; Chu, G.; Wang, W.; Xiang, A.; Yang, G. BAMBI promotes porcine granulosa cell steroidogenesis involving TGF-β signaling. Theriogenology 2017, 100, 24–31. [Google Scholar] [CrossRef]
- Soofi, A.; Wolf, K.I.; Ranghini, E.J.; Amin, M.A.; Dressler, G.R. The kielin/chordin-like protein KCP attenuates nonalcoholic fatty liver disease in mice. Am. J. Physiol. Liver Physiol. 2016, 311, G587–G598. [Google Scholar] [CrossRef] [PubMed]
- Kuroda, K.; Han, H.; Tani, S.; Tanigaki, K.; Tun, T.; Furukawa, T.; Taniguchi, Y.; Kurooka, H.; Hamada, Y.; Toyokuni, S.; et al. Regulation of Marginal Zone B Cell Development by MINT, a Suppressor of Notch/RBP-J Signaling Pathway. Immunity 2003, 18, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Gouw, A.M.; Margulis, K.; Liu, N.S.; Raman, S.J.; Mancuso, A.; Toal, G.G.; Tong, L.; Mosley, A.; Hsieh, A.L.; Sullivan, D.K.; et al. The MYC Oncogene Cooperates with Sterol-Regulated Element-Binding Protein to Regulate Lipogenesis Essential for Neoplastic Growth. Cell Metab. 2019, 30, 556–572.e5. [Google Scholar] [CrossRef]
- Singh, K.B.; Hahm, E.-R.; Kim, S.-H.; Wendell, S.G.; Singh, S.V. A novel metabolic function of Myc in regulation of fatty acid synthesis in prostate cancer. Oncogene 2020, 40, 592–602. [Google Scholar] [CrossRef]

| Gene | Annotation | Log2(foldchange) | Regulation | Possible Functions |
|---|---|---|---|---|
| LOC113810077 | pancreatic lipase-related protein 2-like | 2.055644473 | up | lipid metabolism |
| LOC113805268 | group XV phospholipase A2-like | 1.512752809 | up | |
| LOC113816418 | elongation of very long chain fatty acids protein 6-like | −1.109271264 | down | |
| LOC113807853 | 17-beta-hydroxysteroid dehydrogenase 13-like | −1.278968958 | down | |
| LOC113815038 | sphingomyelin phosphodiesterase-like | −1.520403624 | down | |
| LOC113814209 | TLC domain-containing protein 2-like | −1.529060798 | down | |
| LOC113808685 | CYP450 | −1.642795706 | down | |
| LOC113815027 | apolipoprotein D-like | −1.833408201 | down | |
| LOC113813817 | apolipoprotein D-like | −2.306012563 | down | |
| LOC113825958 | Leukocyte Elastase Inhibitor | 2.011631663 | up | immune-related genes |
| LOC113823643 | hemocyanin C chain-like | 1.497524117 | up | |
| LOC113823622 | hemocyanin C chain-like | 1.108584479 | up | |
| LOC113825585 | cathepsin L1-like | 1.099745679 | up | |
| LOC113823633 | hemocyanin C chain-like | 1.036162219 | up | |
| LOC113807239 | Beta-1,3-glucan-binding protein | −1.589632096 | down | |
| LOC113807240 | Beta-1,3-glucan-binding protein | −1.606268937 | down | |
| LOC113828583 | metabotropic glutamate receptor 3-like | 5.937174763 | up | intracellular signal transduction, and transportation |
| LOC113805618 | sorting nexin-18-like | 2.99536541 | up | |
| LOC113811177 | SLC19A1 | 2.14994886 | up | |
| LOC113807061 | sialin-like | 1.603719878 | up | |
| LOC113800776 | neurexin-1-like | 1.363895445 | up | |
| LOC113810616 | hemocyanin C chain-like | 1.096986803 | up | |
| LOC113805473 | YvkC-like | −1.052007081 | down | |
| LOC113821593 | phosphoenolpyruvate synthase | −1.071866976 | down | sugar synthesis and breakdown |
| LOC113806982 | delta-1-pyrroline-5-carboxylate synthase-like | −1.79551763 | down | |
| LOC113814709 | Hydroxyacid-oxoacid transhydrogenase | −2.835674612 | down | |
| LOC113828937 | period circadian protein-like | 1.590523236 | up | Other metabolism related genes |
| LOC113825869 | trichohyalin-like | 1.283633328 | up | |
| LOC113812271 | methionine synthase-like | −1.283853613 | down | |
| LOC113822192 | SMCT1-like | −1.478478109 | down | |
| LOC113805739 | chymotrypsin BII-like | −1.820489948 | down | protein metabolism |
| LOC113822498 | ubiquitin-40S ribosomal protein S27a-like | −3.724189533 | down | |
| LOC113814568 | kielin/chordin-like protein | 1.127120613 | up | Genes related to the growth and development of organisms |
| LOC113821325 | leucine-rich repeat transmembrane neuronal protein 4-like | −1.036589648 | down | |
| LOC113820707 | protein patched homolog 2-like | −1.599886556 | down | |
| LOC113826172 | HSP70 | −1.132309417 | down | Genes involved in cellular homeostasis |
| LOC113806042 | HSP83/HSP90 | −1.529544744 | down | |
| LOC113813873 | heme-binding protein 2-like | −1.925589424 | down | |
| LOC113828559 | branchpoint-bridging protein-like | −1.31764033 | down | Transcription regulation of related genes |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Niu, R.; Zhang, X.; Yu, Y.; Bao, Z.; Yang, J.; Yuan, J.; Li, F. Identification of Growth-Related Gene BAMBI and Analysis of Gene Structure and Function in the Pacific White Shrimp Litopenaeus vannamei. Animals 2024, 14, 1074. https://doi.org/10.3390/ani14071074
Niu R, Zhang X, Yu Y, Bao Z, Yang J, Yuan J, Li F. Identification of Growth-Related Gene BAMBI and Analysis of Gene Structure and Function in the Pacific White Shrimp Litopenaeus vannamei. Animals. 2024; 14(7):1074. https://doi.org/10.3390/ani14071074
Chicago/Turabian StyleNiu, Ruigang, Xiaojun Zhang, Yang Yu, Zhenning Bao, Junqing Yang, Jianbo Yuan, and Fuhua Li. 2024. "Identification of Growth-Related Gene BAMBI and Analysis of Gene Structure and Function in the Pacific White Shrimp Litopenaeus vannamei" Animals 14, no. 7: 1074. https://doi.org/10.3390/ani14071074
APA StyleNiu, R., Zhang, X., Yu, Y., Bao, Z., Yang, J., Yuan, J., & Li, F. (2024). Identification of Growth-Related Gene BAMBI and Analysis of Gene Structure and Function in the Pacific White Shrimp Litopenaeus vannamei. Animals, 14(7), 1074. https://doi.org/10.3390/ani14071074






