The In Vitro Effects of Carprofen on Lipopolysaccharide-Induced Neutrophil Extracellular Trap Formation in Dairy Cows
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Animals and Management
2.3. Reagents and Materials
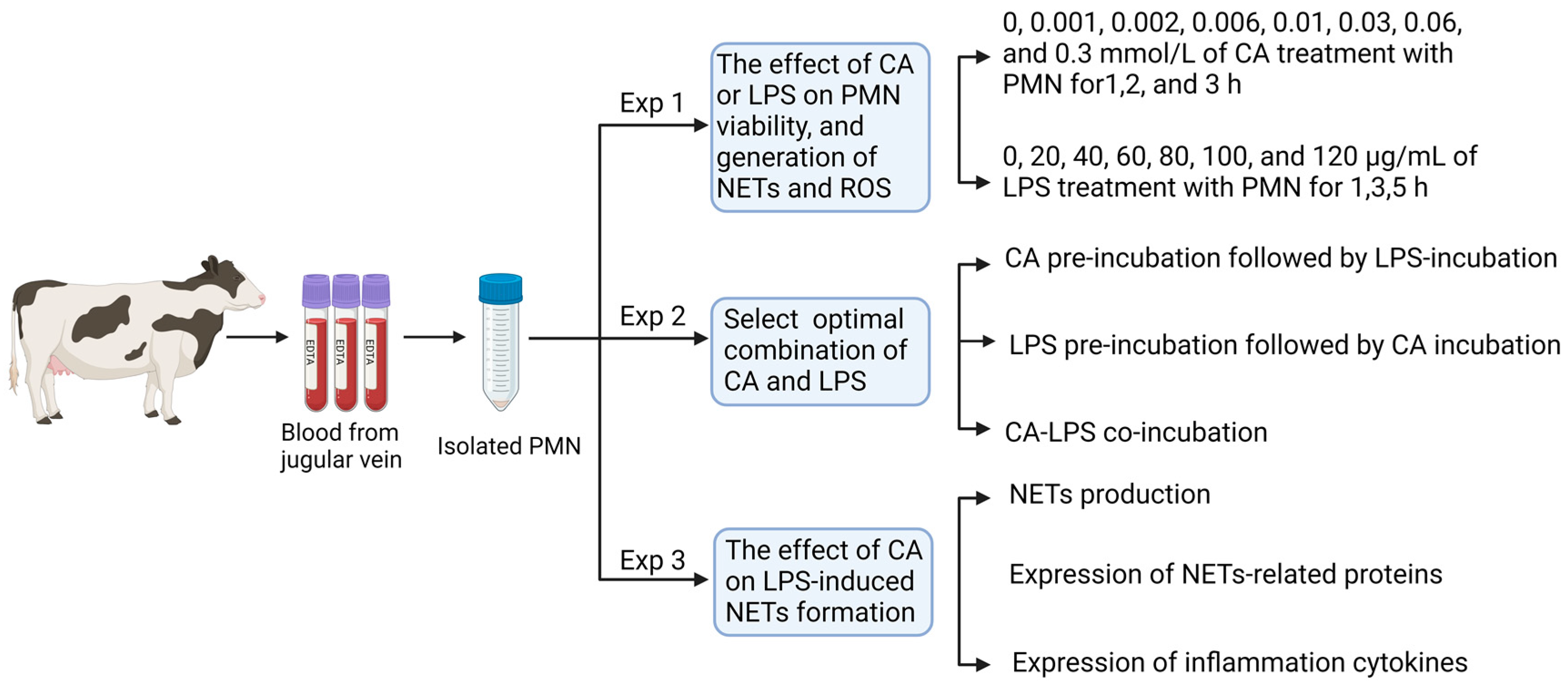
2.4. Experiment 1: Effects of CA and LPS on Circulating PMNs Viability, PMNs Production of NETs and ROS
2.4.1. Blood Sample Collection and PMNs Isolation
2.4.2. Determination of PMNs Viability
2.4.3. Detection of ROS Production
2.4.4. Quantification of NETs
2.5. Experiment 2: Selecting the Optimal Combination of CA and LPS Incubation Based on PMNs Viability
2.6. Experiment 3: The Effects of LPS and CA Co-Incubation (LC-Co) on PMNs’ NETs Formation, Protein and Inflammatory Factors’ Expression
2.6.1. Morphological Observation and Quantification of NETs
2.6.2. Protein Extraction and Western Blotting
2.6.3. RNA Extraction and Quantitative Real-Time PCR
2.7. Statistical Analysis
3. Results
3.1. Experiment 1: Effects of CA and LPS on PMNs Viability, PMNs Production of ROS and NETs
3.2. Experiment 2: The Optimal Combination of CA and LPS Incubation Based on PMNs Viability
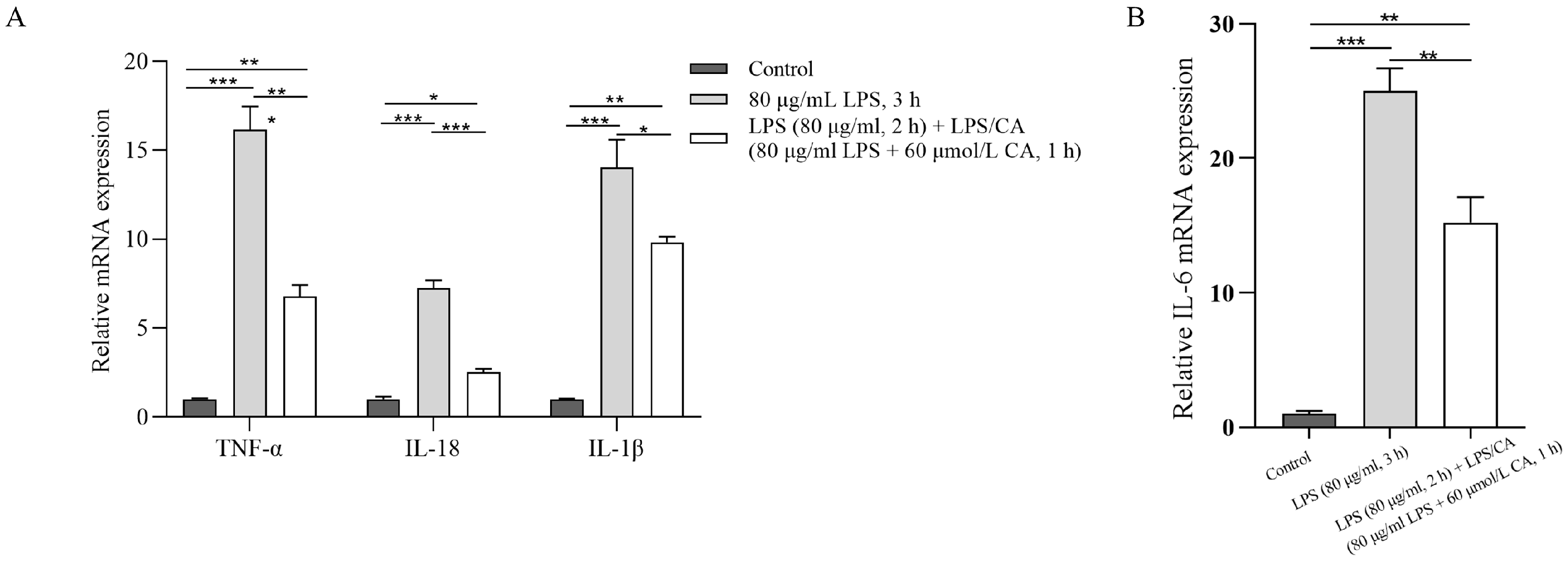
3.3. Experiment 3: The Effects of LPS and CA Co-Incubation (LC-Co) on PMNs’ NETs Formation, Protein and Inflammatory Expression
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bradford, B.J.; Yuan, K.; Farney, J.K.; Mamedova, L.K.; Carpenter, A.J. Invited review: Inflammation during the transition to lactation: New adventures with an old flame. J. Dairy Sci. 2015, 98, 6631–6650. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, M.M.; Yasui, T.; Felippe, M.J.; Overton, T.R. Associations between the degree of early lactation inflammation and performance, metabolism, and immune function in dairy cows. J. Dairy Sci. 2016, 99, 680–700. [Google Scholar] [CrossRef]
- Guimarães-Costa, A.B.; Nascimento, M.T.; Froment, G.S.; Soares, R.P.; Morgado, F.N.; Conceição-Silva, F.; Saraiva, E.M. Leishmania amazonensis promastigotes induce and are killed by neutrophil extracellular traps. Proc. Natl. Acad. Sci. USA 2009, 106, 6748–6753. [Google Scholar] [CrossRef] [PubMed]
- Azzouz, L.; Cherry, A.; Riedl, M.; Khan, M.; Pluthero, F.G.; Kahr, W.H.A.; Palaniyar, N.; Licht, C. Relative antibacterial functions of complement and NETs: NETs trap and complement effectively kills bacteria. Mol. Immunol. 2018, 97, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Green, J.N.; Kettle, A.J.; Winterbourn, C.C. Protein chlorination in neutrophil phagosomes and correlation with bacterial killing. Free Radic. Biol. Med. 2014, 77, 49–56. [Google Scholar] [CrossRef]
- Collins, L.V.; Kristian, S.A.; Weidenmaier, C.; Faigle, M.; Van Kessel, K.P.; Van Strijp, J.A.; Götz, F.; Neumeister, B.; Peschel, A. Staphylococcus aureus strains lacking D-alanine modifications of teichoic acids are highly susceptible to human neutrophil killing and are virulence attenuated in mice. J. Infect. Dis. 2002, 186, 214–219. [Google Scholar] [CrossRef]
- Monteith, A.J.; Miller, J.M.; Maxwell, C.N.; Chazin, W.J.; Skaar, E.P. Neutrophil extracellular traps enhance macrophage killing of bacterial pathogens. Sci. Adv. 2021, 7, eabj2101. [Google Scholar] [CrossRef]
- Sieprawska-Lupa, M.; Mydel, P.; Krawczyk, K.; Wójcik, K.; Puklo, M.; Lupa, B.; Suder, P.; Silberring, J.; Reed, M.; Pohl, J.; et al. Degradation of human antimicrobial peptide LL-37 by Staphylococcus aureus-derived proteinases. Antimicrob. Agents Chemother. 2004, 48, 4673–4679. [Google Scholar] [CrossRef]
- Mulay, S.R.; Anders, H.J. Neutrophils and Neutrophil Extracellular Traps Regulate Immune Responses in Health and Disease. Cells 2020, 9, 2130. [Google Scholar] [CrossRef]
- Xie, L.; Pascottini, O.B.; Zhi, J.; Yang, H.; Opsomer, G.; Dong, Q. In Vitro Production of Neutrophils Extracellular Traps Is Affected by the Lactational Stage of Dairy Cows. Animals 2022, 12, 564. [Google Scholar] [CrossRef]
- Arroyo, R.; Khan, M.A.; Echaide, M.; Pérez-Gil, J.; Palaniyar, N. SP-D attenuates LPS-induced formation of human neutrophil extracellular traps (NETs), protecting pulmonary surfactant inactivation by NETs. Commun. Biol. 2019, 2, 470. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.; Farahvash, A.; Douda, D.N.; Licht, J.C.; Grasemann, H.; Sweezey, N.; Palaniyar, N. JNK Activation Turns on LPS- and Gram-Negative Bacteria-Induced NADPH Oxidase-Dependent Suicidal NETosis. Sci. Rep. 2017, 7, 3409. [Google Scholar] [CrossRef] [PubMed]
- Pijanowski, L.; Golbach, L.; Kolaczkowska, E.; Scheer, M.; Verburg-van Kemenade, B.M.; Chadzinska, M. Carp neutrophilic granulocytes form extracellular traps via ROS-dependent and independent pathways. Fish Shellfish. Immunol. 2013, 34, 1244–1252. [Google Scholar] [CrossRef] [PubMed]
- Revelo, X.S.; Waldron, M.R. In vitro effects of Escherichia coli lipopolysaccharide on the function and gene expression of neutrophils isolated from the blood of dairy cows. J. Dairy Sci. 2012, 95, 2422–2441. [Google Scholar] [CrossRef] [PubMed]
- Munafo, D.B.; Johnson, J.L.; Brzezinska, A.A.; Ellis, B.A.; Wood, M.R.; Catz, S.D. DNase I inhibits a late phase of reactive oxygen species production in neutrophils. J. Innate. Immun. 2009, 1, 527–542. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.L.; Tangsombatvisit, S.; Rosenberg, J.M.; Mandelbaum, G.; Gillespie, E.C.; Gozani, O.P.; Alizadeh, A.A.; Utz, P.J. Specific post-translational histone modifications of neutrophil extracellular traps as immunogens and potential targets of lupus autoantibodies. Arthritis Res. Ther. 2012, 14, R25. [Google Scholar] [CrossRef] [PubMed]
- Rausch, P.G.; Moore, T.G. Granule enzymes of polymorphonuclear neutrophils: A phylogenetic comparison. Blood 1975, 46, 913–919. [Google Scholar] [CrossRef]
- Wang, Y.; Li, M.; Stadler, S.; Correll, S.; Li, P.; Wang, D.; Hayama, P.D.R.; Leonelli, L.; Han, H.; Grigoryev, S.; et al. Histone hypercitrullination mediates chromatin decondensation and neutrophil extracellular trap formation. J. Cell Biol. 2009, 184, 205–213. [Google Scholar] [CrossRef]
- Thiam, H.R.; Wong, S.L.; Wagner, D.D.; Waterman, C.M. Cellular Mechanisms of NETosis. Annu. Rev. Cell Dev. Biol. 2020, 36, 191–218. [Google Scholar] [CrossRef]
- Armstrong, S.; Lees, P. Effects of carprofen (R and S enantiomers and racemate) on the production of IL-1, IL-6 and TNF-alpha by equine chondrocytes and synoviocytes. J. Vet. Pharmacol. Ther. 2002, 25, 145–153. [Google Scholar] [CrossRef]
- Sohn, E.J.; Paape, M.J.; Connor, E.E.; Bannerman, D.D.; Fetterer, R.H.; Peters, R.R. Bacterial lipopolysaccharide stimulates bovine neutrophil production of TNF-alpha, IL-1beta, IL-12 and IFN-gamma. Vet. Res. 2007, 38, 809–818. [Google Scholar] [CrossRef]
- Molina, A.; Solar, M.; Palma, P.; Bustamante, H. Early postpartum treatment with carprofen in a dairy herd with high incidence of clinical metritis—A case study. J. Appl. Anim. Res. 2021, 49, 139–146. [Google Scholar] [CrossRef]
- Lošytė, G.; Navalinskaitė, I.; Prokopavičiūtė, Ž.; Pocevičienė, E.; Antanaitis, R. The effect of carprofen on fresh dairy cows health. Žemės Ūkio Moksl. 2019, 26, 40–46. [Google Scholar] [CrossRef]
- Mitchell, M.A. Carprofen. Semin. Avian Exot. Pet Med. 2005, 14, 61–64. [Google Scholar] [CrossRef]
- Wyns, H.; Meyer, E.; Plessers, E.; Watteyn, A.; van Bergen, T.; Schauvliege, S.; De Baere, S.; Devreese, M.; De Backer, P.; Croubels, S. Modulation by gamithromycin and ketoprofen of in vitro and in vivo porcine lipopolysaccharide-induced inflammation. Vet. Immunol. Immunopathol. 2015, 168, 211–222. [Google Scholar] [CrossRef] [PubMed]
- Mendoza Garcia, F.J.; Gonzalez-De Cara, C.; Aguilera-Aguilera, R.; Buzon-Cuevas, A.; Perez-Ecija, A. Meloxicam ameliorates the systemic inflammatory response syndrome associated with experimentally induced endotoxemia in adult donkeys. J. Vet. Intern. Med. 2020, 34, 1631–1641. [Google Scholar] [CrossRef] [PubMed]
- Caldeira, M.O.; Bruckmaier, R.M.; Wellnitz, O. Meloxicam affects the inflammatory responses of bovine mammary epithelial cells. J. Dairy Sci. 2019, 102, 10277–10290. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.K.; Giaglis, S.; Hasler, P.; Hahn, S. Efficient neutrophil extracellular trap induction requires mobilization of both intracellular and extracellular calcium pools and is modulated by cyclosporine A. PLoS ONE 2014, 9, e97088. [Google Scholar] [CrossRef]
- Zou, Y.; Chen, X.; Xiao, J.; Zhou, D.B.; Lu, X.X.; Li, W.; Xie, B.; Kuang, X.; Chen, Q. Neutrophil extracellular traps promote lipopolysaccharide-induced airway inflammation and mucus hypersecretion in mice. Oncotarget 2018, 9, 13276–13286. [Google Scholar] [CrossRef]
- Xiang, M.; Yin, M.; Xie, S.; Shi, L.; Nie, W.; Shi, B.; Yu, G. The molecular mechanism of neutrophil extracellular traps and its role in bone and joint disease. Heliyon 2023, 9, e22920. [Google Scholar] [CrossRef] [PubMed]
- Ravindran, M.; Khan, M.A.; Palaniyar, N. Neutrophil Extracellular Trap Formation: Physiology, Pathology, and Pharmacology. Biomolecules 2019, 9, 365. [Google Scholar] [CrossRef]
- Chen, J.L.; Tong, Y.; Zhu, Q.; Gao, L.Q.; Sun, Y. Neutrophil extracellular traps induced by Porphyromonas gingivalis lipopolysaccharide modulate inflammatory responses via a Ca(2+)-dependent pathway. Arch. Oral Biol. 2022, 141, 105467. [Google Scholar] [CrossRef]
- Li, T.; Zhang, Z.; Li, X.; Dong, G.; Zhang, M.; Xu, Z.; Yang, J. Neutrophil Extracellular Traps: Signaling Properties and Disease Relevance. Mediat. Inflamm. 2020, 2020, 9254087. [Google Scholar] [CrossRef]
- Huang, Z.; Zhang, H.; Fu, X.; Han, L.; Zhang, H.; Zhang, L.; Zhao, J.; Xiao, D.; Li, H.; Li, P. Autophagy-driven neutrophil extracellular traps: The dawn of sepsis. Pathol. Res. Pract. 2022, 234, 153896. [Google Scholar] [CrossRef]
- Klopf, J.; Brostjan, C.; Eilenberg, W.; Neumayer, C. Neutrophil Extracellular Traps and Their Implications in Cardiovascular and Inflammatory Disease. Int. J. Mol. Sci. 2021, 22, 559. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Pan, B.; Alam, H.B.; Deng, Q.; Wang, Y.; Chen, E.; Liu, B.; Tian, Y.; Williams, A.M.; Duan, X.; et al. Inhibition of peptidylarginine deiminase alleviates LPS-induced pulmonary dysfunction and improves survival in a mouse model of lethal endotoxemia. Eur. J. Pharmacol. 2018, 833, 432–440. [Google Scholar] [CrossRef] [PubMed]
- O’Donoghue, A.J.; Jin, Y.; Knudsen, G.M.; Perera, N.C.; Jenne, D.E.; Murphy, J.E.; Craik, C.S.; Hermiston, T.W. Global substrate profiling of proteases in human neutrophil extracellular traps reveals consensus motif predominantly contributed by elastase. PLoS ONE 2013, 8, e75141. [Google Scholar] [CrossRef] [PubMed]




| a | |||
| Antigen | Host | Manufacturer | Dilution |
| Anti-PAD4 | Rabbit | Abcam (#ab214810) | 1:5000 (WB) |
| Anti-Cathepsin G | Rabbit | Abcam (#ab282105) | 1:5000 (WB) |
| Anti-Histone H3 | Rabbit | Abcam (#ab32356) | 1:5000 (WB) |
| Anti-Histone H3 (citrulline R2 + R8 + R17) | Rabbit | Abcam (#ab5103) | 1:5000 (WB) |
| β-actin | Mouse | Sungene Biotech (KM9001T) | 1:5000 (WB) |
| b | |||
| Antigen | Host | Manufacturer | Dilution |
| HRP-Conjugated Goat Anti-Rabbit | Rabbit | Beyotime (A0208) | 1:2000 (WB) |
| HRP-Conjugated Goat Anti-Mouse | Mouse | Beyotime (A0216) | 1:2000 (WB) |
| Genes | Accession No. | Accession No. | Products (bp) |
|---|---|---|---|
| β-actin | NM_173979.3 | F: TGAGGCTCAGAGCAAGAGAGG R: AGGCATACAGGGACAGCACA | 266 |
| TNF-α | NM_173966.3 | F: CTCCTTCCTCCTGGTTGCAG R: CACCTGGGGACTGCTCTTC | 92 |
| IL-6 | NM_173923.2 | F: AGCGCATGGTCGACAAAATC R: CCCAGATTGGAAGCATCCGT | 143 |
| IL-18 | NM_174091.2 | F: GATTATTGCATCAGCTTTGTGGAAA R: GATCTGATTCCAGGTCTTCATCAT | 88 |
| IL-1β | NM_174093.1 | F: AAAATCCCTGGTGCTGGCTA R: AGCTCATGCAGAACACCACT | 92 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhi, J.; Qiao, K.; Xie, L.; Pascottini, O.B.; Opsomer, G.; Dong, Q. The In Vitro Effects of Carprofen on Lipopolysaccharide-Induced Neutrophil Extracellular Trap Formation in Dairy Cows. Animals 2024, 14, 985. https://doi.org/10.3390/ani14060985
Zhi J, Qiao K, Xie L, Pascottini OB, Opsomer G, Dong Q. The In Vitro Effects of Carprofen on Lipopolysaccharide-Induced Neutrophil Extracellular Trap Formation in Dairy Cows. Animals. 2024; 14(6):985. https://doi.org/10.3390/ani14060985
Chicago/Turabian StyleZhi, Jianbo, Kaixi Qiao, Lei Xie, Osvaldo Bogado Pascottini, Geert Opsomer, and Qiang Dong. 2024. "The In Vitro Effects of Carprofen on Lipopolysaccharide-Induced Neutrophil Extracellular Trap Formation in Dairy Cows" Animals 14, no. 6: 985. https://doi.org/10.3390/ani14060985
APA StyleZhi, J., Qiao, K., Xie, L., Pascottini, O. B., Opsomer, G., & Dong, Q. (2024). The In Vitro Effects of Carprofen on Lipopolysaccharide-Induced Neutrophil Extracellular Trap Formation in Dairy Cows. Animals, 14(6), 985. https://doi.org/10.3390/ani14060985






