SP1 and KROX20 Regulate the Proliferation of Dermal Papilla Cells and Target the CUX1 Gene
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Animals, Cell Isolation, Cell Culture, and Cell Transfection
2.3. Total RNA Extraction, cDNA Synthesis, and qRT-PCR
2.4. Primers for qRT-PCR
2.5. Construction of SP1 and KROX20 Overexpression Vector and CUX1 Promoter Reporter Vector
2.6. Immunofluorescence
2.7. Dual-Luciferase Assay
2.8. CCK-8 Assay
2.9. EdU Assay
2.10. Statistical Analysis
3. Results
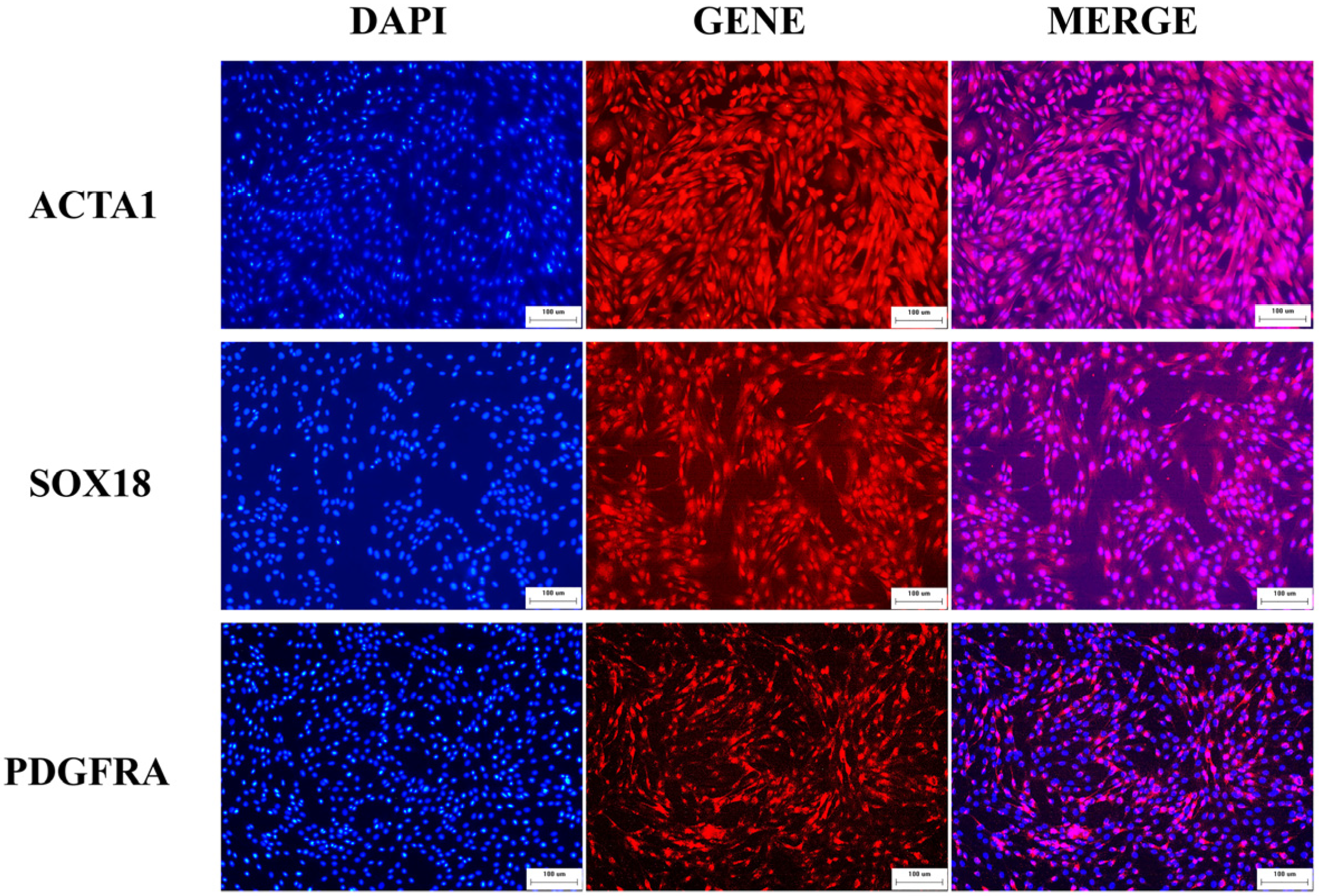
3.1. Identification of Marker Genes in Hu Sheep DPCs
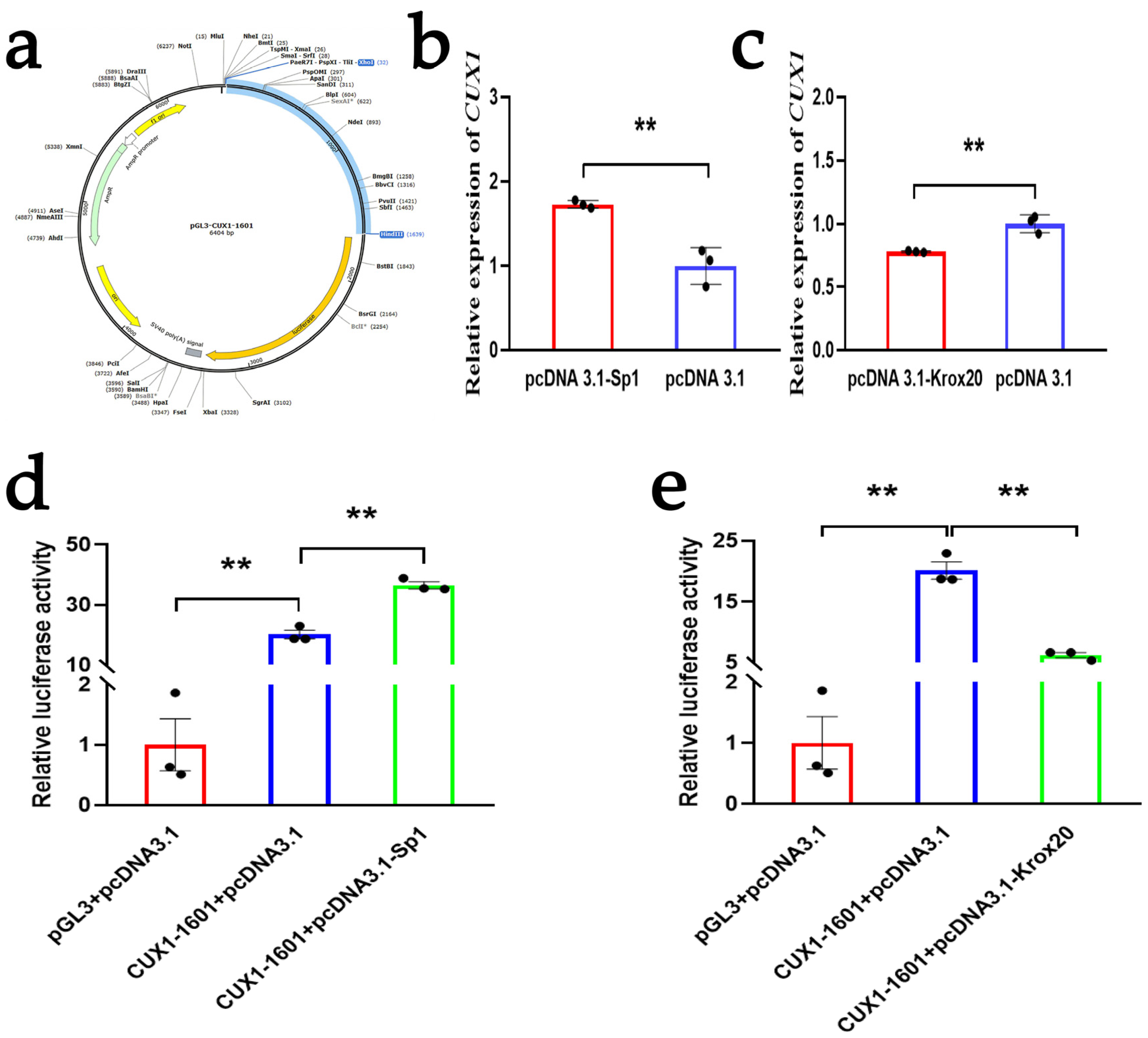
3.2. Analysis of Transcription Factor Binding Sites on the CUX1 Gene
3.3. SP1 and KROX20 Act as Two Transcription Regulatory Factors of the CUX1 Gene
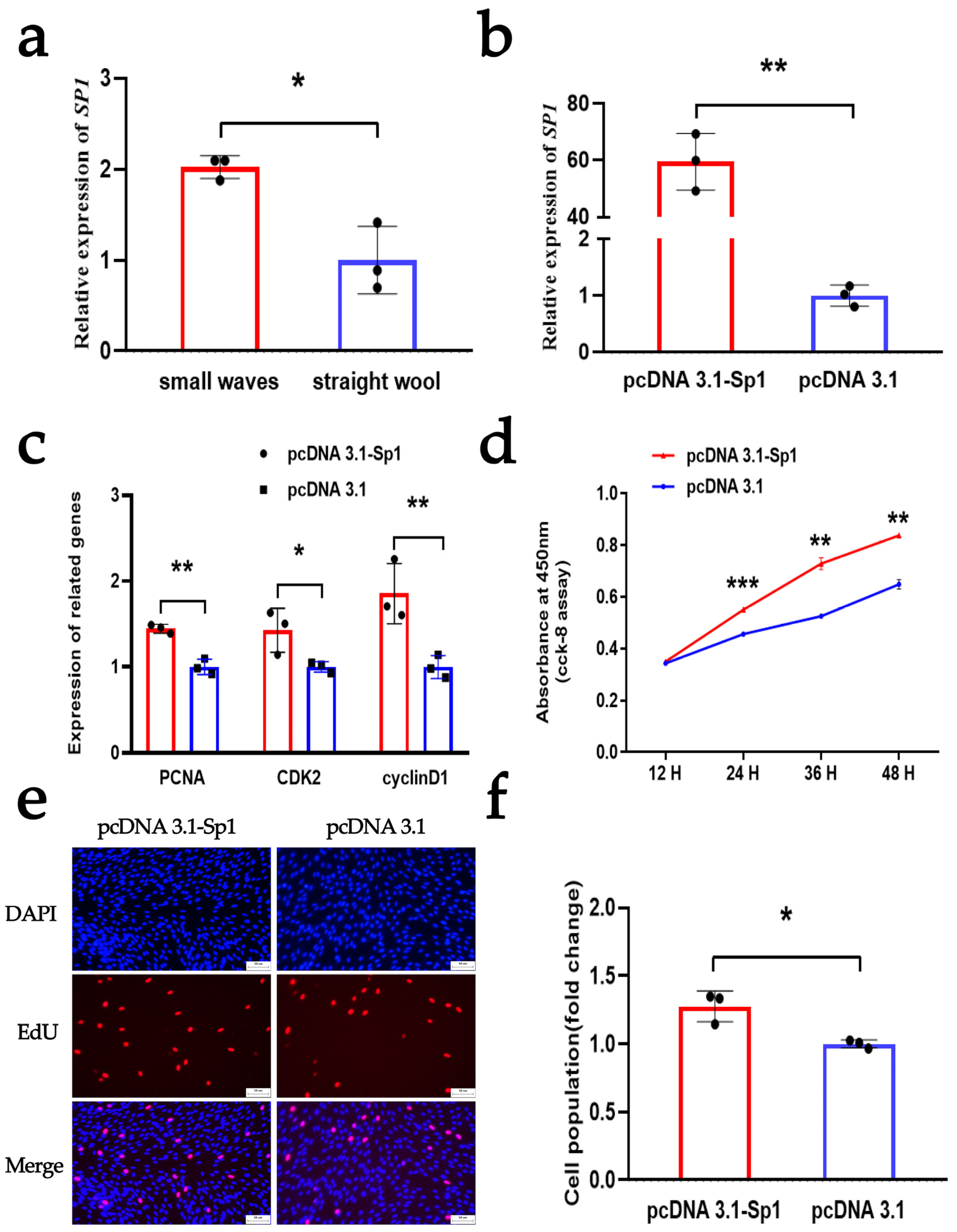
3.4. SP1 Enhances the Proliferation of DPCs
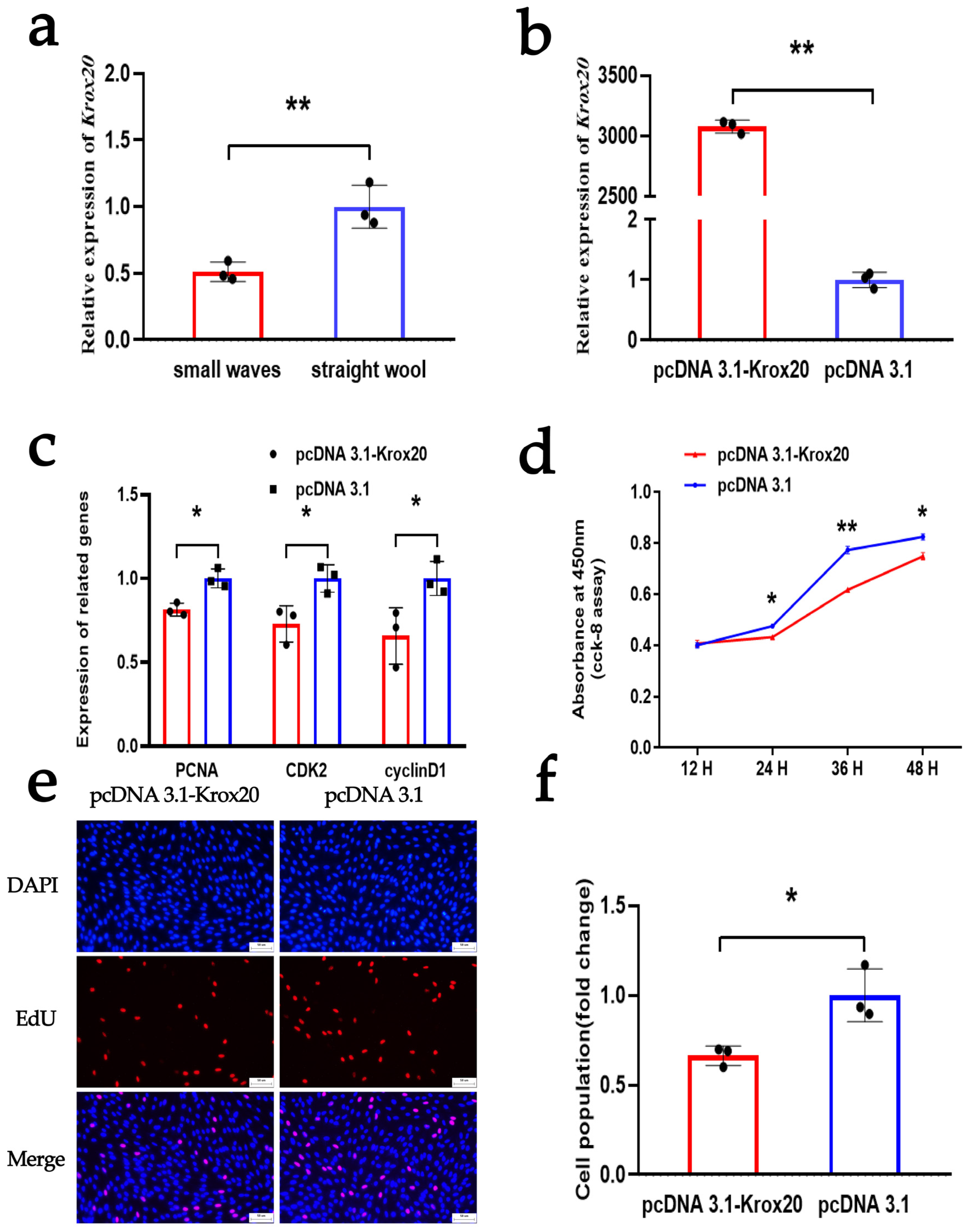
3.5. KROX20 Suppresses the Proliferation of DPCs
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Houschyar, K.S.; Borrelli, M.R.; Tapking, C.; Popp, D.; Puladi, B.; Ooms, M.; Chelliah, M.P.; Rein, S.; Pforringer, D.; Thor, D. Molecular Mechanisms of Hair Growth and Regeneration: Current Understanding and Novel Paradigms. Dermatology 2020, 236, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Mok, K.-W.; Saxena, N.; Heitman, N.; Grisanti, L.; Srivastava, D.; Muraro, M.J.; Jacob, T.; Sennett, R.; Wang, Z.; Su, Y. Dermal Condensate Niche Fate Specification Occurs Prior to Formation and Is Placode Progenitor Dependent. Dev. Cell 2019, 48, 32–48.e35. [Google Scholar] [CrossRef]
- Jahoda, C.A.B.; Christiano, A.M. Niche Crosstalk: Intercellular Signals at the Hair Follicle. Cell 2011, 146, 678–681. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.-C.; Cotsarelis, G. Review of hair follicle dermal cells. J. Dermatol. Sci. 2010, 57, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Nissimov, J.N.; Das Chaudhuri, A.B. Hair curvature: A natural dialectic and review. Biol. Rev. 2014, 89, 723–766. [Google Scholar] [CrossRef] [PubMed]
- Millar, S.E. Molecular mechanisms regulating hair follicle development. J. Investig. Dermatol. 2002, 118, 216–225. [Google Scholar] [CrossRef]
- Schneider, M.R.; Schmidt-Ullrich, R.; Paus, R. The hair follicle as a dynamic miniorgan. Curr. Biol. 2009, 19, R132–R142. [Google Scholar] [CrossRef]
- Paus, R.; Epstein, F.H.; Cotsarelis, G. The Biology of Hair Follicles. New Engl. J. Med. 1999, 341, 491–497. [Google Scholar] [CrossRef]
- Driskell, R.R.; Clavel, C.; Rendl, M.; Watt, F.M. Hair follicle dermal papilla cells at a glance. J. Cell Sci. 2011, 124, 1179–1182. [Google Scholar] [CrossRef]
- Zhou, H.; Huang, S.; Lv, X.; Wang, S.; Cao, X.; Yuan, Z.; Getachew, T.; Mwacharo, J.M.; Haile, A.; Quan, K. Effect of CUX1 on the Proliferation of Hu Sheep Dermal Papilla Cells and on the Wnt/beta-Catenin Signaling Pathway. Genes 2023, 14, 423. [Google Scholar] [CrossRef]
- Ellis, T.; Gambardella, L.; Horcher, M.; Tschanz, S.; Capol, J.; Bertram, P.; Jochum, W.; Barrandon, Y.; Busslinger, M. The transcriptional repressor CDP (Cutl1) is essential for epithelial cell differentiation of the lung and the hair follicle. Genes Dev. 2001, 15, 2307–2319. [Google Scholar] [CrossRef]
- Sinclair, A.M.; Lee, J.A.; Goldstein, A.; Xing, D.; Liu, S.; Ju, R.; Tucker, P.W.; Neufeld, E.J.; Scheuermann, R.H. Lymphoid apoptosis and myeloid hyperplasia in CCAAT displacement protein mutant mice. Blood 2001, 98, 3658–3667. [Google Scholar] [CrossRef]
- Luong, M.X.; van der Meijden, C.M.; Xing, D.; Hesselton, R.; Monuki, E.S.; Jones, S.N.; Lian, J.B.; Stein, J.L.; Stein, G.S.; Neufeld, E.J. Genetic Ablation of the CDP/Cux Protein C Terminus Results in Hair Cycle Defects and Reduced Male Fertility. Mol. Cell. Biol. 2023, 22, 1424–1437. [Google Scholar] [CrossRef]
- Sansregret, L.; Goulet, B.; Harada, R.; Wilson, B.; Leduy, L.; Bertoglio, J.; Nepveu, A. The p110 Isoform of the CDP/Cux Transcription Factor Accelerates Entry into S Phase. Mol. Cell. Biol. 2023, 26, 2441–2455. [Google Scholar] [CrossRef]
- Truscott, M.; Denault, J.-B.; Goulet, B.; Leduy, L.; Salvesen, G.S.; Nepveu, A. Carboxyl-terminal Proteolytic Processing of CUX1 by a Caspase Enables Transcriptional Activation in Proliferating Cells. J. Biol. Chem. 2007, 282, 30216–30226. [Google Scholar] [CrossRef]
- Jing, J.; Xu, P.; Xu, J.-L.; Ding, Y.-X.; Yang, X.-S.; Jin, X.-Q.; Zhou, L.-J.; Chen, Y.-H.; Wu, X.-J.; Lu, Z.-F. Expression and localization of Sox10 during hair follicle morphogenesis and induced hair cycle. Int. J. Med. Sci. 2021, 18, 3498–3505. [Google Scholar] [CrossRef]
- Krieger, K.; Millar, S.E.; Mikuda, N.; Krahn, I.; Kloepper, J.E.; Bertolini, M.; Scheidereit, C.; Paus, R.; Schmidt-Ullrich, R. NF-κB Participates in Mouse Hair Cycle Control and Plays Distinct Roles in the Various Pelage Hair Follicle Types. J. Investig. Dermatol. 2018, 138, 256–264. [Google Scholar] [CrossRef]
- Tomann, P.; Paus, R.; Millar, S.E.; Scheidereit, C.; Schmidt-Ullrich, R. LHX2 is a direct NF-κB target gene that promotes primary hair follicle placode down-growth. Development 2016, 143, 1512–1522. [Google Scholar] [CrossRef]
- Kadaja, M.; Keyes, B.E.; Lin, M.; Pasolli, H.A.; Genander, M.; Polak, L.; Stokes, N.; Zheng, D.; Fuchs, E. SOX9: A stem cell transcriptional regulator of secreted niche signaling factors. Genes. Dev. 2014, 28, 328–341. [Google Scholar] [CrossRef]
- Folgueras, A.R.; Guo, X.; Pasolli, H.A.; Stokes, N.; Polak, L.; Zheng, D.; Fuchs, E. Architectural Niche Organization by LHX2 Is Linked to Hair Follicle Stem Cell Function. Cell Stem Cell 2013, 13, 314–327. [Google Scholar] [CrossRef]
- Kang, X.; Liu, Y.; Zhang, J.; Xu, Q.; Liu, C.; Fang, M. Characteristics and Expression Profile ofKRT71Screened by Suppression Subtractive Hybridization cDNA Library in Curly Fleece Chinese Tan Sheep. DNA Cell Biol. 2017, 36, 552–564. [Google Scholar] [CrossRef]
- Wang, S.; Wu, T.; Sun, J.; Li, Y.; Yuan, Z.; Sun, W. Single-Cell Transcriptomics Reveals the Molecular Anatomy of Sheep Hair Follicle Heterogeneity and Wool Curvature. Front. Cell Dev. Biol. 2021, 9, 800157. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Jahoda, C.A.B.; Reynolds, A.J.; Chaponnier, C.; Forester, J.C.; Gabbiani, G. Smooth muscle α-actin is a marker for hair follicle dermis in vivo and in vitro. J. Cell Sci. 1991, 99, 627–636. [Google Scholar] [CrossRef]
- Wang, S.; Hu, T.; He, M.; Gu, Y.; Cao, X.; Yuan, Z.; Lv, X.; Getachew, T.; Quan, K.; Sun, W. Defining ovine dermal papilla cell markers and identifying key signaling pathways regulating its intrinsic properties. Front. Vet. Sci. 2023, 10, 1127501. [Google Scholar] [CrossRef]
- Dunn, S.M.; Keough, R.A.; Rogers, G.E.; Powell, B.C. Regulation of a hair follicle keratin intermediate filament gene promoter. J. Cell Sci. 1998, 111, 3487–3496. [Google Scholar] [CrossRef]
- Schlake, T. Krox20, a novel candidate for the regulatory hierarchy that controls hair shaft bending. Mech. Dev. 2006, 123, 641–648. [Google Scholar] [CrossRef]
- Duverger, O.; Morasso, M.I. Epidermal patterning and induction of different hair types during mouse embryonic development. Birth Defects Res. Part C Embryo Today Rev. 2009, 87, 263–272. [Google Scholar] [CrossRef]
- Lv, X.; Li, Y.; Chen, W.; Wang, S.; Cao, X.; Yuan, Z.; Getachew, T.; Mwacharo, J.; Haile, A.; Li, Y. Association between DNA Methylation in the Core Promoter Region of the CUT-like Homeobox 1 (CUX1) Gene and Lambskin Pattern in Hu Sheep. Genes 2023, 14, 1873. [Google Scholar] [CrossRef]
- Stenn, K.S.; Cotsarelis, G. Bioengineering the hair follicle: Fringe benefits of stem cell technology. Curr. Opin. Biotechnol. 2005, 16, 493–497. [Google Scholar] [CrossRef] [PubMed]
- Watabe, R.; Yamaguchi, T.; Kabashima-Kubo, R.; Yoshioka, M.; Nishio, D.; Nakamura, M. Leptin controls hair follicle cycling. Exp. Dermatol. 2014, 23, 228–229. [Google Scholar] [CrossRef] [PubMed]
- Avigad Laron, E.; Aamar, E.; Enshell-Seijffers, D. The Mesenchymal Niche of the Hair Follicle Induces Regeneration by Releasing Primed Progenitors from Inhibitory Effects of Quiescent Stem Cells. Cell Rep. 2018, 24, 909–921.e903. [Google Scholar] [CrossRef]
- Yu, D.W.; Yang, T.; Sonoda, T.; Gaffney, K.; Jensen, P.J.; Dooley, T.; Ledbetter, S.; Freedberg, I.M.; Lavker, R.; Sun, T.T. Message of nexin 1, a serine protease inhibitor, is accumulated in the follicular papilla during anagen of the hair cycle. J. Cell Sci. 1995, 108, 3867–3874. [Google Scholar] [CrossRef]
- Driskell, R.R.; Giangreco, A.; Jensen, K.B.; Mulder, K.W.; Watt, F.M. Sox2-positive dermal papilla cells specify hair follicle type in mammalian epidermis. Development 2009, 136, 2815–2823. [Google Scholar] [CrossRef]
- Chi, W.; Wu, E.; Morgan, B.A. Dermal papilla cell number specifies hair size, shape and cycling and its reduction causes follicular decline. Development 2013, 140, 1676–1683. [Google Scholar] [CrossRef]
- Pierard-Franchimont, C.; Paquet, P.; Quatresooz, P.; Pierard, G.E. Mechanobiology and cell tensegrity: The root of ethnic hair curling? J. Cosmet. Dermatol. 2011, 10, 163–167. [Google Scholar] [CrossRef]
- Lv, X.; Sun, W.; Zou, S.; Chen, L.; Mwacharo, J.M.; Wang, J. Characteristics of the BMP7 Promoter in Hu Sheep. Animals 2019, 9, 874. [Google Scholar] [CrossRef]
- Raveh, E.; Cohen, S.; Levanon, D.; Negreanu, V.; Groner, Y.; Gat, U. Dynamic expression of Runx1 in skin affects hair structure. Mech. Dev. 2006, 123, 842–850. [Google Scholar] [CrossRef]
- Raveh, E.; Cohen, S.; Levanon, D.; Groner, Y.; Gat, U. Runx3 is involved in hair shape determination. Dev. Dyn. 2005, 233, 1478–1487. [Google Scholar] [CrossRef]
- Gebhardt, A.; Kosan, C.; Herkert, B.; Möröy, T.; Lutz, W.; Eilers, M.; Elsässer, H.-P. Miz1 is required for hair follicle structure and hair morphogenesis. J. Cell Sci. 2007, 120, 2586–2593. [Google Scholar] [CrossRef] [PubMed][Green Version]

| Gene | Primer Sequence (5′-3′) | Product Size (bp) | Annealing Temperature (°C) | Accession Number |
|---|---|---|---|---|
| SP1 | F: GCAAGACCTCACACCTACGG R: GACACTCAGGGCAGGCAAAT | 162 | 60 | XM_027967251.2 |
| KROX20 | F: CACGTCGGTGACCATTTTCC | 151 | 60 | XM_027962248.2 |
| R: TGAGACCGGAGCAAAGCTG | ||||
| CUX1 | F: GCACGACATTGAGACGGAG | 160 | 60 | XM_012104502.5 |
| R: AGCTATGGTCTCAGCCTGGT | ||||
| PCNA | F: CGAGGGCTTCGACACTTAC | 97 | 60 | XM_004014340.5 |
| R: GTCTTCATTGCCAGCACATT | ||||
| CDK2 | F: AGAAGTGGCTGCATCACAAG | 92 | 60 | NM_001142509.1 |
| R: TCTCAGAATCTCCAGGGAATAG | ||||
| cyclinD1 | F: CCGAGGAGAACAAGCAGATC | 91 | 60 | XM_027959928.2 |
| R: GAGGGTGGGTTGGAAATG | ||||
| GAPDH | F: TCTCAAGGGCATTCTAGGCTAC | 151 | 60 | NM_001190390.1 |
| R: GCCGAATTCATTGTCGTACCAG |
| Primer Name | Primer Sequence (5′-3′) | Product Size (bp) | Annealing Temperature (°C) |
|---|---|---|---|
| pcDNA3.1-SP1 | F: CTAGCGTTTAAACTTAAGCTTATGAGCGACCAAGATCACTCCATG | 2361 | 65 |
| R: TGCTGGATATCTGCAGAATTCTCAGAAGCCATTGCCACTGATATTG | |||
| pcDNA3.1-KROX20 | F: CTAGCGTTTAAACTTAAGCTTATGCGAGTCGGGCTCCCCT | 1458 | 65 |
| R: TGCTGGATATCTGCAGAATTCTCAGGGCGTCCGGGTCC | |||
| pGL3-CUX1-1601 | F: GCGTGCTAGCCCGGGCTCGAGTCCCCTGGACCAGCCTAGG R: CAGTACCGGAATGCCAAGCTTTCTTCCTCACTCCTAGTTGGTTCTG | 1601 | 63 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lv, X.; He, M.; Zhou, H.; Wang, S.; Cao, X.; Yuan, Z.; Getachew, T.; Li, Y.; Sun, W. SP1 and KROX20 Regulate the Proliferation of Dermal Papilla Cells and Target the CUX1 Gene. Animals 2024, 14, 429. https://doi.org/10.3390/ani14030429
Lv X, He M, Zhou H, Wang S, Cao X, Yuan Z, Getachew T, Li Y, Sun W. SP1 and KROX20 Regulate the Proliferation of Dermal Papilla Cells and Target the CUX1 Gene. Animals. 2024; 14(3):429. https://doi.org/10.3390/ani14030429
Chicago/Turabian StyleLv, Xiaoyang, Mingliang He, Hui Zhou, Shanhe Wang, Xiukai Cao, Zehu Yuan, Tesfaye Getachew, Yutao Li, and Wei Sun. 2024. "SP1 and KROX20 Regulate the Proliferation of Dermal Papilla Cells and Target the CUX1 Gene" Animals 14, no. 3: 429. https://doi.org/10.3390/ani14030429
APA StyleLv, X., He, M., Zhou, H., Wang, S., Cao, X., Yuan, Z., Getachew, T., Li, Y., & Sun, W. (2024). SP1 and KROX20 Regulate the Proliferation of Dermal Papilla Cells and Target the CUX1 Gene. Animals, 14(3), 429. https://doi.org/10.3390/ani14030429







