Genome-Wide Microsatellites in Acanthopagrus latus: Development, Distribution, Characterization, and Polymorphism
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
2.2. Genome-Wide Microsatellites Mining
2.3. Characteristics of SSRs in the A. latus Genome and Chromosome
2.4. GO and KEGG Enrichment
2.5. Primer Designing and Validation
2.6. SSR Markers Cross-Species Transferability in Acanthopagrus
3. Results
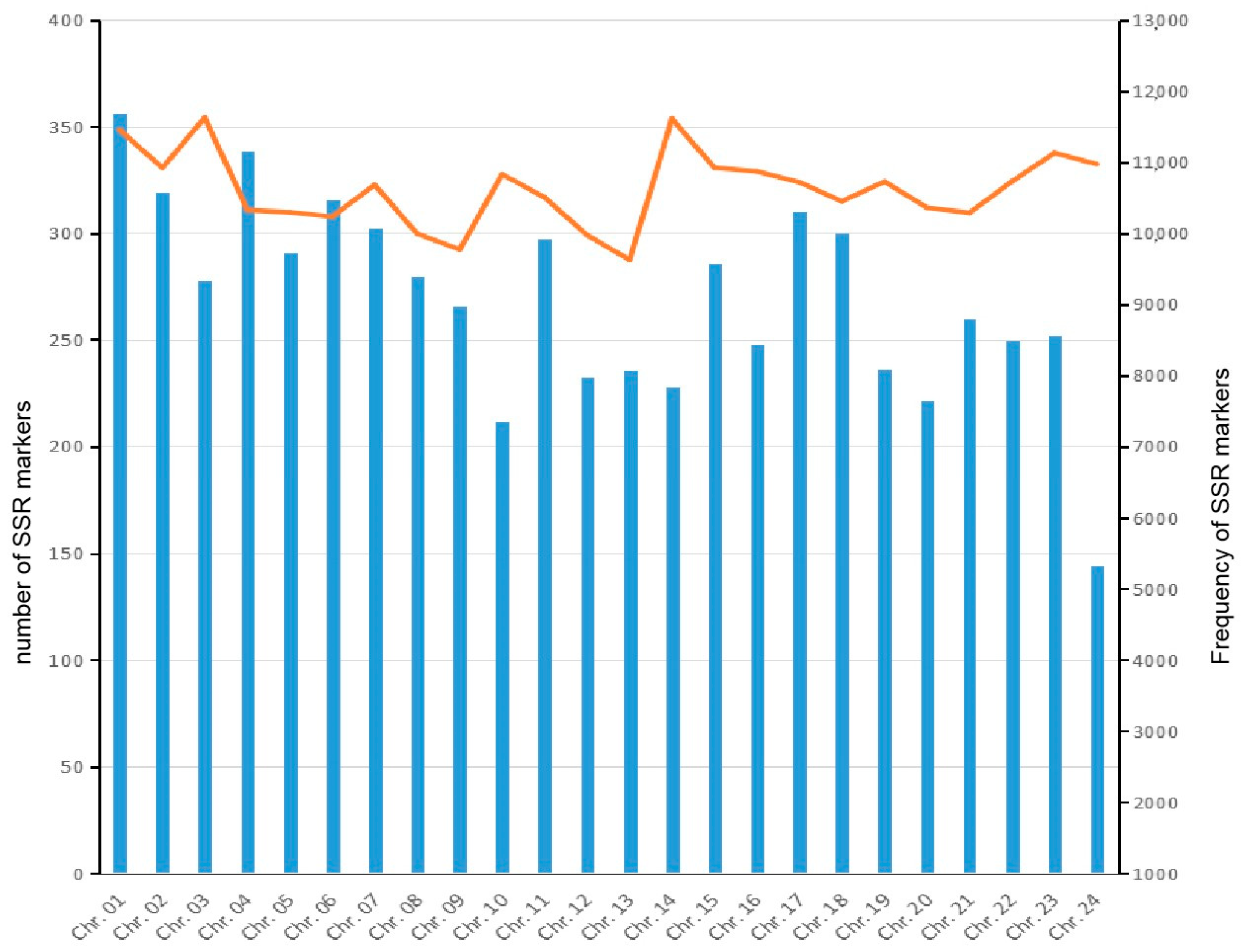
3.1. Features of the SSRs in the A. latus Genome
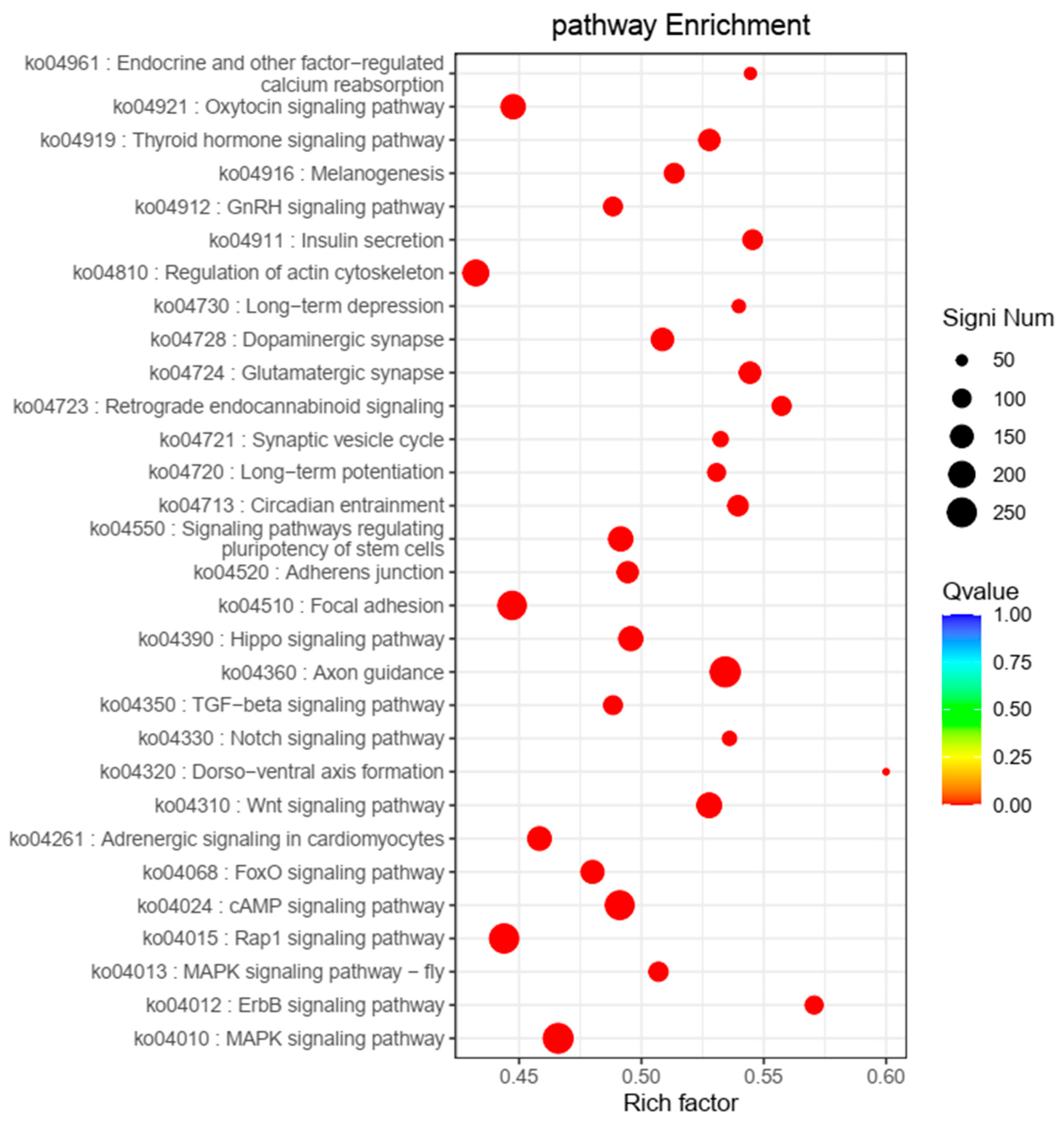
3.2. GO and KEGG Pathway Analyses
3.3. Development of SSR Markers
3.4. Evaluation of the Polymorphism of SSR Markers
3.5. Cross-Species Development of SSR Markers in A. schlegelii
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Morshedi, V.; Hamedi, S.; Pourkhazaei, F.; Torfi Mozanzadeh, M.; Tamadoni, R.; Ebadi, M.; Esmaili, A.; Azodi, M.; Gisbert, E. Larval rearing and ontogeny of digestive enzyme activities in yellowfin seabream (Acanthopagrus latus, Houttuyn 1782). Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2021, 261, 111044. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Guo, Y.; Zhao, N.; Zhang, S.; Pei, K.; Qin, C. Fish eDNA detection and its technical optimization: A case study of Acanthopagrus latus. Mar. Environ. Res. 2022, 176, 105588. [Google Scholar] [CrossRef] [PubMed]
- Bureau of Fisheries; Ministry of Agriculture of China. China Fishery Statistical Yearbook; China Statistics Press: Beijing, China, 2024; p. 22.
- Zhu, K.C.; Zhang, N.; Liu, B.S.; Guo, L.; Guo, H.Y.; Jiang, S.G.; Zhang, D.C. A chromosome-level genome assembly of the yellowfin seabream (Acanthopagrus latus; Hottuyn 1782) provides insights into its osmoregulation and sex reversal. Genomics 2021, 113, 1617–1627. [Google Scholar] [CrossRef]
- Jia, P.Y.; Guo, H.Y.; Zhu, K.C.; Liu, B.S.; Guo, L.; Zhang, N.; Jiang, S.G.; Zhang, D.C. Cryopreservation of sperm of Acanthopagrus latus. S. China Fish. Sci. 2021, 17, 58–65, (Abstract in English Only). [Google Scholar]
- Trochez-Solarte, J.D.; Ruiz-Erazo, X.; Almanza-Pinzon, M.; Zambrano-Gonzalez, G. Role of microsatellites in genetic analysis of Bombyx mori silkworm: A review. F1000Research 2019, 8, 1424. [Google Scholar] [CrossRef]
- Wenne, R. Microsatellites as Molecular Markers with Applications in Exploitation and Conservation of Aquatic Animal Populations. Genes 2023, 14, 808. [Google Scholar] [CrossRef]
- Huang, H.; Fan, S.; Wang, P.; Chen, J.; Zhao, C.; Yan, L.; Qiu, L.; Pan, Y. Genetic diversity analysis of six geographical populations of Lateolabrax maculatus based on microsatellite markers. S. China Fish. Sci. 2022, 18, 99–106, (Abstract in English Only). [Google Scholar]
- Lu, H.Y.; Tian, H.F.; Hu, Q.M.; Li, Z. Parentage assignment of Monopterus albus using multiplex PCR of microsatallites. S. China Fish. Sci. 2023, 19, 97–106, (Abstract in English Only). [Google Scholar]
- Kawakatsu, K.; Yagi, M.; Harada, T.; Yamaguchi, H.; Itoh, T.; Kumagai, M.; Itoh, R.; Numa, H.; Katayose, Y.; Kanamori, H.; et al. Development of an SSR marker-based genetic linkage map and identification of a QTL associated with flowering time in Eustoma. Breed. Sci. 2021, 71, 344–353. [Google Scholar] [CrossRef]
- Ma, Z.; Gao, W.; Liu, L.; Liu, M.; Zhao, N.; Han, M.; Wang, Z.; Jiao, W.; Gao, Z.; Hu, Y.; et al. Identification of QTL for resistance to root rot in sweetpotato (Ipomoea batatas (L.) Lam) SSR Linkages Maps. BMC Genom. 2020, 21, 366. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, C.; Lin, H.H.; Kang, S.R.; Hung, C.Y.; Sun, P.Y.; Yu, C.C.; Toh, K.L.; Yu, P.J.; Ju, Y.T. Development of 16 novel EST-SSR markers for species identification and cross-genus amplification in sambar, sika, and red deer. PLoS ONE 2022, 17, e0265311. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.X.; Zhai, Y.; Xiao, Y.; Niu, S.F.; Zhang, H.R.; Li, X.; Chen, W.Y. Microsatellite marker development for Acanthopagrus latus and cross-species amplification in the family Sparidae. J. Appl. Oceanogr. 2019, 38, 356–364. [Google Scholar]
- Kamarudin, A.S.; Makoto, W.; Takao, O.; Kenichi, O.; Tetsuya, U. Ten novel polymorphic microsatellite loci for yellowfifin black seabream (Acanthopagrus latus), Conser. Genet. Resour. 2012, 4, 909–911. [Google Scholar]
- Hu, T.; Chitnis, N.; Monos, D.; Dinh, A. Next-generation sequencing technologies: An overview. Hum. Immunol. 2021, 82, 801–811. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Li, Q.; Yu, H.; Kong, L. Genome-wide analysis of simple sequence repeats in marine animals-a comparative approach. Mar. Biotech. 2014, 16, 604–619. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Zhang, L.; Chen, S. Microsatellite Genome-Wide Database Development for the Commercial Blackhead Seabream (Acanthopagrus schlegelii). Genes 2023, 14, 620. [Google Scholar] [CrossRef]
- Fan, S.; Huang, H.; Liu, Y.; Wang, P.; Zhao, C.; Yan, L.; Qiao, X.; Qiu, L. Genome-wide identification of microsatellite and development of polymorphic SSR markers for spotted sea bass (Lateolabrax maculatus). Aquac. Rep. 2021, 20, 100677. [Google Scholar]
- Peng, Y.; Li, J.; Wang, T.; Zhang, K.; Ning, X.; Ji, J.; Yin, S. Preliminary study on distribution characteristics and positioning of microsatellites in whole genome of Pelteobagrus vachelli. S. China Fish. Sci. 2022, 18, 90–98, (Abstract in English Only). [Google Scholar]
- Qing, S.; Chen, K.; Liu, H.; Mi, O.; Luo, Q.; Wang, Y.; Xu, S.; Zhao, J. Characteristics of micorsatellites and genetic structure of wild Channa maculata. S. China Fish. Sci. 2020, 16, 47–60, (Abstract in English Only). [Google Scholar]
- Lu, J.; Gao, D.; Sims, Y.; Fang, W.; Collins, J.; Torrance, J.; Lin, G.; Xie, J.; Liu, J.; Howe, K. Chromosome-level Genome Assembly of Acanthopagrus latus Progvides Insights into Salinity Stress Adaptation of Sparidae. Mar. Biotech. 2022, 24, 655–660. [Google Scholar] [CrossRef]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Fang, L.; Zheng, H.; Zhang, Y.; Chen, J.; Zhang, Z.; Wang, J.; Li, S.; Li, R.; Bolund, L.; et al. WEGO: A web tool for plotting GO annotations. Nucleic Acids Res. 2006, 34B, W293–W297. [Google Scholar] [CrossRef] [PubMed]
- Yeh, C.; Boule, T. POPGENE-1.32: A Free Program for the Analysis of Genetic Variation Among and Within Populations Using Co-Dominant and Dominant Markers; University of Alberta: Edmonton, AB, Canada, 2000; Department of Renewable Resources. [Google Scholar]
- Liu, K.; Muse, S.V. PowerMarker: An integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef] [PubMed]
- Mathew, D.; Anju, P.S.; Tom, A.; Johnson, N.; Lidia George, M.; Davis, S.P.; Ravisankar, V.; Asha, K.N. Genome-wide microsatellites and species specific markers in genus Phytophthora revealed through whole genome analysis. 3 Biotech 2020, 10, 442. [Google Scholar] [CrossRef]
- Sahu, B.P.; Majee, P.; Singh, R.R.; Sahoo, N.; Nayak, D. Genome-wide identification and characterization of microsatellite markers within the Avipoxviruses. 3 Biotech 2022, 12, 113. [Google Scholar] [CrossRef]
- Kumari, R.; Wankhede, D.P.; Bajpai, A.; Maurya, A.; Prasad, K.; Gautam, D.; Rangan, P.; Latha, M.; John, K.; Bhat, K.V.; et al. Genome wide identification and characterization of microsatellite markers in black pepper (Piper nigrum): A valuable resource for boosting genomics applications. PLoS ONE 2019, 14, e0226002. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Li, W.; Jian, Z.; Yue, B.; Yan, Y. Genome-wide distribution and organization of microsatellites in six species of birds. Biochem. Syst. Ecol. 2016, 67, 95–102. [Google Scholar] [CrossRef]
- Wang, X.T.; Zhang, Y.J.; Qiao, L.; Chen, B. Comparative analyses of simple sequence repeats (SSRs) in 23 mosquito species genomes: Identification, characterization and distribution (Diptera: Culicidae). Insect. Sci. 2019, 26, 607–619. [Google Scholar] [CrossRef] [PubMed]
- Satam, H.; Joshi, K.; Mangrolia, U.; Waghoo, S.; Zaidi, G.; Rawool, S.; Thakare, R.P.; Banday, S.; Mishra, A.K.; Das, G. Next-generation sequencing technology: Current trends and advancements. Biology 2023, 12, 997. [Google Scholar] [CrossRef] [PubMed]
- Logsdon, G.A.; Vollger, M.R.; Eichler, E.E. Long-read human genome sequencing and its applications. Nat. Rev. Genet. 2020, 21, 597–614. [Google Scholar] [CrossRef] [PubMed]
- Geethanjali, S.; Kadirvel, P.; Anumalla, M.; Sadhana, N.H.; Annamalai, A.; Ali, J. Streamlining of Simple Sequence Repeat Data Mining Methodologies and Pipelines for Crop Scanning. Plants 2024, 13, 2619. [Google Scholar] [CrossRef] [PubMed]
- Pootakham, W. Genotyping by Sequencing (GBS) for Genome-Wide SNP Identification in Plants. Methods Mol. Biol. 2023, 2638, 1–8. [Google Scholar] [PubMed]
- He, J.; Gai, J. Genome-Wide Association Studies (GWAS). Methods Mol. Biol. 2023, 2638, 123–146. [Google Scholar] [PubMed]
- Gaur, R.; Verma, S.; Pradhan, S.; Ambreen, H.; Bhatia, S. A high-density SNP-based linkage map using genotyping-by-sequencing and its utilization for improved genome assembly of chickpea (Cicer arietinum L.). Funct. Integr. Genom. 2020, 20, 763–773. [Google Scholar] [CrossRef] [PubMed]
- Saif-Ur-Rehman, M.; Hassan, F.U.; Reecy, J.; Deng, T. Whole-genome SNP markers reveal runs of homozygosity in indigenous cattle breeds of Pakistan. Anim. Biotechnol. 2023, 34, 1384–1396. [Google Scholar] [CrossRef] [PubMed]
- Fandade, V.; Singh, P.; Singh, D.; Sharma, H.; Thakur, G.; Saini, S.; Kumar, P.; Mantri, S.; Bishnoi, O.P.; Roy, J. Genome-wide identification of microsatellites for mapping, genetic diversity and cross-transferability in wheat (Triticum spp.). Genes 2024, 896, 148039. [Google Scholar] [CrossRef]
- Cui, X.; Li, C.; Qin, S.; Huang, Z.; Gan, B.; Jiang, Z.; Huang, X.; Yang, X.; Li, Q.; Xiang, X.; et al. High-throughput sequencing-based microsatellite genotyping for polyploids to resolve allele dosage uncertainty and improve analyses of genetic diversity, structure and differentiation: A case study of the hexaploid Camellia oleifera. Mol. Ecol. Resour. 2022, 22, 199–211. [Google Scholar] [CrossRef]
- Lepais, O.; Chancerel, E.; Boury, C.; Salin, F.; Manicki, A.; Taillebois, L.; Dutech, C.; Aissi, A.; Bacles, C.F.E.; Daverat, F.; et al. Fast sequence-based microsatellite genotyping development workflow. PeerJ 2020, 8, e9085. [Google Scholar] [CrossRef]
- Wang, Y.; Sha, H.; Li, X.; Zhou, T.; Luo, X.; Zou, G.; Chai, Y.; Liang, H. Microsatellite Characteristics of Silver Carp (Hypophthalmichthys molitrix) Genome and Genetic Diversity Analysis in Four Cultured Populations. Genes 2022, 13, 1267. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.; Zhou, Y.; Price, M.; Song, Z. Genome-wide characterization of microsatellite DNA in fishes: Survey and analysis of their abundance and frequency in genome-specific regions. BMC Genom. 2021, 22, 421. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yuan, J.; Sun, Y.; Li, S.; Gao, Y.; Yu, Y.; Liu, C.; Wang, Q.; Lv, X.; Zhang, X.; et al. Penaeid shrimp genome provides insights into benthic adaptation and frequent molting. Nat. Commun. 2019, 10, 356. [Google Scholar] [CrossRef] [PubMed]
- Qi, W.H.; Jiang, X.M.; Du, L.M.; Xiao, G.S.; Hu, T.Z.; Yue, B.S.; Quan, Q.M. Genome-Wide Survey and Analysis of Microsatellite Sequences in Bovid Species. PLoS ONE 2015, 10, e0133667. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Fu, Y.; Gu, P.; Zhang, Y.; Tu, W.; Chao, Z.; Wu, H.; Cao, J.; Zhou, X.; Liu, B.; et al. Genome-Wide Characterization and Comparative Analyses of Simple Sequence Repeats among Four Miniature Pig Breeds. Animals 2020, 10, 1792. [Google Scholar] [CrossRef]
- Perinchery, G.; Nojima, D.; Goharderakhshan, R.; Tanaka, Y.; Alonzo, J.; Dahiya, R. Microsatellite instability of dinucleotide tandem repeat sequences is higher than trinucleotide, tetranucleotide and pentanucleotide repeat sequences in prostate cancer. Int. J. Oncol. 2000, 16, 1203–1209. [Google Scholar] [CrossRef]
- Chakraborty, R.; Kimmel, M.; Stivers, D.N.; Davison, L.J.; Deka, R. Relative mutation rates at di-, tri-, and tetranucleotide microsatellite loci. Proc. Natl. Acad. Sci. USA 1997, 94, 1041–1046. [Google Scholar] [CrossRef] [PubMed]
- Edwards, Y.J.K.; Elgar, G.; Clark, M.S.; Bishop, M.J. The identification and characterization of microsatellites in the compact genome of the Japanese pufferfish, Fugu rubripes: Perspectives in functional and comparative genomic analyses. J. Mol. Biol. 1998, 278, 843–854. [Google Scholar] [CrossRef]
- Xiong, L.W.; Wang, Q.; Qiu, G.F. Large-scale isolation of microsatellites from Chinese Mitten Crab Eriocheir sinensis via a Solexa Genomic Survey. Int. J. Mol. Sci. 2012, 13, 16333–16345. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Xu, Y.; Li, Z.; Fan, J.; Yang, Y. Genome-wide mining of microsatellites in king cobra (Ophiophagus hannah) and cross-species development of tetranucleotide SSR markers in Chinese cobra (Naja atra). Mol. Biol. Rep. 2019, 46, 6087–6098. [Google Scholar] [CrossRef]
- Castagnone-Sereno, P.; Danchin, E.G.; Deleury, E.; Guillemaud, T.; Malausa, T.; Abad, P. Genome-wide survey and analysis of microsatellites in nematodes, with a focus on the plant-parasitic species Meloidogyne incognita. BMC Genom. 2010, 11, 598. [Google Scholar] [CrossRef]
- Tian, H.F.; Hu, Q.M.; Li, Z. Genome-wide identification of simple sequence repeats and development of polymorphic SSR markers in swamp eel (Monopterus albus). Sci. Prog. 2021, 104, 368504211035597. [Google Scholar] [CrossRef]
- Kong, J.; Gao, H.A. Analysis of tandem repeats in the genome of Chinese shrimp Fenneropenaeus chinensis. Chin. Sci. Bull. 2005, 50, 1462–1469. [Google Scholar] [CrossRef]
- Luo, H.; Wang, X.; Zhan, G.; Wei, G.; Zhou, X.; Zhao, J.; Huang, L.; Kang, Z. Genome-Wide Analysis of Simple Sequence Repeats and Efficient Development of Polymorphic SSR Markers Based on Whole Genome Re-Sequencing of Multiple Isolates of the Wheat Stripe Rust Fungus. PLoS ONE 2015, 10, e0130362. [Google Scholar] [CrossRef] [PubMed]
- Jian, Y.; Yan, W.; Xu, J.; Duan, S.; Li, G.; Jin, L. Genome-wide simple sequence repeat markers in potato: Abundance, distribution, composition, and polymorphism. DNA Res. 2021, 28, dsab020. [Google Scholar] [CrossRef]
- Manee, M.M.; Algarni, A.T.; Alharbi, S.N.; Al-Shomrani, B.M.; Ibrahim, M.A.; Binghadir, S.A.; Al-Fageeh, M.B. Genome-wide characterization and analysis of microsatellite sequences in camelid species. Mammal. Res. 2020, 65, 359–373. [Google Scholar] [CrossRef]
- Bagshaw, A.T.M. Functional Mechanisms of Microsatellite DNA in Eukaryotic Genomes. Genome Biol. Evol. 2017, 9, 2428–2443. [Google Scholar] [CrossRef]
- Yin, B.; Wang, H.; Zhu, P.; Weng, S.; He, J.; Li, C. A Polymorphic (CT)n SSR Influences the Activity of the Litopenaeus vannamei IRF Gene Implicated in Viral Resistance. Front. Genet. 2019, 10, 1257. [Google Scholar] [CrossRef]
- Yin, B.; Wang, H.; Weng, S.; Li, S.; He, J.; Li, C. A simple sequence repeats marker of disease resistance in shrimp Litopenaeus vannamei and its application in selective breeding. Front. Genet. 2023, 14, 1144361. [Google Scholar] [CrossRef]
- Ayuso, P.; Martínez, C.; Pastor, P.; Lorenzo-Betancor, O.; Luengo, A.; Jiménez-Jiménez, F.J.; Alonso-Navarro, H.; Agúndez, J.A.; García-Martín, E. An association study between Heme oxygenase-1 genetic variants and Parkinson’s disease. Front. Cell. Neurosci. 2014, 8, 298. [Google Scholar] [CrossRef]
- Zhang, L.; Song, F.F.; Huang, Y.B.; Zheng, H.; Song, F.J.; Chen, K.X. Association between the (GT)n polymorphism of the HO-1 gene promoter region and cancer risk: A meta-analysis. Asian Pac. J. Cancer Prev. APJCP 2014, 15, 4617–4622. [Google Scholar] [CrossRef]
- Chen, H.Y.; Ma, S.L.; Huang, W.; Ji, L.; Leung, V.H.; Jiang, H.; Yao, X.; Tang, N.L. The mechanism of transactivation regulation due to polymorphic short tandem repeats (STRs) using IGF1 promoter as a model. Sci. Rep. 2016, 6, 38225. [Google Scholar] [CrossRef]
- Geldermann, H.; Mir, M.R.; Kuss, A.W.; Bartenschlager, H. OLA-DRB1 microsatellite variants are associated with ovine growth and reproduction traits. Genet. Sel. Evol. 2006, 38, 431–444. [Google Scholar] [CrossRef][Green Version]
- Harris, R.M.; Hofmann, H.A. Seeing is believing: Dynamic evolution of gene families. Proc. Natl. Acad. Sci. USA 2015, 112, 1252–1253. [Google Scholar] [CrossRef]
- Kim, J.C.; Mirkin, S.M. The balancing act of DNA repeat expansions. Curr. Opin. Genet. Dev. 2013, 23, 280–288. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Qi, X.; Gao, R.; Wang, J.; Dong, B.; Jiang, J.; Chen, S.; Guan, Z.; Fang, W.; Liao, Y.; et al. Microsatellite polymorphism among Chrysanthemum sp. polyploids: The influence of whole genome duplication. Sci. Rep. 2014, 4, 6730. [Google Scholar] [CrossRef]
- Pan, C.; Gao, C.; Chen, T.; Chen, X.; Yang, C.; Zeng, D.; Feng, P.; Jiang, W.; Peng, M. The complete mitochondrial genome of yellowfin seabream, Acanthopagrus latus (Percoiformes, Sparidae) from Beibu Bay. Mitochondrial DNA Part B Resour. 2021, 6, 1313–1314. [Google Scholar] [CrossRef]

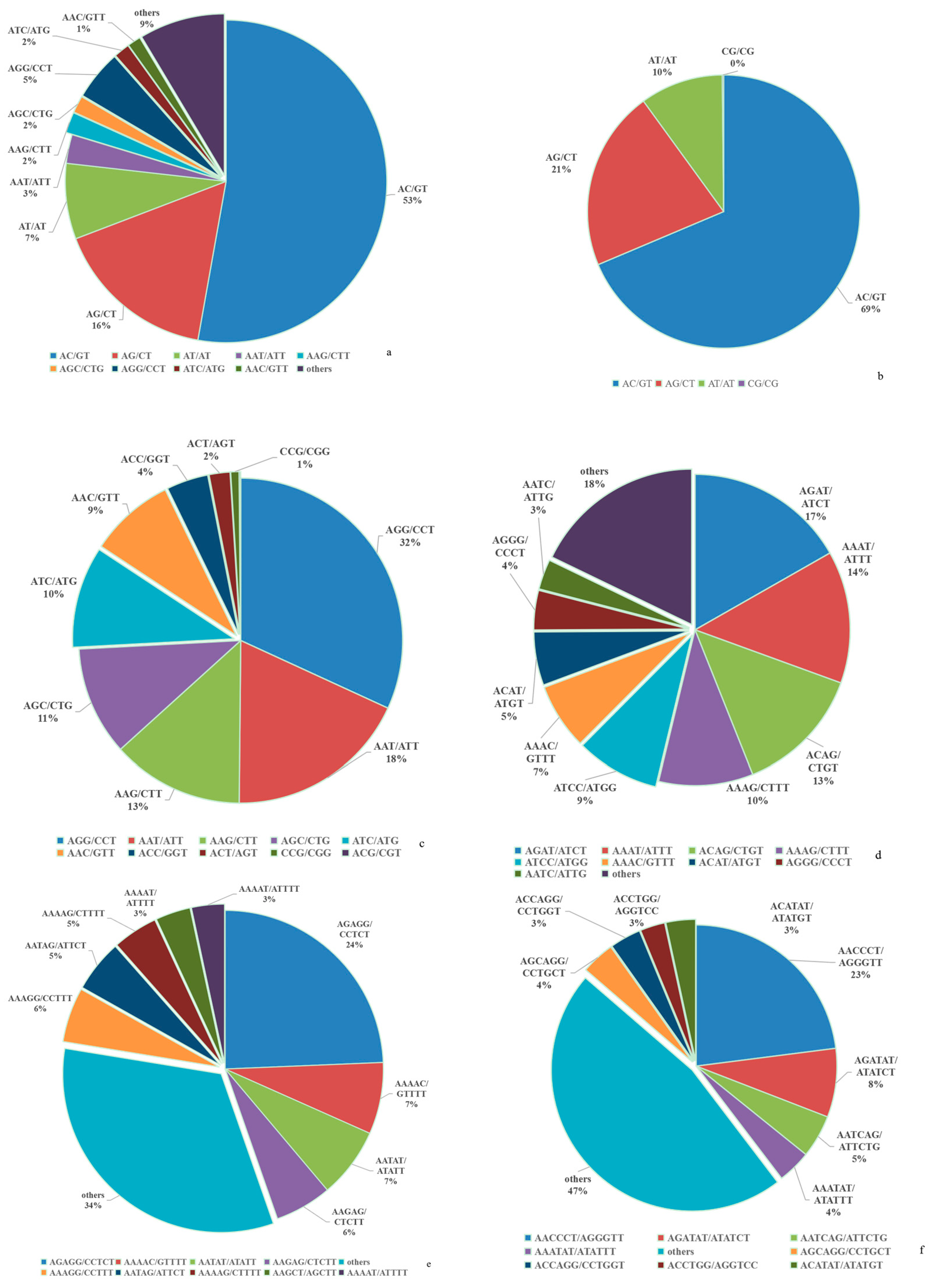
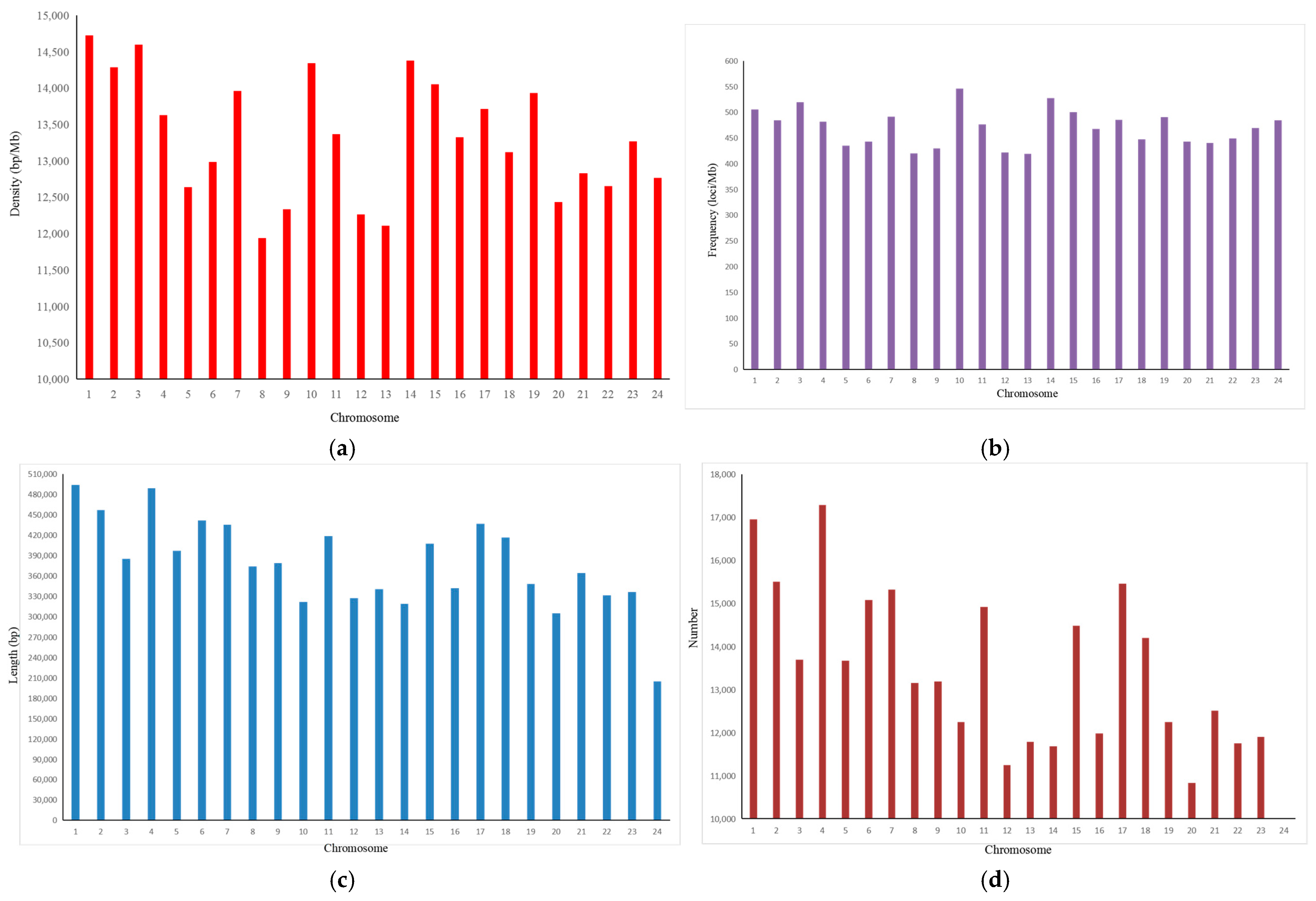
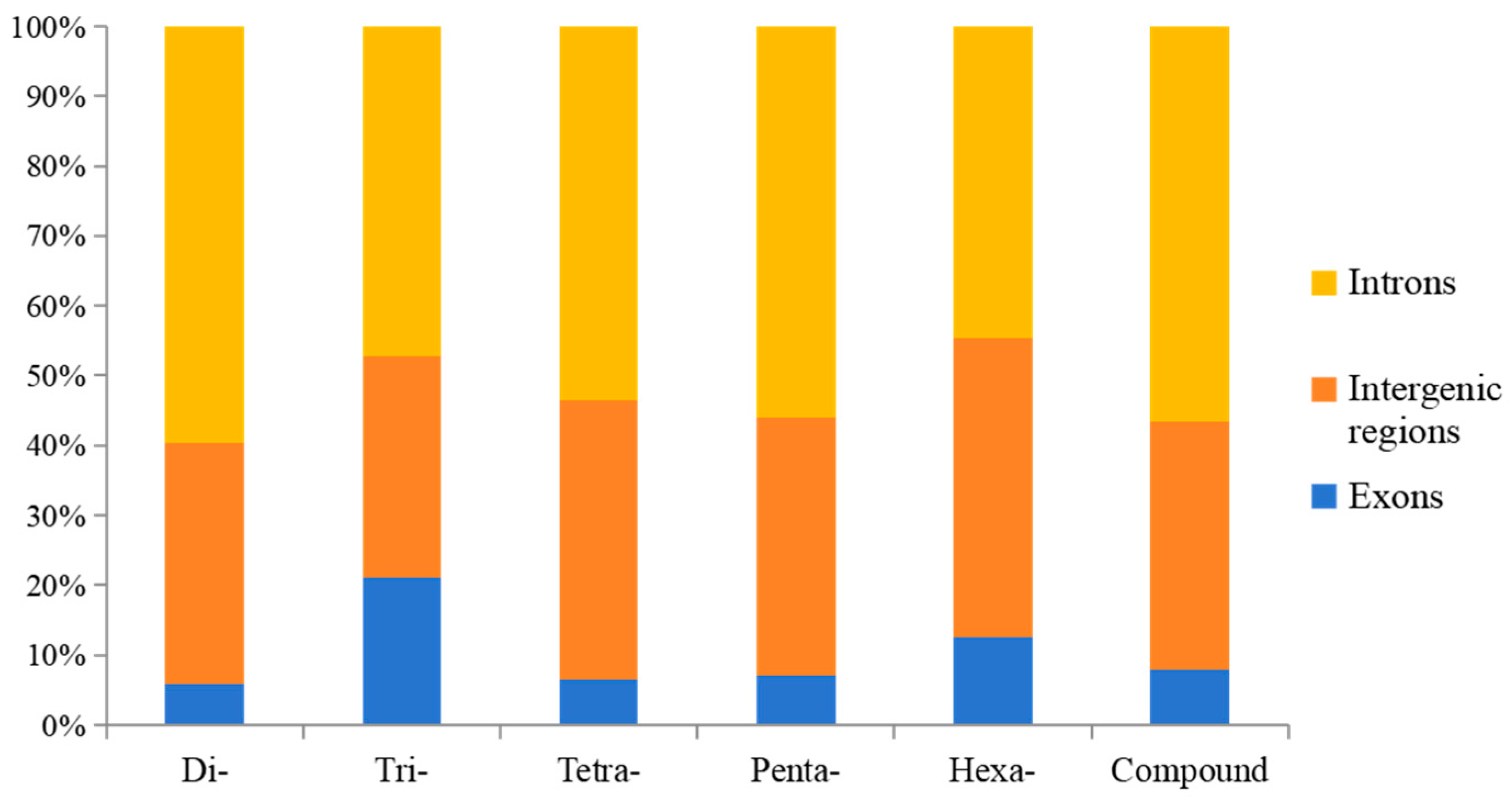

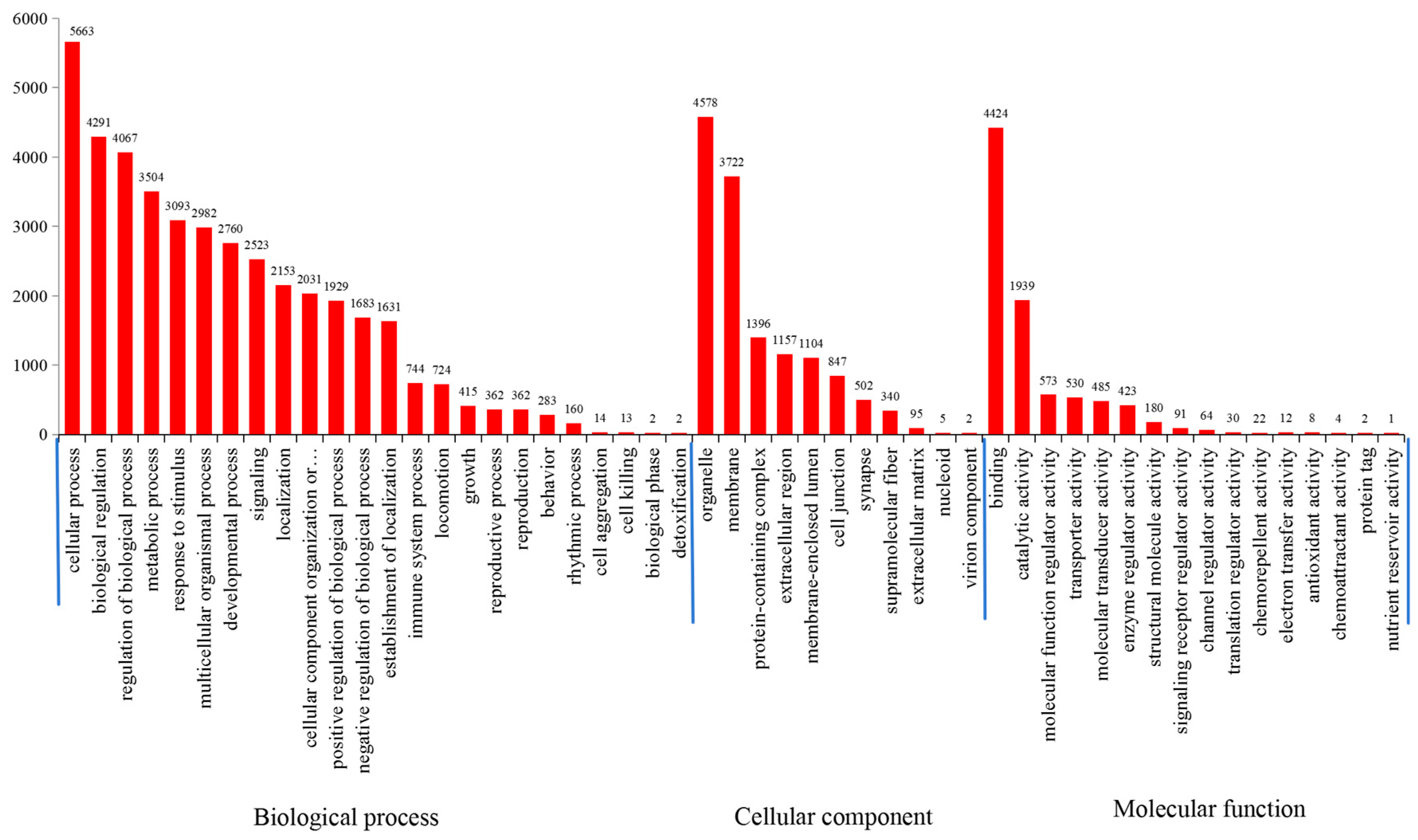
| Repeats | Number | Proportion (%) | Frequency (loci/Mb) | Length (bp) | Density (bp/Mb) |
|---|---|---|---|---|---|
| Di- | 245,271 | 76.92% | 357.99 | 7,045,666 | 10,283.73 |
| Tri- | 50,220 | 15.75% | 73.30 | 1,188,063 | 1734.08 |
| Tetra- | 18,107 | 5.68% | 26.43 | 628,736 | 917.69 |
| Penta- | 4108 | 1.29% | 6.00 | 162,205 | 236.75 |
| Hexa- | 1156 | 0.36% | 1.69 | 45,000 | 65.68 |
| Total | 318,862 | 100% | 468.41 | 9,069,670 | 13,237.93 |
| Categories | Number | Frequency (loci/Mb) | Length (bp) | Density (bp/Mb) |
|---|---|---|---|---|
| AC | 168,390 | 245.78 | 5,004,294 | 7304.18 |
| AG | 52,305 | 76.34 | 1,113,538 | 1625.30 |
| AT | 24,152 | 35.25 | 922,230 | 1346.07 |
| AGG | 15,989 | 23.34 | 346,359 | 505.54 |
| AAT | 9198 | 13.43 | 292,392 | 426.77 |
| AAG | 6610 | 9.65 | 167,997 | 245.21 |
| AGC | 5448 | 7.95 | 111,348 | 162.52 |
| ATC | 5069 | 7.40 | 113,091 | 165.07 |
| AAC | 4268 | 6.23 | 85,422 | 124.68 |
| AGAT | 3038 | 4.43 | 144,423 | 210.80 |
| Genomic Region | Exons | Intergenic Regions | Introns | All |
|---|---|---|---|---|
| Di- | 7909 | 45,439 | 78,991 | 132,339 |
| Tri- | 6655 | 10,027 | 14,969 | 31,651 |
| Tetra- | 761 | 4689 | 6308 | 11,758 |
| Penta- | 207 | 1071 | 1623 | 2901 |
| Hexa- | 70 | 236 | 247 | 553 |
| Compound | 3776 | 16,794 | 26,801 | 47,371 |
| All | 19,378 | 78,256 | 128,939 | 226,573 |
| Percentage | 8.55% | 34.54% | 56.91% |
| Locus | Na | Ne | Ho | He | I | PIC |
|---|---|---|---|---|---|---|
| Hqd1 | 8 | 4.2582 | 0.1724 | 0.7786 | 1.7101 | 0.7370 |
| Hqd2 | 10 | 4.7380 | 0.3103 | 0.8028 | 1.7962 | 0.7601 |
| Hqd3 | 9 | 4.5459 | 0.1034 | 0.7937 | 1.7266 | 0.7482 |
| Hqd6 | 6 | 4.2690 | 0.4138 | 0.7792 | 1.5529 | 0.7270 |
| Hqd8 | 4 | 1.9157 | 0.5172 | 0.4864 | 0.8398 | 0.4145 |
| Hqd12 | 3 | 2.6869 | 0.4483 | 0.6388 | 1.0333 | 0.5488 |
| Hqd14 | 8 | 3.6805 | 0.1379 | 0.7411 | 1.5372 | 0.6870 |
| Hqd32 | 7 | 5.2399 | 0.2069 | 0.8234 | 1.7807 | 0.7843 |
| Hqd33 | 7 | 3.5787 | 0.1724 | 0.7332 | 1.5565 | 0.6907 |
| mean | 6.8889 | 3.8792 | 0.2759 | 0.7308 | 1.5037 | 0.6775 |
| St. Dev | 2.2608 | 1.0450 | 0.1513 | 0.1064 | 0.3396 | 0.1200 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, C.; Luo, C.; Xiang, G.; Huang, J.; Shao, L.; Huang, H.; Fan, S. Genome-Wide Microsatellites in Acanthopagrus latus: Development, Distribution, Characterization, and Polymorphism. Animals 2024, 14, 3709. https://doi.org/10.3390/ani14243709
Peng C, Luo C, Xiang G, Huang J, Shao L, Huang H, Fan S. Genome-Wide Microsatellites in Acanthopagrus latus: Development, Distribution, Characterization, and Polymorphism. Animals. 2024; 14(24):3709. https://doi.org/10.3390/ani14243709
Chicago/Turabian StylePeng, Chao, Congqiang Luo, Guangqing Xiang, Jiezhen Huang, Liye Shao, Haihong Huang, and Sigang Fan. 2024. "Genome-Wide Microsatellites in Acanthopagrus latus: Development, Distribution, Characterization, and Polymorphism" Animals 14, no. 24: 3709. https://doi.org/10.3390/ani14243709
APA StylePeng, C., Luo, C., Xiang, G., Huang, J., Shao, L., Huang, H., & Fan, S. (2024). Genome-Wide Microsatellites in Acanthopagrus latus: Development, Distribution, Characterization, and Polymorphism. Animals, 14(24), 3709. https://doi.org/10.3390/ani14243709




