Simple Summary
This study represents the first investigation of enteric protozoan parasites in hedgehogs in mainland Portugal, focusing on prevalent parasites, including Blastocystis sp., Balantioides coli, Cryptosporidium spp., and Giardia duodenalis. Blastocystis and Cryptosporidium were detected with an occurrence of 0.91% (1/110, 95% CI: 0.02–4.96). These findings highlight the need for continued research and surveillance and mark the first report of Blastocystis and Cryptosporidium occurrences in hedgehogs of Portugal, providing valuable data to address the occurrence and distribution of these parasites. The detection of parasites with zoonotic potential raises concerns about potential transmission to humans, emphasizing the necessity for continuous monitoring. This study suggests the importance of additional research with larger sample sizes and a wider geographical representation. This can contribute to the development of prevention and control strategies, aiming to better understand and manage potential risks in hedgehog populations and their interaction with other animals and humans.
Abstract
Enteric protozoan parasites, such as Blastocystis sp., Balantioides coli, Cryptosporidium spp., and Giardia duodenalis, may have implications for both animal and human health.Transmitted through the fecal–oral route, these parasites cause symptoms such as diarrhea, abdominal pain, and weight loss. This study investigated the presence of these enteric protozoan parasites and genetically characterized them in hedgehogs from Portugal. A total of 110 hedgehog stool samples were collected. Molecular detection methods showed an overall occurrence of protozoa in 1.82% (2/110 95% CI: 0.22–6.41) of hedgehogs, with Blastocystis being found in one hedgehog and Cryptosporidium being found in another. No evidence for the presence of B. coli or G. duodenalis was found. This study suggests that there is a need to stay aware of hedgehogs as potential hosts of enteric protozoa. Ongoing research and surveillance efforts are recommended to explore practical prevention and control strategies. The results contribute to the limited knowledge of these parasites in Portuguese hedgehog populations and underscore their potential relevance to both veterinary and public health.
1. Introduction
Blastocystis sp., Balantioides coli, Cryptosporidium spp., and Giardia duodenalis are common enteric protozoan parasites that share the same fecal–oral infection route and can pose a threat to the health and well-being of many animals [1,2,3,4]. Transmission occurs by ingesting resistant cysts (Blastocystis sp., B. coli, and G. duodenalis) or oocysts (Cryptosporidium spp.) [5,6,7,8]. Infection can be direct (human-to-human, animal-to-human) or indirect when exposed to contaminated water and food [8]. Common symptoms and clinical signs of enteric protozoa found in humans and animals are acute diarrhea, abdominal pain, malabsorption, dehydration, anorexia, and weight loss [5,7,9,10]. However, asymptomatic infections can also occur frequently, depending on the health status of the host and also, particularly, on the protozoan species in question [3,11,12,13].
Blastocystis sp. is characterized by a lack of flagella [4]. This protozoan has a wide range of hosts, including humans, non-human primates, farm animals, and other wild mammals, such as artiodactyls, proboscideans, rodents, marsupials, birds, reptiles, amphibians, and insects [14]. The number of Blastocystis sp. infections is estimated to be 1 billion humans worldwide [15]. Its prevalence can vary from one country to another, with higher rates generally observed in developing countries [16], most likely due to poor hygiene habits and socioeconomic conditions [11]. Currently, there are at least 37 described subtypes (ST) of Blastocystis sp. The most common subtypes in humans are ST1–ST9, although a recent study has suggested the presence of ST12 in humans as well [14,17].
Balantioides coli is the only ciliate that infects humans [3]. Pigs are considered the main reservoirs, but other species, such as camels, cattle, donkeys, sheep, and goats, have been proposed as potential reservoirs [3,18]. Worldwide prevalence in humans typically does not exceed 1% [8]. The parasite has a direct life cycle, with a vegetative form, trophozoites, residing in the host intestine, and a resting cyst that is released into the environment in feces [18].
Cryptosporidium is a protozoan parasite of medical and veterinary importance that causes gastroenteritis in a variety of vertebrate species, including humans, as well as wild and domestic animals [2,9]. A recent study showed that Cryptosporidium infection was the fifth leading cause of diarrhea in children younger than five years, and acute infection caused more than 48,000 deaths [19]. Frequently reported risk factors include overcrowding, household diarrhea, poor-quality drinking water, animal contact, and open defecation/lack of toilet [20]. At least 44 Cryptosporidium species are considered taxonomically valid [21]. Of these, 19 are found in humans. Cryptosporidium hominis and C. parvum are responsible for approximately 95% of infections in humans, followed by C. meleagridis, C. felis, C. canis, and C. ubiquitum. Other species, such as C. cuniculus, C. viatorum, C. muris, C. andersoni, C. erinacei, C. tyzzeri, C. bovis, C. suis, C. scrofarum, C. occultus, C. xiaoi, C. fayeri, C. ditrichi, and the Cryptosporidium horse genotype, have also been found in humans [22,23,24].
Giardia duodenalis (syn Giardia lamblia and Giardia intestinalis) is a major cause of parasite-induced diarrheal disease infecting humans and animals [25]. Its life cycle consists of two stages: a fecally–orally transmitted cyst and a disease-causing trophozoite [26]. G. duodenalis is categorized into eight distinct assemblages (genotypes A–H), distinguished by host specificity and genetic variations [8,25]. Assemblages A and B specifically infect humans, and certain sub-genotypes exhibit zoonotic potential [25]. Following ingestion by a new host, acidic conditions in the host’s stomach facilitate excystation. Each cyst produces two trophozoites, which migrate to the duodenum and proximal jejunum. At this location, they adhere to the mucosal wall through a ventral adhesive disk and replicate via binary fission [5].
Hedgehogs are frequent dwellers of suburban environments. This proximity to humans raises the risk of exposure to infected hedgehog feces, particularly for individuals engaged in hedgehog rescue and rehabilitation efforts, providing the opportunity for zoonotic infections by enteric pathogens [27]. Hedgehogs can be hosts of enteric zoonotic protozoa such as Blastocystis sp., Cryptosporidium spp., and G. duodenalis [27,28,29,30]. Despite their potential role as hosts for enteric parasites, little data about their occurrence and molecular diversity are available in the scientific literature, with no studies having been conducted in Portugal. As such, this study aims to determine the occurrence and genetic diversity of Blastocystis sp., B. coli, Cryptosporidium spp., and G. duodenalis in hedgehogs from different areas of mainland Portugal.
2. Materials and Methods
2.1. Sample Collection
A total of 110 individual stool samples from hedgehogs were collected from Centro de Recuperação de Fauna do Parque Biológico de Gaia (CRFPBG; n = 30) and from Centro de Recuperação e Interpretação do Ouriço (CRIDO; n = 80), centers dedicated to rescuing hedgehogs from various regions, for further release into their natural environment.
All hedgehog stool samples from the CRFPBG were from European hedgehogs (Erinaceus europaeus) and were obtained between September 2021 and April 2023. Stool samples from CRIDO included European hedgehogs, except for four individuals, namely three four-toed hedgehogs (Atelerix albiventris) and one long-eared hedgehog (Hemiechinus auritus). CRIDO samples were collected between December 2022 and May 2023. Most samples (n = 94) were derived from hedgehogs found in the Porto district, with smaller numbers from Braga (n = 6), Lisboa (n = 4), Setúbal (n = 2), Aveiro (n = 2), Coimbra (n = 1), and Viana do Castelo (n = 1) (Figure 1).
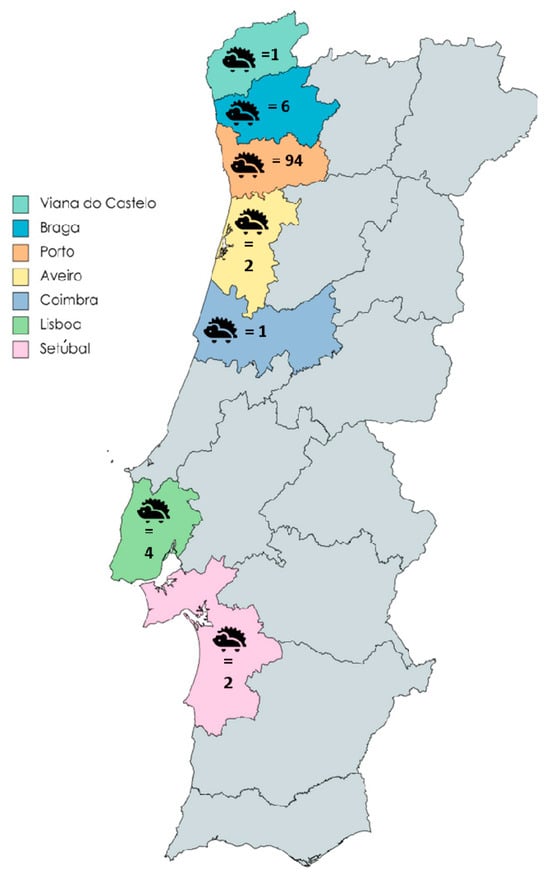
Figure 1.
Geographic distribution of Portuguese hedgehogs tested for Blastocystis sp., Balantioides coli, Cryptosporidium spp., and Giardia duodenalis. Each district is distinguished by a unique color, followed by the corresponding number of hedgehogs obtained from from that district.
Stool samples were obtained from the cages where hedgehogs were kept, and all presented a well-structured texture indicative of absence of gastrointestinal disease. Importantly, no hedgehogs were sacrificed for the purpose of this study. The stool samples were quickly stored at 4 °C, maintained at this temperature during transportation to the laboratory, and then preserved at −20 °C until the DNA extraction process.
2.2. DNA Extraction
Fecal samples for DNA extraction were prepared by suspending fecal matter (10%) in phosphate-buffered saline (PBS) at pH 7.2. The suspensions were then centrifuged at 8000× g for 5 min. The supernatants were then used for DNA extraction and purification. DNA extraction and purification were performed using the QIAamp Cador Pathogen Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The QIAcube® automated platform (Qiagen) was used to process the extracted DNA. The eluted DNA was stored at −80 °C in RNase-free water.
2.3. Molecular Detection of Blastocystis sp., Balantioides coli, Cryptosporidium spp., and Giardia duodenalis
Specific molecular techniques were employed for the individual detection of Blastocystis sp., Balantioides coli, Cryptosporidium spp., and Giardia duodenalis. For Blastocystis sp., a direct PCR approach was employed, targeting a 600 bp segment of the SSU rRNA gene. This method followed the protocol described by Scicluna et al. (2006) [31] and utilized the primer set RD5/BhRDr with an annealing temperature of 59 °C.
For Balantioides coli, a direct PCR was performed targeting the complete ITS1–5.8s-rRNA–ITS2 region and the last 117 bp (3′ end) of the SSU rRNA gene, amplifying a 400 bp product. This procedure follows the method outlined by Ponce-Gordo et al. (2011) [32] and employs the primer set B5D/RD5 with an annealing temperature of 60 °C.
For identification of Cryptosporidium spp., a nested PCR was performed, targeting the 587 bp fragment of the SSU rRNA gene. This method utilized primer sets CR-P1/CRP2 and CR-P3/CPB-DIAGR for both rounds of amplification, maintaining a consistent annealing temperature of 50 °C, as described in Tiangtip and Jong-Wutiwes (2002) [33].
For Giardia duodenalis, a nested PCR approach was implemented. The initial RH11-derivates/Gia2150c primer pair was used to amplify a 497 bp product with an annealing temperature of 55 °C. Subsequently, a secondary RH11-derivates/RH4-derivates primer pair was employed to amplify a 293 bp fragment with an annealing temperature of 59 °C, replicating the strategy described in Helmy et al. (2018) [34].
2.4. PCR
The molecular detection of targeted parasites was performed through a series of PCR reactions, including endpoint, nested, and semi-nested protocols. Each PCR was carried out on the T100 thermocycler provided by Bio-Rad (Neuried, Germany). The reaction mixtures included the Fast PCR Mastermix (GRiSP®, Dortmund, Germany) and the 2× Xpert Fast Hotstart Mastermix (GRiSP®), ensuring optimal conditions for DNA amplification.
Following PCR amplification, DNA fragments, specific for each targeted parasite, were carefully separated and visualized through electrophoresis on 1.5% agarose gels. These gels were stained with Xpert Green Safe DNA gel dye (GRiSP®).
Electrophoresis was carried out at a constant voltage of 120 V for 30 min. For the confirmation of the results, the agarose gels were irradiated with UV light.
2.5. Statistical Analysis
Data processing and initial analysis were carried out using Microsoft® Excel® for Microsoft 365 MSO (version 2312 Build 16. 0. 17126. 20132) 64-bit. Descriptive analyses were conducted using IBM SPSS version 28.0.0.0 software for Windows (SPSS, Chicago, IL, USA). Throughout all analyses, a confidence interval (CI) of 95% was applied.
2.6. Sanger Sequencing and Phylogenetic Analysis
To isolate and purify amplicons, GRS PCR and Gel Band Purification Kit (GRiSP®) technology was employed. This method effectively separates positive amplicons that match the expected size from the surrounding non-target DNA fragments. Subsequently, the Sanger sequencing method was used, employing specific internal primers designed for the target gene. The bidirectional sequencing process generated two complementary sequences, which were then aligned and compared with existing entries in the NCBI (GenBank) nucleotide database. This comparative analysis was performed using the BioEdit Sequence Alignment Editor v7.1.9 software package (version 2.1), relying on the database accessed on 20 November 2023. For a more in-depth analysis and interpretation of the obtained sequences, the MEGA version X software was utilized [35]. Applying the Hasegawa–Kishino–Yano model, maximum likelihood (ML) bootstrap values with 1000 replicates were estimated for statistical robustness. The selection of this model, recognized as the most effective replacement, was determined through MEGA version X [35]. The sequences from this study have been deposited in the GenBank® database, with each assigned a unique accession number: OR835985 (Blastocystis sp.) and OR880424 (Cryptosporidium parvum).
3. Results
Occurrence of Enteric Protozoan Parasites in Hedgehogs
From the analysis of the 110 stool samples for enteric protozoa, 1.82% (2/110 95% CI: 0.22–6.41) were positive for at least one protozoan. Among these, one sample (0.91% 1/110, 95% CI: 0.02–4.96) tested positive for Blastocystis and another for Cryptosporidium (0.91% 1/110, 95% CI: 0.02–4.96), as we can see in Table 1. All protozoa-positive stools were found exclusively in European hedgehogs from Centro de Recuperação de Fauna do Parque Biológico de Gaia in the Porto district of the northern region of mainland Portugal.

Table 1.
An overview of the occurrence of each enteric protozoan parasite in European hedgehogs (Erinaceus europaeus) from Portugal.
Regarding the characterization of Blastocystis, the phylogenetic tree illustrated in Figure 2 showed clustering of the sequence of Blastocystis sp. obtained in our study (highlighted in bold) with the sequences OR660650 and MN339604 isolated from a bovine and a dog in China, respectively. BLAST tool analysis established a 100% identity between these sequences. The reference sequence OR660650, originating from a bovine host, belongs to subtype ST3. This association, along with the 100% match, suggests that the sequence present in this study might also belong to the ST3 subtype.

Figure 2.
Phylogenetic analysis of the small-subunit ribosomal RNA (SSU-rRNA) gene was conducted for Blastocystis sp. found in European hedgehogs. Inference was performed using the maximum likelihood method and the Tamura 3-parameter model. The analyses were performed with the assistance of MEGA X software, followed by tree editing using Interactive Tree of Life (iTOL). The isolates obtained in this study are highlighted in bold with their respective accession numbers, while the 21 additional Blastocystis species strains obtained from GenBank are shown without bold formatting, followed by their respective accession numbers, species, hosts, and country of origin.
As for the genetic characterization of the detected Cryptosporidium, the phylogenetic analysis in Figure 3 and BLAST search revealed that the sequence obtained in this study exhibited 100% identity with a previously identified C. parvum sequence from an alpaca isolate in Peru (JN812214), suggesting that the sequence obtained in the current study represents a C. parvum strain.

Figure 3.
Phylogenetic analysis of the small-subunit ribosomal RNA (SSU-rRNA) gene was conducted for Cryptosporidium spp. found in European hedgehogs. Inference was performed using the maximum likelihood method and the Tamura 3-parameter model. The analyses were performed with the assistance of MEGA X software, followed by tree editing using Interactive Tree of Life (iTOL). The isolates obtained in this study are highlighted in bold with their respective accession numbers, while the 19 additional Cryptosporidium species strains obtained from GenBank are shown without bold formatting, followed by their respective accession numbers, species, hosts, and country of origin.
4. Discussion
This study represents the first investigation performed in Portugal focusing on the molecular detection and phylogenetic analysis of intestinal protozoa in hedgehogs. Consequently, it represents the first report of Blastocystis and Cryptosporidium in hedgehogs from Portugal. Blastocystis sp. and Cryptosporidium spp. are among the most common agents responsible for enteric diseases globally, affecting both animals and humans [8]. Considering this information, the surveillance and identification of hedgehogs as possible hosts of these pathogenic protozoa is important for understanding the role of these agents.
In the present study, Blastocystis sp. subtype ST3 was identified in one hedgehog, representing an occurrence of 0.91% (1/110, 95% CI: 0.02–4.96). Noteworthily, Blastocystis sp. subtype ST3 has been previously found in humans [14]. There is a lack of information about the presence of Blastocystis in hedgehogs. To date and to the best of our knowledge, the only study available on the topic was conducted by Kaczmarek and Salamatin in 2018 [28] (Table 2), revealing an occurrence of 10.2%. It is crucial to note that the sample size in the previously mentioned study was approximately half of the size used in the current investigation. The identification of the subtype ST3 emphasizes the need for further research to understand the role of hedgehogs as possible reservoirs and their role in the transmission of Blastocystis sp. to both humans and animals.

Table 2.
Study on Blastocystis sp. in European hedgehogs (Erinaceus europaeus).
Cryptosporidium was found in one isolate with the occurrence of 0.91% (1/110, 95% CI: 0.02–4.96). Cryptosporidium species have been reported in European hedgehogs such as C. erinacei, C. parvum, and C. hominis [27,36]. Compared to other studies performed with European hedgehogs (Table 3), the occurrence obtained in this study is lower than in those in the UK (8%) [37], Germany (29.8%) [30], Denmark (5.2%) [29], Czech Republic (73%) [36], and the Netherlands (9%) [27]. The disparities observed between our findings and previous research can be attributed to variations in sample size and the employment of diverse detection methods, including conventional microscopy and coproantigen detection techniques. Several studies have underscored the efficacy of molecular methods as highly sensitive and specific analytical tools, enabling simultaneous diagnosis and characterization of infections and offering more reliable data compared to serological and parasitological approaches [38]. Even when comparing studies employing PCR, the implementation of distinct oligonucleotide primers targeting different regions may contribute to disparities in sensitivities. The isolate was classified as C. parvum, one of the most common species of Cryptosporidium found in humans, with a global prevalence of 7.6% [39]. Given the synanthropic presence of hedgehogs in urban areas and their proximity to humans, concerns about their possible role as hosts for C. parvum should be considered. This underscores the need for further research, surveillance, and initiatives to mitigate the potential transmission of C. parvum from hedgehogs to humans and other animals.

Table 3.
Studies on Cryptosporidium spp. in European hedgehogs (Erinaceus europaeus).
Data on the occurrence of G. duodenalis and B. coli in European hedgehogs are limited. While the presence of B. coli has not yet been confirmed in hedgehogs, its recent detection in wild animals from Portugal, including wild boars [40] and wild cervids [41], suggests the potential for this parasite to exist in the country’s wildlife population. While G. duodenalis was not detected in the present study, its presence has been documented in other investigations. For instance, prevalence studies of G. duodenalis have been conducted in Germany, Russia, Denmark, New Zealand, and the Netherlands. The prevalence of G. duodenalis was found to be 0% in Germany, Russia, and Denmark, while the prevalence in New Zealand and the Netherlands was 33% and 11%, respectively (Table 4).

Table 4.
Studies on Giardia duodenalis in European hedgehogs (Erinaceus europaeus).
The detection of zoonotic enteric protozoan parasites, including Blastocystis and Cryptosporidium, could raise concerns for human and animal health. This emphasizes the need for monitoring and research to actively contain and prevent potential outbreaks in the hedgehog population.
5. Conclusions
In conclusion, this study represents the first investigation into the molecular detection and phylogenetic analysis of enteric protozoan parasites in European hedgehogs (Erinaceus europaeus) across mainland Portugal. This research showed a relatively low overall prevalence of these parasites within the hedgehog population. This study marks the initial identification of Blastocystis and Cryptosporidium in Portuguese hedgehogs, providing valuable data to the limited regional knowledge of these parasites. The detection of zoonotic parasites, particularly Cryptosporidium and Blastocystis, raises concerns about a possible transmission to humans. The surveillance, coupled with further research involving larger sample sizes and broader geographical representation, could be effective for the development of prevention and control strategies to mitigate the risk of outbreaks in hedgehog populations.
Author Contributions
Conceptualization, S.G.-G. and J.R.M.; methodology, S.G.-G., S.S.-S., A.V.S.C., P.B., and J.R.M.; resources C.R., V.S., and J.R.M.; writing—original draft preparation, S.G.-G.; writing—review and editing, S.G.-G., S.S.-S., A.V.S.C., P.B., C.R., V.S., and J.R.M.; funding acquisition, J.R.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Fundação para Ciência e Tecnologia (FCT), grant numbers 2021.09461.BD and 2022.15408.BD, also financed by national funds through the FCT—Fundação para a Ciência e a Tecnologia, I.P.—under the projects UIDB/04750/2020 and LA/P/0064/2020.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Acknowledgments
Sérgio Santos-Silva and Andreia V. S. Cruz thank the Fundação para a Ciência e a Tecnologia (FCT) for the financial support of their PhD work under the 2021 and 2022 Maria de Sousa program.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Santin, M. Cryptosporidium and Giardia in Ruminants. Vet. Clin. N. Am. Food Anim. Pract. 2020, 36, 223–238. [Google Scholar] [CrossRef] [PubMed]
- Mamedova, S.; Karanis, P. Cryptosporidium spp. Infections in Livestock and Wild Animals in Azerbaijan Territory. J. Water Health 2021, 19, 545–562. [Google Scholar] [CrossRef]
- Ponce-Gordo, F.; García-Rodríguez, J.J. Balantioides coli. Res. Vet. Sci. 2021, 135, 424–431. [Google Scholar] [CrossRef] [PubMed]
- Zanetti, A.D.S.; Malheiros, A.F.; de Matos, T.A.; Longhi, F.G.; Moreira, L.M.; Silva, S.L.; Castrillon, S.K.I.; Ferreira, S.M.B.; Ignotti, E.; Espinosa, O.A. Prevalence of Blastocystis sp. Infection in Several Hosts in Brazil: A Systematic Review and Meta-Analysis. Parasit. Vectors 2020, 13, 30. [Google Scholar] [CrossRef] [PubMed]
- Kucik, C.J.; Martin, G.L.; Sortor, B. V Common Intestinal Parasites. Am. Fam. Physician 2004, 69, 1161–1168. [Google Scholar] [PubMed]
- Hassan, E.M.; Örmeci, B.; DeRosa, M.C.; Dixon, B.R.; Sattar, S.A.; Iqbal, A. A Review of Cryptosporidium spp. and Their Detection in Water. Water Sci. Technol. J. Int. Assoc. Water Pollut. Res. 2021, 83, 1–25. [Google Scholar] [CrossRef]
- Calero-Bernal, R.; Santín, M.; Maloney, J.G.; Martín-Pérez, M.; Habela, M.A.; Fernández-García, J.L.; Figueiredo, A.; Nájera, F.; Palacios, M.J.; Mateo, M.; et al. Blastocystis sp. Subtype Diversity in Wild Carnivore Species from Spain. J. Eukaryot. Microbiol. 2020, 67, 273–278. [Google Scholar] [CrossRef]
- Figueiredo, A.M.; Köster, P.C.; Dashti, A.; Torres, R.T.; Fonseca, C.; Mysterud, A.; Bailo, B.; Carvalho, J.; Ferreira, E.; Hipólito, D.; et al. Molecular Detection and Distribution of Giardia duodenalis and Cryptosporidium spp. Infections in Wild and Domestic Animals in Portugal. Transbound. Emerg. Dis. 2023, 2023, 5849842. [Google Scholar] [CrossRef]
- Bouzid, M.; Hunter, P.R.; Chalmers, R.M.; Tyler, K.M. Cryptosporidium Pathogenicity and Virulence. Clin. Microbiol. Rev. 2013, 26, 115–134. [Google Scholar] [CrossRef]
- Byun, J.-W.; Park, J.-H.; Moon, B.-Y.; Lee, K.; Lee, W.-K.; Kwak, D.; Lee, S.-H. Identification of Zoonotic Balantioides coli in Pigs by Polymerase Chain Reaction-Restriction Fragment Length Polymorphism (PCR-RFLP) and Its Distribution in Korea. Animal 2021, 11, 2659. [Google Scholar] [CrossRef]
- Malatyalı, E.; Ertabaklar, H.; Ertuğ, S. Subtype Distribution of Blastocystis in Türkiye. Turk. Parazitoloji. Derg. 2023, 47, 184–189. [Google Scholar] [CrossRef] [PubMed]
- Crawford, F.G.; Vermund, S.H. Human Cryptosporidiosis. Crit. Rev. Microbiol. 1988, 16, 113–159. [Google Scholar] [CrossRef] [PubMed]
- Einarsson, E.; Ma’ayeh, S.; Svärd, S.G. An Up-Date on Giardia and Giardiasis. Curr. Opin. Microbiol. 2016, 34, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Hernandez, F.; Martinez-Ibarra, J.A.; Lopez-Escamilla, E.; Villanueva-Garcia, C.; Muñoz-Garcia, C.I.; Rendon-Franco, E.; Maravilla, P.; Villalobos, G. Molecular Genotyping of Blastocystis spp. in Wild Mammals from Mexico. Parasitol. Res. 2020, 119, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Gentekaki, E.; Curtis, B.A.; Stairs, C.W.; Klimeš, V.; Eliáš, M.; Salas-Leiva, D.E.; Herman, E.K.; Eme, L.; Arias, M.C.; Henrissat, B.; et al. Extreme Genome Diversity in the Hyper-Prevalent Parasitic Eukaryote Blastocystis. PLoS Biol. 2017, 15, e2003769. [Google Scholar] [CrossRef] [PubMed]
- Alfellani, M.A.; Stensvold, C.R.; Vidal-Lapiedra, A.; Onuoha, E.S.U.; Fagbenro-Beyioku, A.F.; Clark, C.G. Variable Geographic Distribution of Blastocystis Subtypes and Its Potential Implications. Acta Trop. 2013, 126, 11–18. [Google Scholar] [CrossRef]
- Santin, M.; Figueiredo, A.; Molokin, A.; George, N.S.; Köster, P.C.; Dashti, A.; González-Barrio, D.; Carmena, D.; Maloney, J.G. Division of Blastocystis ST10 into Three New Subtypes: ST42–ST44. J. Eukaryot. Microbiol. 2023, 71, e12998. [Google Scholar] [CrossRef]
- García-Rodríguez, J.J.; Köster, P.C.; Ponce-Gordo, F. Cyst Detection and Viability Assessment of Balantioides coli in Environmental Samples: Current Status and Future Needs. Food Waterborne Parasitol. 2022, 26, e00143. [Google Scholar] [CrossRef]
- Khalil, I.A.; Troeger, C.; Rao, P.C.; Blacker, B.F.; Brown, A.; Brewer, T.G.; Colombara, D.V.; De Hostos, E.L.; Engmann, C.; Guerrant, R.L.; et al. Morbidity, Mortality, and Long-Term Consequences Associated with Diarrhoea from Cryptosporidium Infection in Children Younger than 5 Years: A Meta-Analyses Study. Lancet Glob. Health 2018, 6, e758–e768. [Google Scholar] [CrossRef]
- Bouzid, M.; Kintz, E.; Hunter, P.R. Risk Factors for Cryptosporidium Infection in Low and Middle Income Countries: A Systematic Review and Meta-Analysis. PLoS Negl. Trop. Dis. 2018, 12, e0006553. [Google Scholar] [CrossRef]
- Elmahallawy, E.K.; Köster, P.C.; Dashti, A.; Alghamdi, S.Q.; Saleh, A.; Gareh, A.; Alrashdi, B.M.; Hernández-Castro, C.; Bailo, B.; Lokman, M.S.; et al. Molecular Detection and Characterization of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi Infections in Dromedary Camels (Camelus dromedaries) in Egypt. Front. Vet. Sci. 2023, 10, 1139388. [Google Scholar] [CrossRef] [PubMed]
- Köster, P.C.; González-Barrio, D.; Carmena, D. Editorial for the Special Issue: Diagnosis, Epidemiology and Transmission Dynamics of Cryptosporidium spp. and Giardia duodenalis. Pathogens 2022, 11, 141. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Chen, M.; He, Y.; Chen, H.; Huang, M.; Li, N.; Ryan, U.; Kváč, M.; Feng, Y.; Xiao, L.; et al. Cryptosporidium Equin. sp. (Apicomplexa: Cryptosporidiidae): Biological and Genetic Characterisations. Int. J. Parasitol. 2023, 53, 545–554. [Google Scholar] [CrossRef] [PubMed]
- Ryan, U.; Zahedi, A.; Feng, Y.; Xiao, L. An Update on Zoonotic Cryptosporidium Species and Genotypes in Humans. Animal 2021, 11, 3307. [Google Scholar] [CrossRef] [PubMed]
- Rojas-López, L.; Marques, R.C.; Svärd, S.G. Giardia duodenalis. Trends Parasitol. 2022, 38, 605–606. [Google Scholar] [CrossRef]
- Adam, R.D. Giardia duodenalis: Biology and Pathogenesis. Clin. Microbiol. Rev. 2021, 34, e00024-19. [Google Scholar] [CrossRef]
- Krawczyk, A.I.; van Leeuwen, A.D.; Jacobs-Reitsma, W.; Wijnands, L.M.; Bouw, E.; Jahfari, S.; van Hoek, A.H.A.M.; van der Giessen, J.W.B.; Roelfsema, J.H.; Kroes, M.; et al. Presence of Zoonotic Agents in Engorged Ticks and Hedgehog Faeces from Erinaceus europaeus in (Sub) Urban Areas. Parasit. Vectors 2015, 8, 210. [Google Scholar] [CrossRef]
- Kaczmarek, A.; Sałamatin, R. First record of Blastocystis cf. hominis (Eukaryota: Stramenopiles) in European hedgehog (Erinaceus europaeus) from Poland. In Proceedings of the 14th International Congress of Parasitology—ICOPA 2018 at: EXCO, Daegu, Republic of Korea, 19–24 August 2018; ICOPA: Daegu, Republic of Korea, 2018; p. 1. P.1-105. [Google Scholar]
- Rasmussen, S.L.; Hallig, J.; van Wijk, R.E.; Petersen, H.H. An Investigation of Endoparasites and the Determinants of Parasite Infection in European Hedgehogs (Erinaceus europaeus) from Denmark. Int. J. Parasitol. Parasites Wildl. 2021, 16, 217–227. [Google Scholar] [CrossRef]
- Dyachenko, V.; Kuhnert, Y.; Schmaeschke, R.; Etzold, M.; Pantchev, N.; Daugschies, A. Occurrence and Molecular Characterization of Cryptosporidium spp. Genotypes in European Hedgehogs (Erinaceus europaeus L.) in Germany. Parasitology 2010, 137, 205–216. [Google Scholar] [CrossRef]
- Scicluna, S.M.; Tawari, B.; Clark, C.G. DNA Barcoding of Blastocystis. Protist 2006, 157, 77–85. [Google Scholar] [CrossRef]
- Ponce-Gordo, F.; Fonseca-Salamanca, F.; Martínez-Díaz, R.A. Genetic Heterogeneity in Internal Transcribed Spacer Genes of Balantidium coli (Litostomatea, Ciliophora). Protist 2011, 162, 774–794. [Google Scholar] [CrossRef] [PubMed]
- Tiangtip, R.; Jongwutiwes, S. Molecular Analysis of Cryptosporidium Species Isolated from HIV-Infected Patients in Thailand. Trop. Med. Int. Health 2002, 7, 357–364. [Google Scholar] [CrossRef] [PubMed]
- Helmy, Y.A.; Spierling, N.G.; Schmidt, S.; Rosenfeld, U.M.; Reil, D.; Imholt, C.; Jacob, J.; Ulrich, R.G.; Aebischer, T.; Klotz, C. Occurrence and Distribution of Giardia Species in Wild Rodents in Germany. Parasit. Vectors 2018, 11, 213. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar] [CrossRef]
- Hofmannová, L.; Hauptman, K.; Huclová, K.; Květoňová, D.; Sak, B.; Kváč, M. Cryptosporidium erinacei and C. parvum in a Group of Overwintering Hedgehogs. Eur. J. Protistol. 2016, 56, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Sangster, L.; Blake, D.P.; Robinson, G.; Hopkins, T.C.; Sa, R.C.C.; Cunningham, A.A.; Chalmers, R.M.; Lawson, B. Detection and Molecular Characterisation of Cryptosporidium parvum in Britis European Hedgehogs (Erinaceus europaeus). Vet. Parasitol. 2016, 217, 39–44. [Google Scholar] [CrossRef]
- Gomes-de-Sá, S.; Santos-Silva, S.; Moreira, A.d.S.; Barradas, P.F.; Amorim, I.; Cardoso, L.; Mesquita, J.R. Assessment of the Circulation of Dirofilaria immitis in Dogs from Northern Portugal through Combined Analysis of Antigens, DNA and Parasite Forms in Blood. Acta Trop. 2023, 239, 106799. [Google Scholar] [CrossRef]
- Menon, V.K.; Okhuysen, P.C.; Chappell, C.L.; Mahmoud, M.; Mahmoud, M.; Meng, Q.; Doddapaneni, H.; Vee, V.; Han, Y.; Salvi, S.; et al. Fully Resolved Assembly of Cryptosporidium parvum. Gigascience 2022, 11, giac010. [Google Scholar] [CrossRef]
- Santos-Silva, S.; Moraes, D.F.d.S.D.; López-López, P.; Palmeira, J.D.; Torres, R.T.; São José Nascimento, M.; Dashti, A.; Carmena, D.; Rivero-Juarez, A.; Mesquita, J.R. Survey of Zoonotic Diarrheagenic Protist and Hepatitis E Virus in Wild Boar (Sus scrofa) of Portugal. Animal 2023, 13, 256. [Google Scholar] [CrossRef]
- Mega, J.; Santos-Silva, S.; Loureiro, A.; Palmeira, J.D.; Torres, R.T.; Rivero-Juarez, A.; Carmena, D.; Mesquita, J. Balantioides Coli Fecal Excretion in Hunted Wild Cervids (Cervus elaphus and Dama dama) from Portugal. Pathogens 2022, 11, 1242. [Google Scholar] [CrossRef]
- Epe, C.; Ising-Volmer, S.; Stoye, M. Parasitological fecal studies of equids, dogs, cats and hedgehogs during the years 1984–1991. Dtsch. Tierarztl. Wochenschr. 1993, 100, 426–428. [Google Scholar] [PubMed]
- Chilvers, B.; Cowan, P.; Waddington, D.; Kelly, P.; Brown, T. The Prevalence of Infection of Giardia spp. and Cryptosporidium spp. in Wild Animals on Farmland, Southeastern North Island, New Zealand. Int. J. Environ. Health Res. 1998, 8, 59–64. [Google Scholar] [CrossRef]
- Epe, C.; Coati, N.; Schnieder, T. Results of parasitological examinations of faecal samples from horses, ruminants, pigs, dogs, cats, hedgehogs and rabbits between 1998 and 2002. Dtsch. Tierarztl. Wochenschr. 2004, 111, 243–247. [Google Scholar] [PubMed]
- Pantchev, N.; Globokar-Vrhovec, M.; Beck, W. Endoparasitosen Bei Kleinsäugern Aus Privater Haltung Und Igeln. Tierärztliche Prax. Ausg. K Kleintiere/Heimtiere 2005, 33, 296–306. [Google Scholar] [CrossRef]
- Raue, K.; Heuer, L.; Böhm, C.; Wolken, S.; Epe, C.; Strube, C. 10-Year Parasitological Examination Results (2003 to 2012) of Faecal Samples from Horses, Ruminants, Pigs, Dogs, Cats, Rabbits and Hedgehogs. Parasitol. Res. 2017, 116, 3315–3330. [Google Scholar] [CrossRef]
- Kurnosova, O.P.; Arisov, M.V.; Odoyevskaya, I.M. Intestinal Parasites of Pets and Other House-Kept Animals in Moscow. Helminthologia 2019, 56, 108–117. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).