Bile Acid Metabolic Profiles and Their Correlation with Intestinal Epithelial Cell Proliferation and Barrier Integrity in Suckling Piglets
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Approval
2.2. Animals and Study Design
2.3. Sample Preparation
2.4. Detection of Serum Biochemical Parameters and Hepatic Lipids
2.5. Real-Time Quantitative PCR (RT-qPCR)
2.6. Determination of BA Composition in Serum
2.7. Statistical Analysis
3. Results
3.1. Serum Biochemical Parameters of Newborn and Suckling Piglets
3.2. Lipid and Total BA Contents in Sera and Livers of Newborn and Suckling Piglets
3.3. Contents and Composition of BAs in the Sera of Newborn and Suckling Piglets
3.4. Differences in BA Metabolism between Newborn Piglets and Suckling Piglets
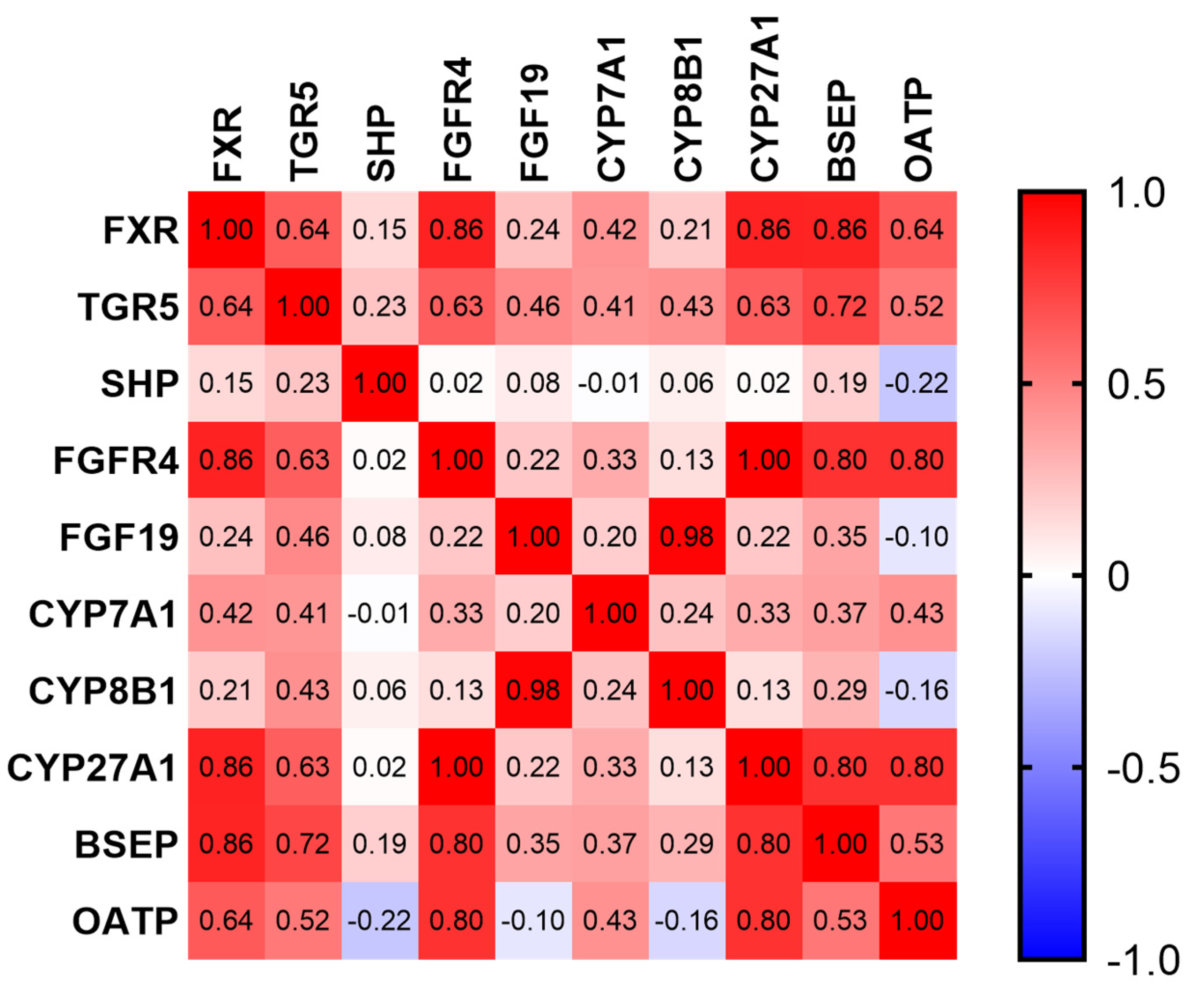
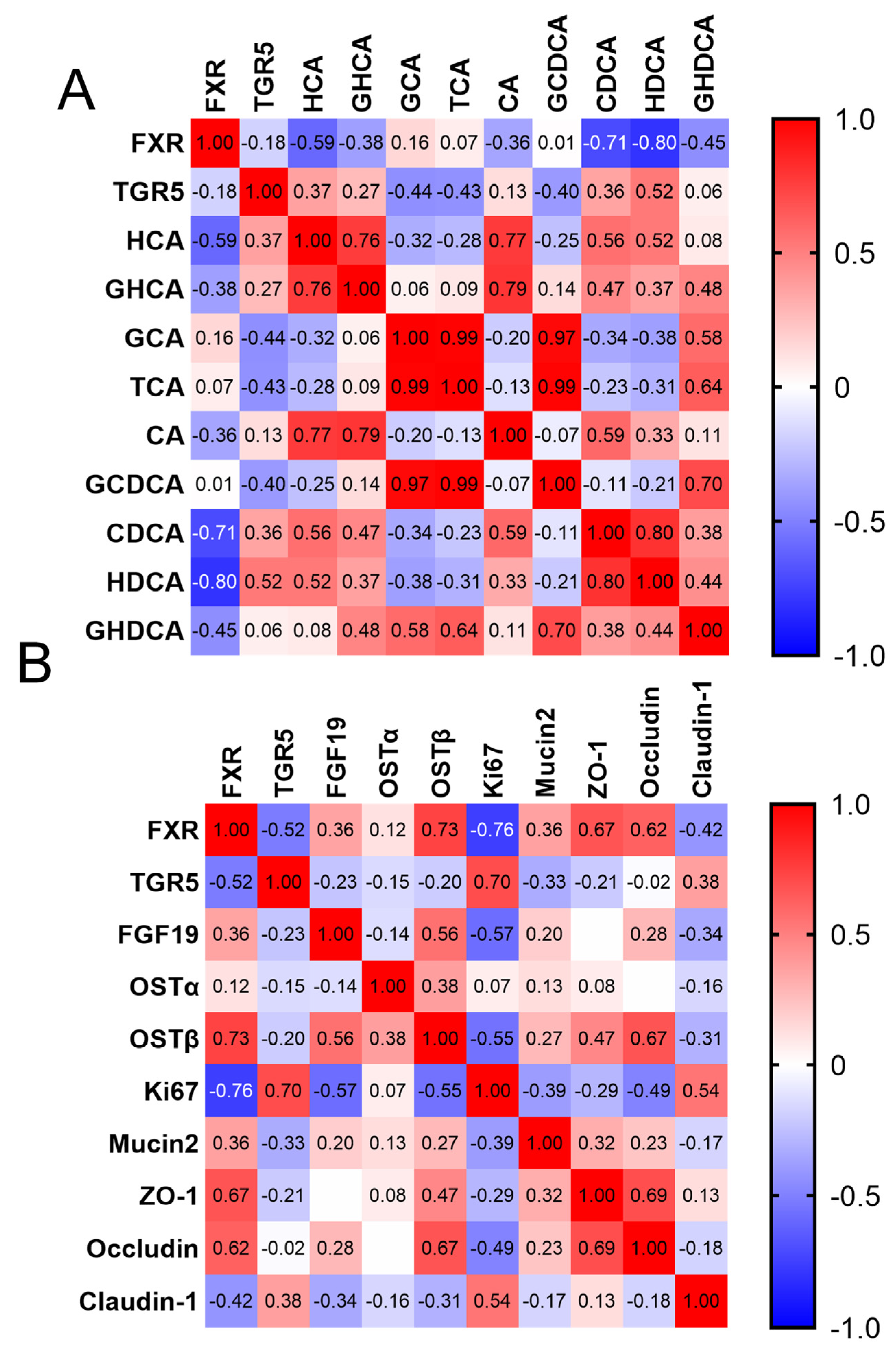
3.5. Correlation between BAs and Intestinal Barrier Development
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Heuss, E.M.; Proll-Cornelissen, M.J.; Neuhoff, C.; Tholen, E.; Grosse-Brinkhaus, C. Invited review: Piglet survival: Benefits of the immunocompetence. Animal 2019, 13, 2114–2124. [Google Scholar] [CrossRef] [PubMed]
- Pluske, J.R.; Turpin, D.L.; Kim, J.C. Gastrointestinal tract (gut) health in the young pig. Anim. Nutr. 2018, 4, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Xiao, Y.; Li, J.; Qi, M.; Tan, B. Serum biochemical parameters and amino acids metabolism are altered in piglets by early-weaning and proline and putrescine supplementations. Anim. Nutr. 2021, 7, 334–345. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zeng, L.; Tan, B.; Li, G.; Huang, B.; Xiong, X.; Li, F.; Kong, X.; Liu, G.; Yin, Y. Developmental changes in intercellular junctions and Kv channels in the intestine of piglets during the suckling and post-weaning periods. J. Anim. Sci. Biotechnol. 2016, 7, 4. [Google Scholar] [CrossRef] [PubMed]
- Agus, A.; Clement, K.; Sokol, H. Gut microbiota-derived metabolites as central regulators in metabolic disorders. Gut 2021, 70, 1174–1182. [Google Scholar] [CrossRef] [PubMed]
- Chiang, J.; Ferrell, J.M. Bile acid receptors FXR and TGR5 signaling in fatty liver diseases and therapy. Am. J. Physiol. Gastrointest. Liver Physiol. 2020, 318, G554–G573. [Google Scholar] [CrossRef] [PubMed]
- Van Best, N.; Rolle-Kampczyk, U.; Schaap, F.G.; Basic, M.; Olde, D.S.; Bleich, A.; Savelkoul, P.; von Bergen, M.; Penders, J.; Hornef, M.W. Bile acids drive the newborn’s gut microbiota maturation. Nat. Commun. 2020, 11, 3692. [Google Scholar] [CrossRef]
- Ward, J.B.J.; Lajczak, N.K.; Kelly, O.B.; O’Dwyer, A.M.; Giddam, A.K.; Gabhann, J.N.; Franco, P.; Tambuwala, M.M.; Jefferies, C.A.; Keely, S.; et al. Ursodeoxycholic acid and lithocholic acid exert anti-inflammatory actions in the colon. Am. J. Physiol.-Gastroint. Liver Physiol. 2017, 312, G550–G558. [Google Scholar] [CrossRef]
- Liu, Y.; Azad, M.; Ding, S.; Zhu, Q.; Blachier, F.; Yu, Z.; Gao, H.; Kong, X. Dietary bile acid supplementation in weaned piglets with intrauterine growth retardation improves colonic microbiota, metabolic activity, and epithelial function. J. Anim. Sci. Biotechnol. 2023, 14, 99. [Google Scholar] [CrossRef]
- Pi, Y.; Wu, Y.; Zhang, X.; Lu, D.; Han, D.; Zhao, J.; Zheng, X.; Zhang, S.; Ye, H.; Lian, S.; et al. Gut microbiota-derived ursodeoxycholic acid alleviates low birth weight-induced colonic inflammation by enhancing M2 macrophage polarization. Microbiome 2023, 11, 19. [Google Scholar] [CrossRef]
- Song, M.; Ye, J.; Zhang, F.; Su, H.; Yang, X.; He, H.; Liu, F.; Zhu, X.; Wang, L.; Gao, P.; et al. Chenodeoxycholic Acid (CDCA) Protects against the Lipopolysaccharide-Induced Impairment of the Intestinal Epithelial Barrier Function via the FXR-MLCK Pathway. J. Agric. Food. Chem. 2019, 67, 8868–8874. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Yang, X.; Yuan, P.; Yang, J.; Wang, P.; Zhong, H.; Zhang, X.; Che, L.; Feng, B.; Li, J.; et al. Undernutrition Shapes the Gut Microbiota and Bile Acid Profile in Association with Altered Gut-Liver FXR Signaling in Weaning Pigs. J. Agric. Food Chem. 2019, 67, 3691–3701. [Google Scholar] [CrossRef]
- Song, M.; Yang, Q.; Zhang, F.; Chen, L.; Su, H.; Yang, X.; He, H.; Liu, F.; Zheng, J.; Ling, M.; et al. Hyodeoxycholic acid (HDCA) suppresses intestinal epithelial cell proliferation through FXR-PI3K/AKT pathway, accompanied by alteration of bile acids metabolism profiles induced by gut bacteria. FASEB J. 2020, 34, 7103–7117. [Google Scholar] [CrossRef]
- Zong, E.; Yan, S.; Wang, M.; Yin, L.; Wang, Q.; Yin, J.; Li, J.; Li, Y.; Ding, X.; Huang, P.; et al. The effects of dietary supplementation with hyodeoxycholic acid on the differentiation and function of enteroendocrine cells and the serum biochemical indices in weaned piglets. J. Anim. Sci. 2019, 97, 1796–1805. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Azad, M.; Zhang, W.; Xiong, L.; Blachier, F.; Yu, Z.; Kong, X. Intrauterine growth retardation affects liver bile acid metabolism in growing pigs: Effects associated with the changes of colonic bile acid derivatives. J. Anim. Sci. Biotechnol. 2022, 13, 117. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Niu, K.; Wang, R.; Liang, X.; Lin, C.; Wu, X.; Zhai, Z. Taurochenodeoxycholic acid inhibits intestinal epithelial cell proliferation and induces apoptosis independent of the farnesoid X receptor. Food Funct. 2023, 14, 5277–5289. [Google Scholar] [CrossRef]
- Sorrentino, G.; Perino, A.; Yildiz, E.; El, A.G.; Bou, S.M.; Gioiello, A.; Pellicciari, R.; Schoonjans, K. Bile Acids Signal via TGR5 to Activate Intestinal Stem Cells and Epithelial Regeneration. Gastroenterology 2020, 159, 956–968.e8. [Google Scholar] [CrossRef]
- Zhai, Z.; Niu, K.M.; Liu, Y.; Lin, C.; Wu, X. The Gut Microbiota-Bile Acids-TGR5 Axis Mediates Eucommia ulmoides Leaf Extract Alleviation of Injury to Colonic Epithelium Integrity. Front. Microbiol. 2021, 12, 727681. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Z.; Niu, K.M.; Liu, H.; Lin, C.; Tu, Y.; Liu, Y.; Cai, L.; Ouyang, K.; Liu, J. Policosanol alleviates hepatic lipid accumulation by regulating bile acids metabolism in C57BL6/mice through AMPK-FXR-TGR5 cross-talk. J. Food Sci. 2021, 86, 5466–5478. [Google Scholar] [CrossRef]
- Tan, C.; Zhai, Z.; Ni, X.; Wang, H.; Ji, Y.; Tang, T.; Ren, W.; Long, H.; Deng, B.; Deng, J.; et al. Metabolomic Profiles Reveal Potential Factors that Correlate with Lactation Performance in Sow Milk. Sci. Rep. 2018, 8, 10712. [Google Scholar] [CrossRef]
- Wahlstrom, A.; Sayin, S.I.; Marschall, H.U.; Backhed, F. Intestinal Crosstalk between Bile Acids and Microbiota and Its Impact on Host Metabolism. Cell Metab. 2016, 24, 41–50. [Google Scholar] [CrossRef]
- Jia, W.; Wei, M.; Rajani, C.; Zheng, X. Targeting the alternative bile acid synthetic pathway for metabolic diseases. Protein Cell 2021, 12, 411–425. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Zheng, X.; Ma, X.; Jiang, R.; Zhou, W.; Zhou, S.; Zhang, Y.; Lei, S.; Wang, S.; Kuang, J.; et al. Theabrownin from Pu-erh tea attenuates hypercholesterolemia via modulation of gut microbiota and bile acid metabolism. Nat. Commun. 2019, 10, 4971. [Google Scholar] [CrossRef] [PubMed]
- Karpen, S.J.; Kelly, D.; Mack, C.; Stein, P. Ileal bile acid transporter inhibition as an anticholestatic therapeutic target in biliary atresia and other cholestatic disorders. Hepatol. Int. 2020, 14, 677–689. [Google Scholar] [CrossRef]
- Guzior, D.V.; Quinn, R.A. Review: Microbial transformations of human bile acids. Microbiome 2021, 9, 140. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Tsai, T.; Deng, F.; Wei, X.; Chai, J.; Knapp, J.; Apple, J.; Maxwell, C.V.; Lee, J.A.; Li, Y.; et al. Longitudinal investigation of the swine gut microbiome from birth to market reveals stage and growth performance associated bacteria. Microbiome 2019, 7, 109. [Google Scholar] [CrossRef]
- Pi, Y.; Mu, C.; Gao, K.; Liu, Z.; Peng, Y.; Zhu, W. Increasing the Hindgut Carbohydrate/Protein Ratio by Cecal Infusion of Corn Starch or Casein Hydrolysate Drives Gut Microbiota-Related Bile Acid Metabolism to Stimulate Colonic Barrier Function. Msystems 2020, 5, e00176-20. [Google Scholar] [CrossRef] [PubMed]
- Ethanic, M.; Stanimirov, B.; Pavlovic, N.; Golocorbin-Kon, S.; Al-Salami, H.; Stankov, K.; Mikov, M. Pharmacological Applications of Bile Acids and Their Derivatives in the Treatment of Metabolic Syndrome. Front. Pharmacol. 2018, 9, 1382. [Google Scholar]
- Zheng, X.; Chen, T.; Jiang, R.; Zhao, A.; Wu, Q.; Kuang, J.; Sun, D.; Ren, Z.; Li, M.; Zhao, M.; et al. Hyocholic acid species improve glucose homeostasis through a distinct TGR5 and FXR signaling mechanism. Cell Metab. 2021, 33, 791–803.e7. [Google Scholar] [CrossRef]
- Lavelle, A.; Sokol, H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 223–237. [Google Scholar] [CrossRef]
- Zhai, Z.; Liu, J.; Niu, K.M.; Lin, C.; Tu, Y.; Liu, Y.; Cai, L.; Liu, H.; Ouyang, K. Integrated Metagenomics and Metabolomics to Reveal the Effects of Policosanol on Modulating the Gut Microbiota and Lipid Metabolism in Hyperlipidemic C57BL/6 Mice. Front Endocrinol. 2021, 12, 722055. [Google Scholar] [CrossRef]
- Fu, T.; Coulter, S.; Yoshihara, E.; Oh, T.G.; Fang, S.; Cayabyab, F.; Zhu, Q.; Zhang, T.; Leblanc, M.; Liu, S.; et al. FXR Regulates Intestinal Cancer Stem Cell Proliferation. Cell 2019, 176, 1098–1112.e18. [Google Scholar] [CrossRef]
- Girisa, S.; Rana, V.; Parama, D.; Dutta, U.; Kunnumakkara, A.B. Differential roles of farnesoid X receptor (FXR) in modulating apoptosis in cancer cells. Adv. Protein Chem. Struct. Biol. 2021, 126, 63–90. [Google Scholar] [PubMed]

| Item | Content (Gestation) | Content (Lactation) |
|---|---|---|
| Ingredients | ||
| Corn | 51.85 | 53.10 |
| Soybean meal | 10.80 | 23.50 |
| Wheat bran | 19.70 | 9.75 |
| Corn protein powder | 1.50 | 1.50 |
| Extruded Soybeans | 8.00 | 2.00 |
| Soybean oil | 3.00 | 5.00 |
| CaHPO4·2H2O | 2.00 | 2.00 |
| Limestone | 1.35 | 1.35 |
| Salt | 0.30 | 0.30 |
| Sodium sulfate | 0.30 | 0.30 |
| Lysine sulfate (70%) | 0.20 | 0.20 |
| Premix 1 | 1.00 | 1.00 |
| Total | 100.00 | 100.00 |
| Nutrient composition 2 | ||
| DE, MCal/kg | 3.24 | 3.41 |
| CP | 15.95 | 18.72 |
| EE | 6.98 | 7.06 |
| CF | 3.42 | 3.59 |
| Ca | 1.07 | 1.09 |
| Lys | 0.93 | 1.21 |
| Met + Cys | 0.54 | 0.64 |
| Thr | 0.58 | 0.75 |
| Trp | 0.19 | 0.24 |
| Items 1 | Groups 2 NPs (d1) | Groups SPs (d14) | SEM | p-Value |
|---|---|---|---|---|
| ABW (kg) | 1.47 | 3.48 | 0.09 | <0.001 |
| Litter weight (kg) | 17.82 | 30.43 | 1.51 | <0.001 |
| ADG (kg) | - | 0.14 | 0.02 | - |
| Backfat thickness of sows (mm) | 19.67 | 17.92 | 0.88 | 0.378 |
| Items 1 | Groups 2 NPs (d1) | Groups SPs (d14) | SEM | p-Value |
|---|---|---|---|---|
| TP (g/L) | 15.86 | 31.16 | 2.39 | <0.001 |
| ALB (g/L) | 6.27 | 23.33 | 2.18 | <0.001 |
| ALT (U/L) | 7.96 | 16.70 | 1.42 | 0.001 |
| AST (U/L) | 22.21 | 39.08 | 4.28 | 0.046 |
| ALP (U/L) | 1024.18 | 598.75 | 198.50 | 0.024 |
| BUN (mmol/L) | 2.84 | 2.23 | 0.22 | 0.176 |
| GLU (mmol/L) | 1.93 | 5.19 | 0.53 | 0.002 |
| Items 1 | Groups 2 NPs (d1) | Groups SPs (d14) | SEM | p-Value |
|---|---|---|---|---|
| Serum TG (mmol/L) | 0.29 | 1.18 | 0.12 | <0.001 |
| Serum TC (mmol/L) | 0.96 | 2.69 | 0.21 | <0.001 |
| Serum LDL-C (mmol/L) | 0.38 | 1.37 | 0.14 | <0.001 |
| Serum HDL-C (mmol/L) | 0.33 | 1.34 | 0.16 | <0.001 |
| Serum TBA (μmol/L) | 14.15 | 33.88 | 4.44 | 0.026 |
| Liver TG (mmol/g prot) | 1.21 | 0.78 | 0.11 | 0.044 |
| Liver TC (mmol/g prot) | 0.41 | 0.39 | 0.02 | 0.487 |
| Liver TBAs (μmol/g prot) | 5.92 | 6.28 | 0.20 | 0.288 |
| Items 1 | Groups 2 NPs (d1) | Groups SPs (d14) | SEM | p-Value |
|---|---|---|---|---|
| HCA | 6.67 | 11,137.29 | 1939.51 | 0.002 |
| GHCA | 1030.56 | 3512.35 | 609.82 | 0.041 |
| CA | 18.73 | 145.73 | 30.78 | 0.004 |
| TCA | 39.26 | 6.70 | 15.02 | 0.818 |
| GCA | 130.12 | 20.52 | 40.21 | 0.093 |
| CDCA | 64.16 | 2252.73 | 405.32 | 0.002 |
| TCDCA | 524.24 | 428.04 | 168.80 | 0.937 |
| GCDCA | 852.88 | 410.58 | 512.82 | 0.818 |
| HDCA | 63.36 | 3146.25 | 543.81 | 0.002 |
| THDCA | 142.14 | 874.12 | 182.89 | 0.009 |
| GHDCA | 488.31 | 769.68 | 144.40 | 0.180 |
| DCA | 2.24 | 57.03 | 15.48 | 0.002 |
| LCA | 10.97 | 56.72 | 11.85 | 0.041 |
| 7-ketoLCA | 12.92 | 47.82 | 10.98 | 0.041 |
| 3-DHCA | - | 46.02 | 8.35 | 0.002 |
| 12-DHCA | - | 63.65 | 26.42 | 0.015 |
| 7-KHCA | - | 9.30 | 2.11 | 0.065 |
| isoLCA | - | 8.152 | 2.37 | 0.065 |
| αMCA | - | 27.20 | 5.40 | 0.002 |
| UDCA | - | 11.34 | 3.04 | 0.015 |
| PBA | 2666.60 | 17,941.12 | 2705.34 | 0.004 |
| SBA | 719.93 | 5090.08 | 780.78 | 0.004 |
| FBA | 179.03 | 17,009.20 | 1379.50 | 0.002 |
| CBA | 3207.50 | 6021.96 | 1368.83 | 0.240 |
| Biological Function | Items 1 | Groups 2 NPs (d1) | Groups SPs (d14) | SEM | p-Value |
|---|---|---|---|---|---|
| Bile acid synthesis | Liver-CYP7A1 | 1.00 | 1.27 | 0.29 | 0.657 |
| Liver-CYP8B1 | 1.00 | 0.60 | 0.12 | 0.101 | |
| Liver-CYP27A1 | 1.00 | 1.98 | 0.27 | 0.003 | |
| Liver-BSEP | 1.00 | 1.26 | 0.19 | 0.297 | |
| Liver-OATP | 1.00 | 6.58 | 0.69 | 0.000 | |
| Ileum-OSTα | 1.00 | 1.14 | 0.14 | 0.623 | |
| Ileum-OSTβ | 1.00 | 0.45 | 0.11 | 0.008 | |
| Bile acid hepatic–intestinal circulation and bile acid receptor | Liver-FXR | 1.00 | 1.32 | 0.08 | 0.051 |
| Liver-TGR5 | 1.00 | 1.49 | 0.12 | 0.044 | |
| Liver-SHP | 1.00 | 0.78 | 0.08 | 0.355 | |
| Liver-FGFR4 | 1.00 | 1.98 | 0.17 | 0.003 | |
| Liver-FGF19 | 1.00 | 0.66 | 0.12 | 0.169 | |
| Ileum-FXR | 1.00 | 0.35 | 0.08 | <0.001 | |
| Ileum-TGR5 | 1.00 | 2.24 | 0.23 | 0.005 | |
| Ileum-FGF19 | 1.00 | 0.17 | 0.15 | 0.004 | |
| Ileum-Ki-67 | 1.00 | 5.39 | 0.53 | <0.001 | |
| Ileum-MUC2 | 1.00 | 0.93 | 0.16 | 0.11 | |
| Ileum-ZO-1 | 1.00 | 0.71 | 0.07 | 0.16 | |
| Intestinal barrier function | Ileum-Occludin | 1.00 | 0.42 | 0.10 | 0.005 |
| Ileum-Claudin1 | 1.00 | 1.51 | 0.12 | 0.04 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, M.; Lin, C.; Niu, K.; Liu, Y.; Zeng, W.; Wang, R.; Guo, X.; Zhai, Z. Bile Acid Metabolic Profiles and Their Correlation with Intestinal Epithelial Cell Proliferation and Barrier Integrity in Suckling Piglets. Animals 2024, 14, 287. https://doi.org/10.3390/ani14020287
Zhu M, Lin C, Niu K, Liu Y, Zeng W, Wang R, Guo X, Zhai Z. Bile Acid Metabolic Profiles and Their Correlation with Intestinal Epithelial Cell Proliferation and Barrier Integrity in Suckling Piglets. Animals. 2024; 14(2):287. https://doi.org/10.3390/ani14020287
Chicago/Turabian StyleZhu, Min, Chong Lin, Kaimin Niu, Yichun Liu, Weirong Zeng, Ruxia Wang, Xiongchang Guo, and Zhenya Zhai. 2024. "Bile Acid Metabolic Profiles and Their Correlation with Intestinal Epithelial Cell Proliferation and Barrier Integrity in Suckling Piglets" Animals 14, no. 2: 287. https://doi.org/10.3390/ani14020287
APA StyleZhu, M., Lin, C., Niu, K., Liu, Y., Zeng, W., Wang, R., Guo, X., & Zhai, Z. (2024). Bile Acid Metabolic Profiles and Their Correlation with Intestinal Epithelial Cell Proliferation and Barrier Integrity in Suckling Piglets. Animals, 14(2), 287. https://doi.org/10.3390/ani14020287





