Comparative Blood Transcriptome Analysis of Semi-Natural and Controlled Environment Populations of Yangtze Finless Porpoise
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Sample Collection
2.2. Total RNA Extraction and cDNA Library Preparation
2.3. mRNA Sequencing and Analysis
2.4. Real-Time Quantitative PCR Validation
2.5. Supporting Information
3. Results
3.1. Sequencing Data and Alignment Analysis
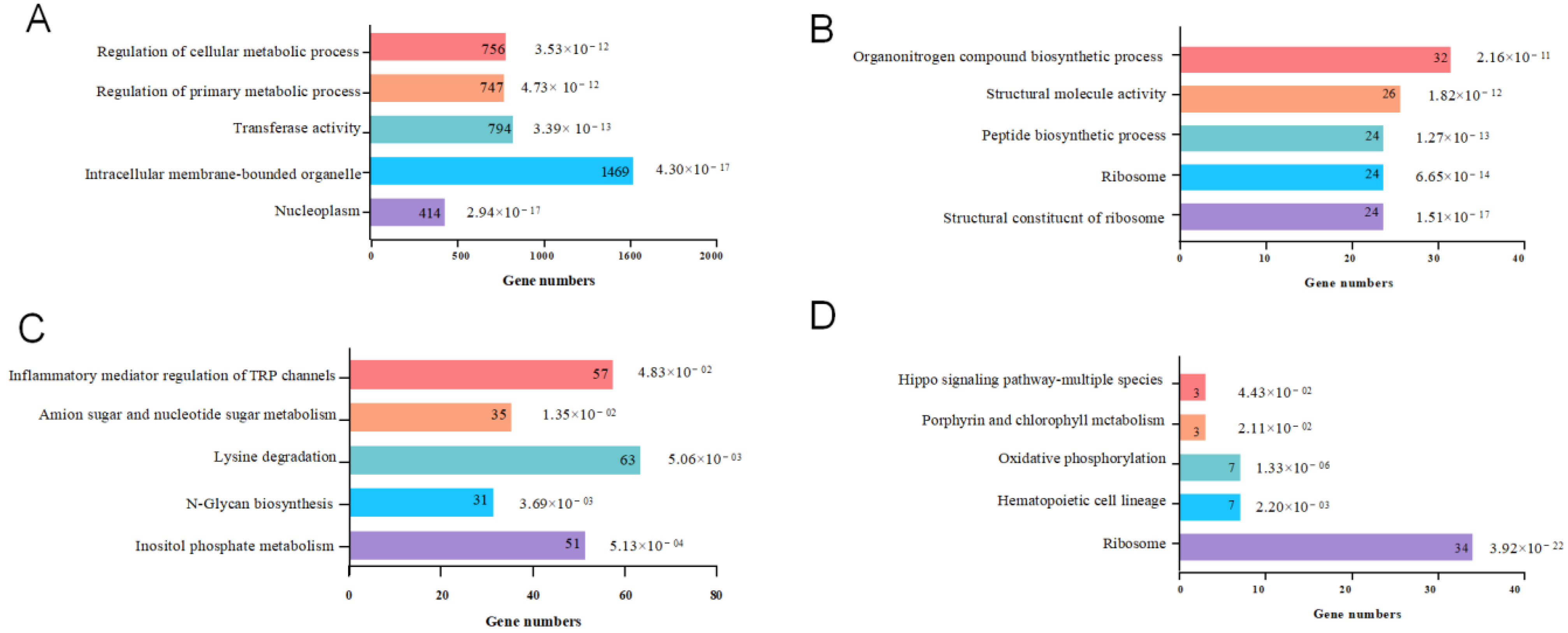
3.2. Analysis and Functional Annotation of Differentially Expressed Genes
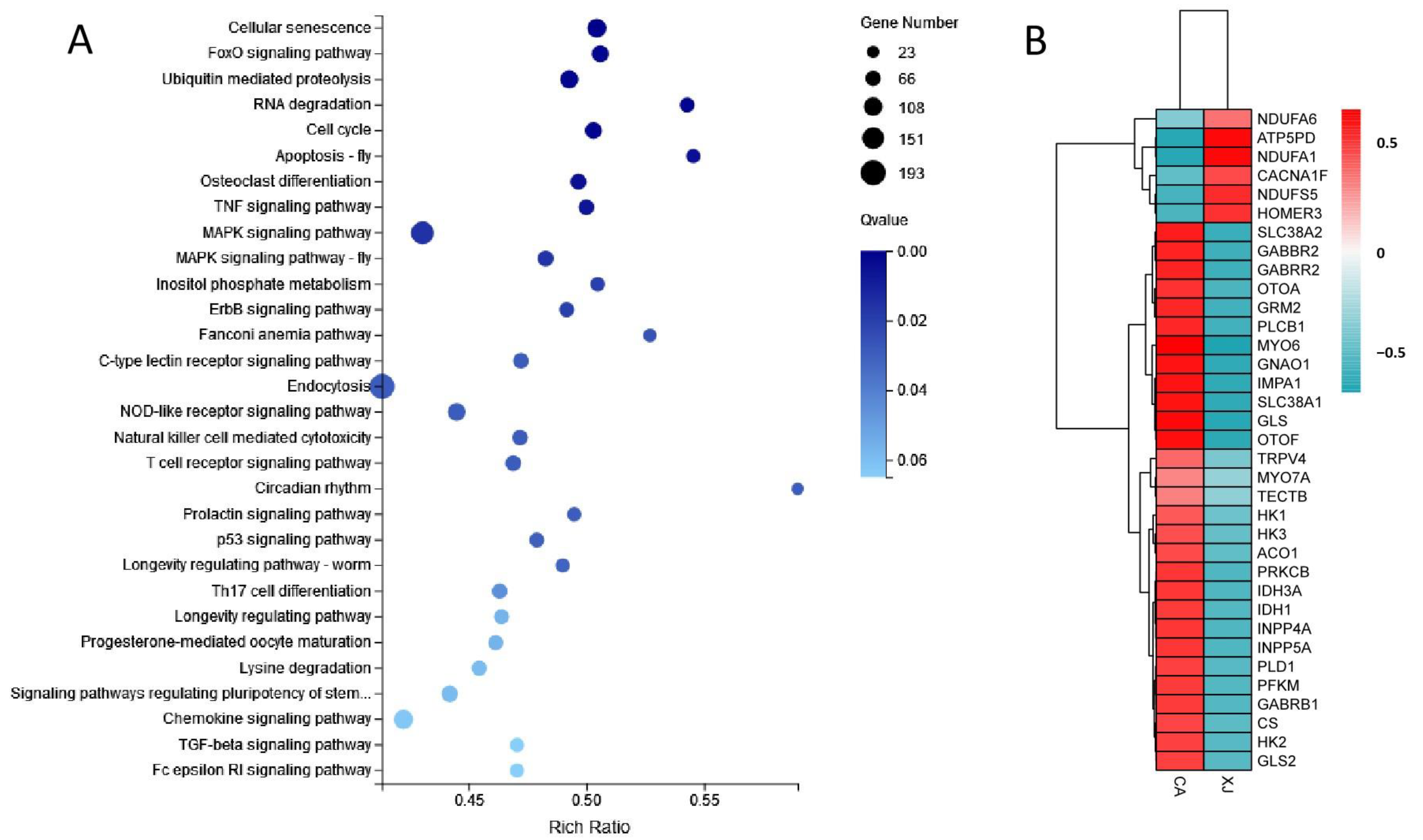
3.3. Enrichment Analysis of Differentially Expressed Genes
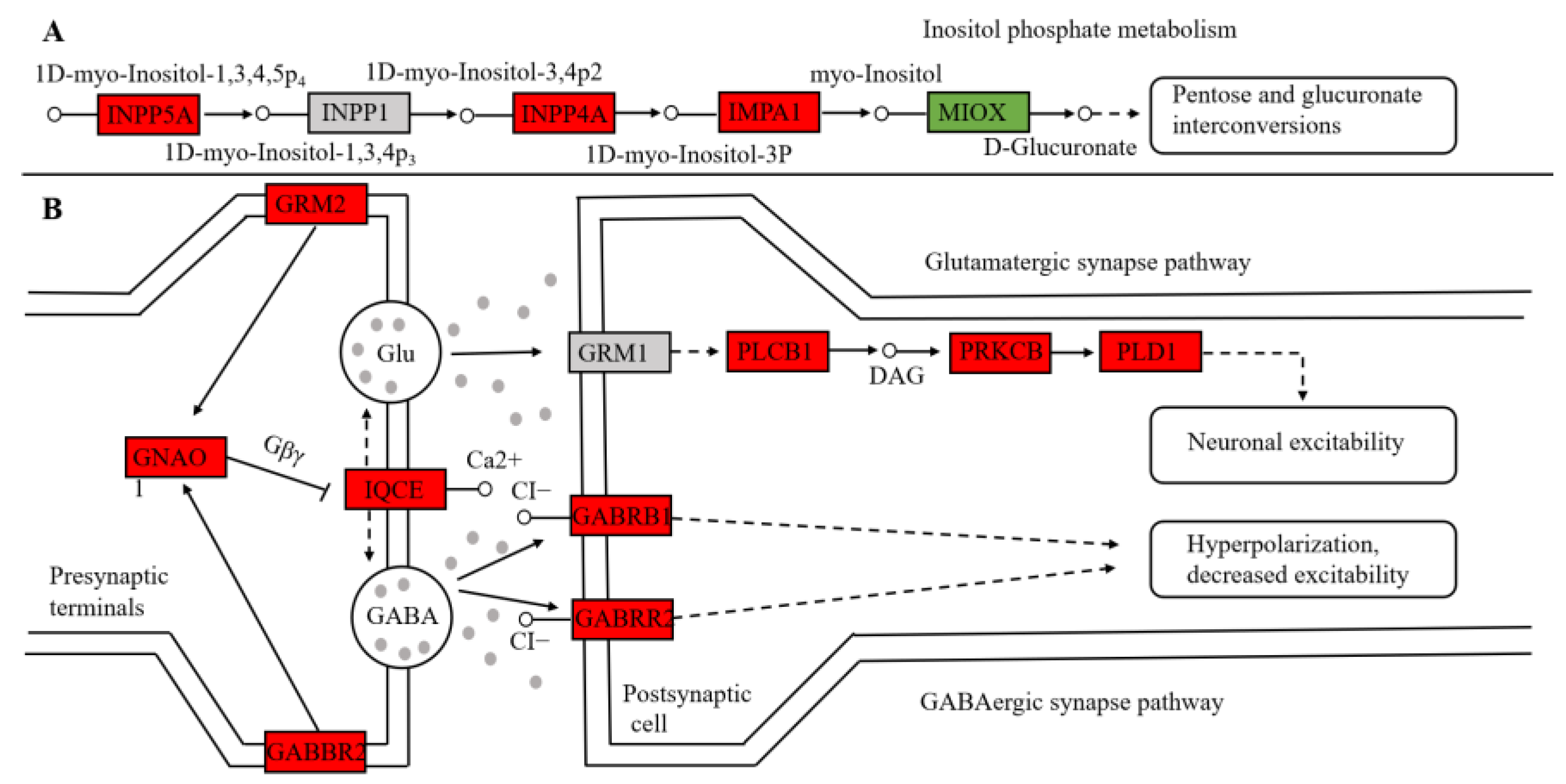
3.4. Analysis of Key Genes
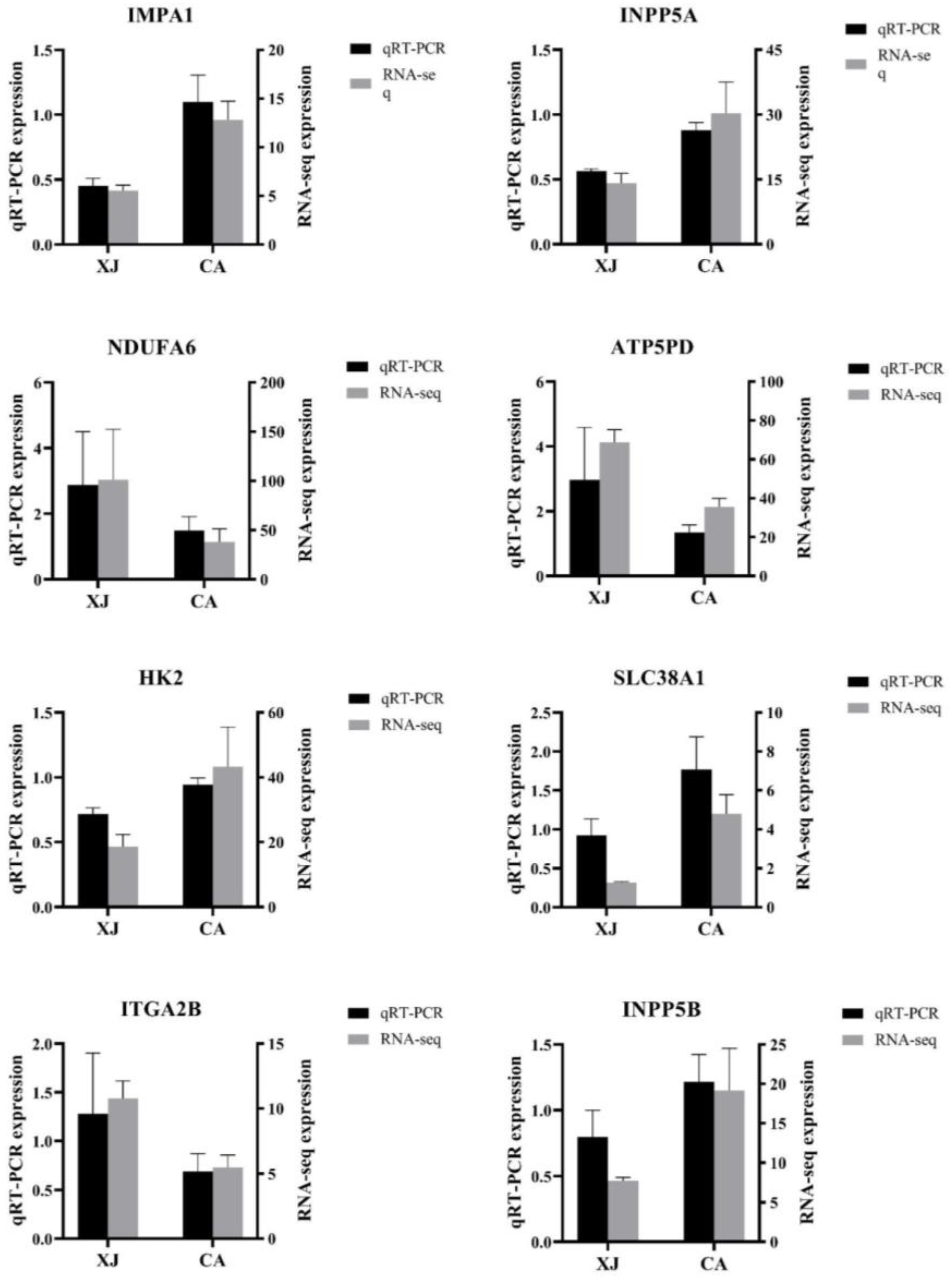
3.5. Validation of RNA-Seq Results by qRT-PCR
4. Discussion
4.1. Differences in Carbohydrate and Amino Acid Metabolism in Different Environments of the Yangtze Finless Porpoise
4.2. Hearing Differences of Yangtze Finless Porpoises in Different Environments
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhao, X.; Jay, B.; Taylor, B.L.; Pitman, R.L.; Wang, K.; Wei, Z.; Stewart, B.S.; Turvey, S.T.; Tomonari, A.; Reeves, R.; et al. Abundance and conservation status of the Yangtze finless porpoise in the Yangtze River, China. Biol. Conserv. 2008, 141, 3006–3018. [Google Scholar] [CrossRef]
- Mei, Z.; Huang, S.L.; Hao, Y.; Turvey, S.T.; Gong, W.; Ding, W.J.B.C. Accelerating population decline of Yangtze finless porpoise (Neophocaena asiaeorientalis asiaeorientalis). Biol. Conserv. 2012, 153, 192–200. [Google Scholar] [CrossRef]
- Nabi, G.; Hao, Y.; Zeng, X.; Jinsong, Z.; McLaughlin, R.W.; Wang, D. Hematologic and biochemical differences between two free ranging Yangtze finless porpoise populations: The implications of habitat. PLoS ONE 2017, 12, e0188570. [Google Scholar] [CrossRef] [PubMed]
- Nabi, G.; Robeck, T.R.; Yujiang, H.; Tang, B.; Zheng, J.; Wang, K.; Wang, D. Circulating concentrations of thyroid hormones and cortisol in wild and semi-natural Yangtze finless porpoise (Neophocaena asiaeorientalis). Conserv. Physiol. 2021, 9, coab034. [Google Scholar] [CrossRef] [PubMed]
- Akamatsu, T.; Wang, D.; Nakamura, K.; Wang, K. Echolocation range of captive and free-ranging baiji (Lipotes vexillifer), finless porpoise (Neophocaena phocaenoides), and bottlenose dolphin (Tursiops truncatus). J. Acoust. Soc. Am. 1998, 104, 2511–2516. [Google Scholar] [CrossRef] [PubMed]
- van Dijk, E.L.; Jaszczyszyn, Y.; Naquin, D.; Thermes, C. The Third Revolution in Sequencing Technology. Trends Genet. 2018, 34, 666–681. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Huang, Y.; Wu, H.; Li, C.; Ling, S.; Sun, J.; Shen, H.; Yue, B.; Zhang, X. Blood transcriptome analysis revealed the immune changes and immunological adaptation of wildness training giant pandas. Mol. Genet. Genom. 2022, 297, 227–239. [Google Scholar] [CrossRef]
- Yin, D.; Lin, D.; Guo, H.; Gu, H.; Ying, C.; Zhang, Y.; Zhang, J.; Liu, K.; Tang, W. Integrated analysis of blood mRNAs and microRNAs reveals immune changes with age in the Yangtze finless porpoise (Neophocaena asiaeorientalis). Comp. Biochem. Physiol. Part B 2021, 256, 110635. [Google Scholar] [CrossRef]
- Wang, Y.; Guo, J.; Wang, L.; Tian, H.; Sui, J. Transcriptome analysis revealed potential mechanisms of differences in physiological stress responses between caged male and female magpies. BMC Genom. 2019, 20, 447. [Google Scholar] [CrossRef]
- Li, R.; Li, Y.; Kristiansen, K.; Wang, J. SOAP: Short oligonucleotide alignment program. Bioinformatics 2008, 24, 713–714. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Yin, D.; Lin, D.; Yan, Y.; Zhu, X.; Ying, C.; Zhang, J.; Xu, P.; Liu, K. Blood Transcriptome Analysis Reveals Gene Expression Differences between Yangtze Finless Porpoises from Two Habitats: Natural and Ex Situ Protected Waters. Fishes 2022, 7, 96. [Google Scholar] [CrossRef]
- Diepenbroek, C.; Serlie, M.J.; Fliers, E.; Kalsbeek, A.; la Fleur, S.E. Brain areas and pathways in the regulation of glucose metabolism. BioFactors 2013, 39, 505–513. [Google Scholar] [CrossRef] [PubMed]
- Chatree, S.; Thongmaen, N.; Tantivejkul, K.; Sitticharoon, C.; Vucenik, I. Role of Inositols and Inositol Phosphates in Energy Metabolism. Molecules 2020, 25, 5079. [Google Scholar] [CrossRef] [PubMed]
- Monte, T.D.; Pilleri, G.; Pilleri, G. Some blood chemistry values obtained from Stenella coeruleoalba (Meyen, 1833) and Delphinus delphis (Linnaeus, 1758) from the western Mediterranean. Invest. Cetacea 1977, 8, 231–232. [Google Scholar]
- Hou, Y.; Han, X. Studies on the biochemical constituents of serum of finless porpoises (Neophocaena phocaenoides) from the changjiang river. J. Nanjing Univ. (Nat. Sci.) 1993, 29, 512–516. [Google Scholar]
- Jin, Q.G. Effects of Carbohydrate Supplement on Serum Amino Acids Concentrations in Heavy-load Training Rats. J. Xi’an Inst. Phys. Educ. 2005, 22, 74–77. [Google Scholar]
- Lehninger, A.L.; Wadkins, C.L. Oxidative phosphorylation. Annu. Rev. Biochem. 1962, 31, 47–78. [Google Scholar] [CrossRef]
- Nabi, G.; Li, Y.; McLaughlin, R.W.; Mei, Z.; Wang, K.; Hao, Y.; Zheng, J.; Wang, D. Immune Responses of the Critically Endangered Yangtze Finless Porpoises (Neophocaena asiaeorientalis ssp. asiaeorientalis) to Escalating Anthropogenic Stressors in the Wild and Seminatural Environments. Front. Physiol. 2019, 10, 1594. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, J.; Persons, J.L.; Peuß, R.; Hassan, H.; Kenzior, A.; Xiong, S.; Olsen, L.; Maldonado, E.; Kowalko, J.E.; Rohner, N. Comparative transcriptome analysis of wild and lab populations of Astyanax mexicanus uncovers differential effects of environment and morphotype on gene expression. J. Exp. Zool. Part B 2020, 334, 530–539. [Google Scholar] [CrossRef] [PubMed]
- Nummela, S.; Thewissen, J.G.; Bajpai, S.; Hussain, T.; Kumar, K. Sound transmission in archaic and modern whales: Anatomical adaptations for underwater hearing. Anat. Rec. 2007, 290, 716–733. [Google Scholar] [CrossRef] [PubMed]
- Zwaenepoel, I.; Mustapha, M.; Leibovici, M.; Verpy, E.; Goodyear, R.; Liu, X.Z.; Nouaille, S.; Nance, W.E.; Kanaan, M.; Avraham, K.B.; et al. Otoancorin, an inner ear protein restricted to the interface between the apical surface of sensory epithelia and their overlying acellular gels, is defective in a autosomal recessive deafness DFNB22. Proc. Natl. Acad. Sci. USA 2002, 99, 6240–6245. [Google Scholar] [CrossRef]
- Chai, S.; Tian, R.; Rong, X.; Li, G.; Chen, B.; Ren, W.; Xu, S.; Yang, G. Evidence of Echolocation in the Common Shrew from Molecular Convergence with Other Echolocating Mammals. Zool. Stud. 2020, 59, e4. [Google Scholar] [CrossRef] [PubMed]
- Friedman, T.B.; Belyantseva, I.A.; Frolenkov, G.I. Myosins and Hearing. Adv. Exp. Med. Biol. 2020, 1239, 317–330. [Google Scholar] [CrossRef]
- Lalayants, M.R.; Mironovich, O.L.; Bliznets, E.A.; Markova, T.G.; Polyakov, A.V.; Tavartkiladze, G.A. OTOF-related auditory neuropathy spectrum disorder. Vestn. Otorinolaringol. 2020, 85, 21–25. [Google Scholar] [CrossRef]
- Ghaffari, R.; Aranyosi, A.J.; Richardson, G.P.; Freeman, D.M. Tectorial membrane travelling waves underlie abnormal hearing in Tectb mutant mice. Nat. Commun. 2010, 1, 96. [Google Scholar] [CrossRef]
- Wang, D. A preliminary study on sound and acoustic behavior of the Yangtze river finless porpoise, Neophocaena phocaenoides. Acta Hydrobiol. Sin. 1996, 2, 127–133. [Google Scholar]
- Sanchez, J.T.; Wang, Y.; Rubel, E.W.; Barria, A. Development of glutamatergic synaptic transmission in binaural auditory neurons. J. Neurophysiol. 2010, 104, 1774–1789. [Google Scholar] [CrossRef]
- Wu, S.H. Synaptic excitation in the dorsal nucleus of the lateral lemniscus. Prog. Neurobiol. 1999, 57, 357–375. [Google Scholar] [CrossRef] [PubMed]
- Coate, T.M.; Scott, M.K.; Gurjar, M. Current concepts in cochlear ribbon synapse formation. Synapse 2019, 73, e22087. [Google Scholar] [CrossRef] [PubMed]
- Lu, N.; Chen, B.; Qing, J.; Lei, J.; Wang, T.; Shi, H.; Wang, J. Transcriptome Analyses Provide Insights into the Auditory Function in Trachemys scripta elegans. Anim. Open Access J. MDPI 2022, 12, 2410. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Wang, X.; Liu, Y.; Fu, Q.; Tian, C.; Wu, C.; Shi, H.; Yuan, Z.; Tan, S.; Liu, S.; et al. Transcriptome analysis reveals enrichment of genes associated with auditory system in swimbladder of channel catfish. Comp. Biochem. Physiol. Part D 2018, 27, 30–39. [Google Scholar] [CrossRef]
- Cao, Z.; Yin, D.; Li, Z.; Yan, Y.; Zhang, P.; Zhang, S.; Lin, D.; Hua, Z.; Zhang, J.; Ying, C.; et al. Blood Transcriptome Analysis Provides Responsive Changes in Gene Expression between Ex Situ and Captive Yangtze Finless Porpoises (Neophocaena asiaeorientalis asiaeorientalis). Fishes 2023, 8, 593. [Google Scholar] [CrossRef]
| Sample | Total Raw Reads (M) | Total Clean Reads (M) | Total Clean Bases (Gb) | Clean Reads Q20 (%) | Clean Reads Q30 (%) | Total Mapping (%) | Uniquely Mapping (%) |
|---|---|---|---|---|---|---|---|
| CA_1 | 128.75 | 126.62 | 12.66 | 97.98 | 93.98 | 89.39 | 68.60 |
| CA_2 | 75.36 | 73.54 | 7.35 | 97.94 | 93.85 | 75.64 | 39.80 |
| CA_3 | 88.15 | 86.37 | 8.64 | 98.00 | 94.06 | 84.83 | 60.10 |
| XJ_1 | 66.32 | 64.62 | 6.46 | 98.02 | 94.09 | 77.62 | 41.93 |
| XJ_2 | 69.75 | 67.55 | 6.76 | 98.09 | 94.28 | 73.90 | 34.36 |
| XJ_3 | 85.73 | 83.27 | 8.33 | 98.09 | 94.28 | 77.75 | 43.38 |
| Pathway ID | Pathway Name | Functional Classification | p Value |
|---|---|---|---|
| Significantly enriched pathways of DEGs related to metabolism | |||
| ko00562 | inositol phosphate metabolism | carbohydrate metabolism | 1.42 × 10−28 |
| ko00520 | amino sugar and nucleotide sugar metabolism | 5.78 × 10−18 | |
| ko00010 | glycolysis/gluconeogenesis | 3.32 × 10−12 | |
| ko00020 | citrate cycle (TCA cycle) | 1.12 × 10−10 | |
| ko00620 | pyruvate metabolism | 1.37 × 10−10 | |
| ko00510 | N-glycan biosynthesis | glycan biosynthesis and metabolism | 1.28 × 10−17 |
| ko00330 | arginine and proline metabolism | amino acid metabolism | 1.19 × 10−14 |
| ko00270 | cysteine and methionine metabolism | 1.28 × 10−12 | |
| ko00563 | valine, leucine and isoleucine degradation | 1.84 × 10−9 | |
| ko00471 | D-glutamine and D-glutamate metabolism | 1.19 × 10−14 | |
| Significantly enriched pathways of DEGs related to environmental adaptation | |||
| ko04714 | thermogenesis | environmental adaptation | 6.53 × 10−106 |
| ko04912 | GnRH signaling pathway | endocrine system | 4.31 × 10−25 |
| ko04921 | oxytocin signaling pathway | 4.67 × 10−34 | |
| ko04725 | cholinergic synapse | nervous system | 7.43 × 10−31 |
| ko04728 | dopaminergic synapse | 1.06 × 10−29 | |
| ko04723 | retrograde endocannabinoid signaling | 3.00 × 10−28 | |
| ko04724 | glutamatergic synapse | 3.96 × 10−21 | |
| ko04727 | GABAergic synapse | 3.72 × 10−13 | |
| ko04750 | inflammatory mediator regulation of TRP channels | sensory system | 1.16 × 10−19 |
| ko04745 | phototransduction-fly | 1.84 × 10−13 | |
| Gene Name | Description | Gene ID | log2FC (CA/XJ) |
|---|---|---|---|
| metabolism-related differentially expressed genes | |||
| HK1 | hexokinase 1 | 112413526 | 1.03 |
| HK2 | hexokinase 2 | 112403280 | 1.33 |
| HK3 | hexokinase 3 | 112397869 | 1.11 |
| PFKM | phosphofructokinase | 112394271 | 1.09 |
| CS | citrate synthase | 112394084 | 1.07 |
| IDH1 | isocitrate dehydrogenase 1 | 112395270 | 1.42 |
| IDH3A | isocitrate dehydrogenase 3 (NAD(+)) alpha | 112405774 | 1.47 |
| ACO1 | aconitase 1 | 112405425 | 1.13 |
| GLUD1 | glutamate dehydrogenase 1 | 112390428 | 1.45 |
| GLS | glutaminase | 112392138 | 2.02 |
| GLS2 | glutaminase 2 | 112394275 | 1.59 |
| INPP5A | inositol polyphosphate-5-phosphatase A | 112411270 | 1.32 |
| INPP4A | inositol polyphosphate-4-phosphatase type I A | 112398954 | 1.62 |
| IMPA1 | inositol monophosphatase 1 | 112390609 | 1.26 |
| NDUFA1 | NADH:ubiquinone oxidoreductase subunit A1 | 112404044 | −1.18 |
| NDUFA6 | NADH:ubiquinone oxidoreductase subunit A6 | 112394690 | −1.69 |
| NDUFS5 | NADH:ubiquinone oxidoreductase subunit S5 | 112395486 | −1.15 |
| ATP5PD | ATP synthase peripheral stalk subunit d | 112410517 | −1.06 |
| MIOX | myo-inositol oxygenase | 112394676 | −1.25 |
| auditory-related differentially expressed genes | |||
| MYO6 | myosin VI | 112390806 | 1.77 |
| MYO7A | myosin VIIA | 112402650 | 1.07 |
| OTOF | otoferlin | 112412779 | 2.69 |
| OTOA | otoancorin | 112399369 | 1.53 |
| TECTB | tectorin beta | 112406974 | 1.72 |
| TRPV4 | transient receptor potential cation channel subfamily V member 4 | 112397312 | 1.77 |
| GNAO1 | G protein subunit alpha o1 | 112403775 | 1.76 |
| GRM2 | glutamate metabotropic receptor 2 | 112400596 | 1.76 |
| IQCE | IQ motif containing E | 112412075 | 1.12 |
| SLC38A1 | solute carrier family 38 member 1 | 112394242 | 1.86 |
| SLC38A2 | solute carrier family 38 member 2 | 112394243 | 2.00 |
| PLCB1 | phospholipase C beta 1 | 112412530 | 1.68 |
| PRKCB | protein kinase C beta | 112399411 | 1.19 |
| PLD1 | phospholipase D1 | 112392372 | 1.54 |
| GABRR2 | gamma-aminobutyric acid type A receptor rho2 subunit | 112401210 | 1.16 |
| GABRB1 | gamma-aminobutyric acid type A receptor beta1 subunit | 112390625 | 2.92 |
| GABBR2 | gamma-aminobutyric acid type B receptor subunit 2 | 112392619 | 1.45 |
| CACNA1F | calcium voltage-gated channel subunit alpha1 F | 112399841 | −2.18 |
| HOMER3 | homer scaffold protein 3 | 112414903 | −1.06 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, W.; Yin, D.; Li, Z.; Zhu, X.; Zhang, S.; Zhang, P.; Lin, D.; Hua, Z.; Cao, Z.; Zhang, H.; et al. Comparative Blood Transcriptome Analysis of Semi-Natural and Controlled Environment Populations of Yangtze Finless Porpoise. Animals 2024, 14, 199. https://doi.org/10.3390/ani14020199
Liu W, Yin D, Li Z, Zhu X, Zhang S, Zhang P, Lin D, Hua Z, Cao Z, Zhang H, et al. Comparative Blood Transcriptome Analysis of Semi-Natural and Controlled Environment Populations of Yangtze Finless Porpoise. Animals. 2024; 14(2):199. https://doi.org/10.3390/ani14020199
Chicago/Turabian StyleLiu, Wang, Denghua Yin, Zhanwei Li, Xiaoyan Zhu, Sigang Zhang, Peng Zhang, Danqing Lin, Zhong Hua, Zhichen Cao, Han Zhang, and et al. 2024. "Comparative Blood Transcriptome Analysis of Semi-Natural and Controlled Environment Populations of Yangtze Finless Porpoise" Animals 14, no. 2: 199. https://doi.org/10.3390/ani14020199
APA StyleLiu, W., Yin, D., Li, Z., Zhu, X., Zhang, S., Zhang, P., Lin, D., Hua, Z., Cao, Z., Zhang, H., Zhang, J., Ying, C., Xu, P., Dong, G., & Liu, K. (2024). Comparative Blood Transcriptome Analysis of Semi-Natural and Controlled Environment Populations of Yangtze Finless Porpoise. Animals, 14(2), 199. https://doi.org/10.3390/ani14020199





