Isolation of Lactic Acid Bacteria (LAB) from Salmonids for Potential Use as Probiotics: In Vitro Assays and Toxicity Assessment of Salmo trutta Embryonated Eggs
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation of LAB and Pathogens from Fish
2.2. Genomic Bacterial DNA Extraction
2.3. PCR Identification of Presumptive Fish Pathogenic Bacteria from the Isolates
2.4. Molecular Identification of the Potential LAB Probiotic Isolates and of Presumptive Fish Pathogenic Bacteria by Partial Sequencing of the 16S rRNA gene and ITS-1 Intergenic Spacer
2.5. Identification of Presumptive Fish Pathogenic Bacteria from the Isolates by MALDI-TOF
2.6. Antibiotic Susceptibility Test Using Disks on LAB Probiotic Candidates
2.7. Antagonistic In Vitro Assay by Agar Plug Diffusion Method of LAB
2.8. Antagonistic In Vitro Assay by Agar Well Diffusion Method with Supernatants from Pure LAB Cultures
2.9. Toxicity Assay of Live Bacteria on Embryonic Eggs of Salmo trutta
2.10. Statistical Analysis
2.11. Ethics
3. Results
3.1. Isolation and Molecular Identificacion of Fish Pathogens and LAB Probiotic Candidates
3.2. Antibiotic Susceptibility Test on LAB Probiotic Candidates
3.3. Antagonistic In Vitro Assay by Agar Plug Diffusion Method
3.4. Antagonistic In Vitro Assay by Agar Well Diffusion Method
3.5. Toxicity Assay of Live Bacteria on Embryonic Eggs of Salmo trutta
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aquaculture Business Association of Spain (APROMAR). Aquaculture in Spain 2023 APROMAR 2023. Available online: https://apromar.es/wp-content/uploads/2023/10/Aquaculture_in_Spain_2023_APROMAR.pdf (accessed on 13 November 2023).
- Fečkaninová, A.; Koščová, J.; Mudroňová, D.; Schusterová, P.; Cingeľová Maruščáková, I.; Popelka, P. Characterization of Two Novel Lactic Acid Bacteria Isolated from the Intestine of Rainbow Trout (Oncorhynchus Mykiss, Walbaum) in Slovakia. Aquaculture 2019, 506, 294–301. [Google Scholar] [CrossRef]
- Balcázar, J.L.; Vendrell, D.; De Blas, I.; Ruiz-Zarzuela, I.; Gironés, O.; Múzquiz, J.L. Quantitative Detection of Aeromonas Salmonicida in Fish Tissue by Real-Time PCR Using Self-Quenched, Fluorogenic Primers. J. Med. Microbiol. 2007, 56, 323–328. [Google Scholar] [CrossRef] [PubMed]
- Keeling, S.E.; Johnston, C.; Wallis, R.; Brosnahan, C.L.; Gudkovs, N.; McDonald, W.L. Development and Validation of Real-Time PCR for the Detection of Yersinia Ruckeri: Yersinia Ruckeri Real-Time PCR. J. Fish Dis. 2012, 35, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Mohsina, K.; Kaur, M.; Bowman, J.P.; Powell, S.; Tamplin, M.L. qPCR Quantification of Carnobacterium Maltaromaticum, Brochothrix Thermosphacta, and Serratia Liquefaciens Growth Kinetics in Mixed Culture. J. Microbiol. Methods 2020, 175, 105961. [Google Scholar] [CrossRef] [PubMed]
- Raza, S.; Koh, Y.; Yoon, S.-S.; Woo, S.-Y.; Ahn, K.-S.; Kim, H.-L.; Kim, H.-N. Identification of Novel Carnobacterium Maltaromaticum Strains in Bone Marrow Samples of Patients with Acute Myeloid Leukemia Using a Metagenomic Binning Approach. Int. Microbiol. 2023, 26, 1033–1040. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.A.; Newman, S.J.; Harrison, C.E.; Loch, T.P. First Isolation of Carnobacterium Maltaromaticum from Farmed Rainbow Trout in Virginia. J. Aquat. Anim. Health 2023, 35, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Marancik, D.P.; Wiens, G.D. A Real-Time Polymerase Chain Reaction Assay for Identification and Quantification of Flavobacterium Psychrophilum and Application to Disease Resistance Studies in Selectively Bred Rainbow Trout Oncorhynchus Mykiss. FEMS Microbiol. Lett. 2013, 339, 122–129. [Google Scholar] [CrossRef]
- Torres-Corral, Y.; Fernández-Álvarez, C.; Santos, Y. High-throughput Identification and Quantification of Vagococcus Salmoninarum by SYBR Green I-based Real-time PCR Combined with Melting Curve Analysis. J. Fish Dis. 2019, 42, 1359–1368. [Google Scholar] [CrossRef]
- Chapela, M.-J.; Ferreira, M.; Varela, C.; Arregui, L.; Garrido-Maestu, A. Development of a Multiplex Real-Time PCR Method for Early Diagnosis of Three Bacterial Diseases in Fish: A Real-Case Study in Trout Aquaculture. Aquaculture 2018, 496, 255–261. [Google Scholar] [CrossRef]
- Seghouani, H.; Garcia-Rangel, C.-E.; Füller, J.; Gauthier, J.; Derome, N. Walleye Autochthonous Bacteria as Promising Probiotic Candidates against Flavobacterium Columnare. Front. Microbiol. 2017, 8, 1349. [Google Scholar] [CrossRef]
- Bondad-Reantaso, M.G.; MacKinnon, B.; Karunasagar, I.; Fridman, S.; Alday-Sanz, V.; Brun, E.; Le Groumellec, M.; Li, A.; Surachetpong, W.; Karunasagar, I.; et al. Review of Alternatives to Antibiotic Use in Aquaculture. Rev. Aquac. 2023, 15, 1421–1451. [Google Scholar] [CrossRef]
- Pérez-Sánchez, T.; Mora-Sánchez, B.; Balcázar, J.L. Biological Approaches for Disease Control in Aquaculture: Advantages, Limitations and Challenges. Trends Microbiol. 2018, 26, 896–903. [Google Scholar] [CrossRef] [PubMed]
- Pérez, T.; Balcázar, J.L.; Peix, A.; Valverde, A.; Velázquez, E.; de Blas, I.; Ruiz-Zarzuela, I. Lactococcus Lactis Subsp. Tructae Subsp. Nov. Isolated from the Intestinal Mucus of Brown Trout (Salmo Trutta) and Rainbow Trout (Oncorhynchus Mykiss). Int. J. Syst. Evol. Microbiol. 2011, 61, 1894–1898. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Sánchez, T.; Balcázar, J.L.; García, Y.; Halaihel, N.; Vendrell, D.; de Blas, I.; Merrifield, D.L.; Ruiz-Zarzuela, I. Identification and Characterization of Lactic Acid Bacteria Isolated from Rainbow Trout, Oncorhynchus Mykiss (Walbaum), with Inhibitory Activity against Lactococcus Garvieae: Trout Endogenous LAB Antagonise L. Garvieae. J. Fish Dis. 2011, 34, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Abid, A.; Davies, S.J.; Waines, P.; Emery, M.; Castex, M.; Gioacchini, G.; Carnevali, O.; Bickerdike, R.; Romero, J.; Merrifield, D.L. Dietary Synbiotic Application Modulates Atlantic Salmon (Salmo Salar) Intestinal Microbial Communities and Intestinal Immunity. Fish Shellfish Immunol. 2013, 35, 1948–1956. [Google Scholar] [CrossRef] [PubMed]
- EFSA Panel on Biological Hazards (BIOHAZ); Koutsoumanis, K.; Allende, A.; Alvarez-Ordóñez, A.; Bolton, D.; Bover-Cid, S.; Chemaly, M.; De Cesare, A.; Hilbert, F.; Lindqvist, R.; et al. Statement on How to Interpret the QPS Qualification on ‘Acquired Antimicrobial Resistance Genes’. EFS2 2023, 21, e08323. [Google Scholar] [CrossRef]
- EFSA Panel on Biological Hazards (BIOHAZ); Koutsoumanis, K.; Allende, A.; Alvarez-Ordóñez, A.; Bolton, D.; Bover-Cid, S.; Chemaly, M.; Davies, R.; De Cesare, A.; Hilbert, F.; et al. Update of the List of QPS-recommended Biological Agents Intentionally Added to Food or Feed as Notified to EFSA 13: Suitability of Taxonomic Units Notified to EFSA until September 2020. EFS2 2021, 19, e06377. [Google Scholar] [CrossRef]
- EFSA Panel on Additives and Products or Substances Used in Animal Feed (FEEDAP); Rychen, G.; Aquilina, G.; Azimonti, G.; Bampidis, V.; de Lourdes Bastos, M.; Bories, G.; Chesson, A.; Cocconcelli, P.S.; Flachowsky, G.; et al. Guidance on the Characterisation of Microorganisms Used as Feed Additives or as Production Organisms. EFS2 2018, 16, e05206. [Google Scholar] [CrossRef]
- EFSA Panel on Additives and Products or Substances Used in Animal Feed (FEEDAP); Rychen, G.; Aquilina, G.; Azimonti, G.; Bampidis, V.; de Lourdes Bastos, M.; Bories, G.; Chesson, A.; Cocconcelli, P.S.; Flachowsky, G.; et al. Guidance on the Assessment of the Safety of Feed Additives for the Target Species. EFS2 2017, 15, e05021. [Google Scholar] [CrossRef]
- EFSA Panel on Additives and Products or Substances Used in Animal Feed (FEEDAP); Rychen, G.; Aquilina, G.; Azimonti, G.; Bampidis, V.; de Lourdes Bastos, M.; Bories, G.; Chesson, A.; Cocconcelli, P.S.; Flachowsky, G.; et al. Guidance on the Identity, Characterisation and Conditions of Use of Feed Additives. EFS2 2017, 15, e05023. [Google Scholar] [CrossRef]
- Miller, J.L.; Miller, M.J.; De Vlaming, V.; Larsen, K.; Smith, E.; Reece, K. Selection and Application of a Rainbow Trout Toxicity Testing Procedure for Screening Sacramento River Watershed, California Samples. Env. Monit. Assess. 2009, 150, 333. [Google Scholar] [CrossRef]
- Rigaud, C.; Härme, J.; Vehniäinen, E.-R. Salmo Trutta Is More Sensitive than Oncorhynchus Mykiss to Early-Life Stage Exposure to Retene. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2022, 252, 109219. [Google Scholar] [CrossRef]
- Hoitsy, G.; András, W.; Thomas, M.-P. Guide to the Small Scale Artificial Propagation of Trout; The FAO, Regional Office for Europe and Central Asia: Budapest, Hungary, 2012. [Google Scholar]
- Wilkins, L.G.E.; Rogivue, A.; Schütz, F.; Fumagalli, L.; Wedekind, C. Increased Diversity of Egg-Associated Bacteria on Brown Trout (Salmo Trutta) at Elevated Temperatures. Sci. Rep. 2015, 5, 17084. [Google Scholar] [CrossRef]
- Jimenez Reyes, M.; Yany, G.; Romero, J. Protocolo para obtencion de alevines axenicos de trucha arcoiris (Oncorhynchus mykiss). Lajar 2017, 45, 1064–1069. [Google Scholar] [CrossRef]
- Donati, V.L.; Dalsgaard, I.; Runtuvuori-Salmela, A.; Kunttu, H.; Jørgensen, J.; Castillo, D.; Sundberg, L.-R.; Middelboe, M.; Madsen, L. Interactions between Rainbow Trout Eyed Eggs and Flavobacterium Spp. Using a Bath Challenge Model: Preliminary Evaluation of Bacteriophages as Pathogen Control Agents. Microorganisms 2021, 9, 971. [Google Scholar] [CrossRef]
- Pérez-Pascual, D.; Vendrell-Fernández, S.; Audrain, B.; Bernal-Bayard, J.; Patiño-Navarrete, R.; Petit, V.; Rigaudeau, D.; Ghigo, J.-M. Gnotobiotic Rainbow Trout (Oncorhynchus Mykiss) Model Reveals Endogenous Bacteria That Protect against Flavobacterium Columnare Infection. PLoS Pathog. 2021, 17, e1009302. [Google Scholar] [CrossRef]
- Padeniya, U.; Larson, E.T.; Septriani, S.; Pataueg, A.; Kafui, A.R.; Hasan, E.; Mmaduakonam, O.S.; Kim, G.; Kiddane, A.T.; Brown, C.L. Probiotic Treatment Enhances Pre-feeding Larval Development and Early Survival in Zebrafish Danio Rerio. J. Aquat. Anim. Health 2022, 34, 3–11. [Google Scholar] [CrossRef]
- Aquatic Animal Health & Vaccines Centre of Excellence. Field Sampling of Fish for Disease Investigation and Health Monitoring, Guidelines and Procedures Manual; Department of Primary Industries Parks, Water & Environment: Hobart, TAS, Australia, 2017.
- Chase, D.M.; Elliott, D.G.; Pascho, R.J. Detection and Quantification of Renibacterium Salmoninarum DNA in Salmonid Tissues by Real-Time Quantitative Polymerase Chain Reaction Analysis. J. VET Diagn. Investig. 2006, 18, 375–380. [Google Scholar] [CrossRef]
- Elliott, D.G.; Applegate, L.J.; Murray, A.L.; Purcell, M.K.; McKibben, C.L. Bench-Top Validation Testing of Selected Immunological and Molecular Renibacterium Salmoninarum Diagnostic Assays by Comparison with Quantitative Bacteriological Culture. J. Fish Dis. 2013, 36, 779–809. [Google Scholar] [CrossRef]
- Bartkova, S.; Kokotovic, B.; Skall, H.F.; Lorenzen, N.; Dalsgaard, I. Detection and Quantification of Aeromonas Salmonicida in Fish Tissue by Real-Time PCR. J. Fish Dis. 2017, 40, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Sepúlveda, D.; Bohle, H.; Labra, Á.; Grothusen, H.; Marshall, S.H. Design and Evaluation of a Unique RT-qPCR Assay for Diagnostic Quality Control Assessment That Is Applicable to Pathogen Detection in Three Species of Salmonid Fish. BMC Vet. Res. 2013, 9, 183. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S Ribosomal DNA Amplification for Phylogenetic Study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef]
- Yoon, S.-H.; Ha, S.-M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A Taxonomically United Database of 16S rRNA Gene Sequences and Whole-Genome Assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Kumar, S.; Tamura, K.; Nei, M. MEGA: Molecular Evolutionary Genetics Analysis Software for Microcomputers. Bioinformatics 1994, 10, 189–191. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Bouzaine, T.; Dauphin, R.D.; Thonart, P.; Urdaci, M.C.; Hamdi, M. Adherence and Colonization Properties of Lactobacillus Rhamnosus TB1, a Broiler Chicken Isolate. Lett. Appl. Microbiol. 2005, 40, 391–396. [Google Scholar] [CrossRef]
- Li, T.; Teng, D.; Mao, R.; Hao, Y.; Wang, X.; Wang, J. A Critical Review of Antibiotic Resistance in Probiotic Bacteria. Food Res. Int. 2020, 136, 109571. [Google Scholar] [CrossRef] [PubMed]
- Anokyewaa, M.A.; Amoah, K.; Li, Y.; Lu, Y.; Kuebutornye, F.K.A.; Asiedu, B.; Seidu, I. Prevalence of Virulence Genes and Antibiotic Susceptibility of Bacillus Used in Commercial Aquaculture Probiotics in China. Aquac. Rep. 2021, 21, 100784. [Google Scholar] [CrossRef]
- Elleuch, L.; Shaaban, M.; Smaoui, S.; Mellouli, L.; Karray-Rebai, I.; Fourati-Ben Fguira, L.; Shaaban, K.A.; Laatsch, H. Bioactive Secondary Metabolites from a New Terrestrial Streptomyces Sp. TN262. Appl. Biochem. Biotechnol. 2010, 162, 579–593. [Google Scholar] [CrossRef] [PubMed]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in Vitro Evaluating Antimicrobial Activity: A Review. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef]
- Hindler, J.A.; Richter, S.S. Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria: M45, Documents/Clinical and Laboratory Standards Institute, 3rd ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2016; ISBN 978-1-56238-918-5. [Google Scholar]
- James, L.S., II. M100 Performance Standards for Antimicrobial; Clinical and Laboratory Standards Institute: Berwyn, PA, USA, 2022; ISBN 978-1-68440-134-5. [Google Scholar]
- Araújo, C.; Muñoz-Atienza, E.; Pérez-Sánchez, T.; Poeta, P.; Igrejas, G.; Hernández, P.E.; Herranz, C.; Ruiz-Zarzuela, I.; Cintas, L.M. Nisin Z Production by Lactococcus Lactis Subsp. Cremoris WA2-67 of Aquatic Origin as a Defense Mechanism to Protect Rainbow Trout (Oncorhynchus Mykiss, Walbaum) Against Lactococcus Garvieae. Mar. Biotechnol. 2015, 17, 820–830. [Google Scholar] [CrossRef]
- Araújo, C.; Muñoz-Atienza, E.; Poeta, P.; Igrejas, G.; Hernández, P.; Herranz, C.; Cintas, L. Characterization of Pediococcus Acidilactici Strains Isolated from Rainbow Trout (Oncorhynchus Mykiss) Feed and Larvae: Safety, DNA Fingerprinting, and Bacteriocinogenicity. Dis. Aquat. Org. 2016, 119, 129–143. [Google Scholar] [CrossRef]
- Amin, M.; Adams, M.; Bolch, C.J.S.; Burke, C.M. In Vitro Screening of Lactic Acid Bacteria Isolated from Gastrointestinal Tract of Atlantic Salmon (Salmo Salar) as Probiont Candidates. Aquacult. Int. 2017, 25, 485–498. [Google Scholar] [CrossRef]
- Meyburgh, C.; Bragg, R.; Boucher, C. Lactococcus Garvieae: An Emerging Bacterial Pathogen of Fish. Dis. Aquat. Org. 2017, 123, 67–79. [Google Scholar] [CrossRef]
- Rösch, R.M.; Buschmann, K.; Brendel, L.; Schwanz, T.; Vahl, C.-F. Lactococcus Garvieae Endocarditis in a Prosthetic Aortic Valve: A Case Report and Literature Review. J. Investig. Med. High Impact Case Rep. 2019, 7, 232470961983205. [Google Scholar] [CrossRef]
- Tena, D.; Martínez, N.M.; Losa, C.; Fernández, C.; Medina, M.J.; Sáez-Nieto, J.A. Acute Acalculous Cholecystitis Complicated with Peritonitis Caused by Lactobacillus Plantarum. Diagn. Microbiol. Infect. Dis. 2013, 76, 510–512. [Google Scholar] [CrossRef]
- Biesiada, G.; Krycińska, R.; Czepiel, J.; Stażyk, K.; Kędzierska, J.; Garlicki, A. Meningoencephalitis Caused by Lactobacillus Plantarum—Case Report. Int. J. Neurosci. 2019, 129, 715–718. [Google Scholar] [CrossRef]
- Usta-Atmaca, H.; Akbas, F.; Karagoz, Y.; Piskinpasa, M.E. A Rarely Seen Cause for Empyema: Leuconostoc Mesenteroıdes. J. Infect. Dev. Ctries 2015, 9, 425–427. [Google Scholar] [CrossRef]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-Resistant, Extensively Drug-Resistant and Pandrug-Resistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Borges, N.; Keller-Costa, T.; Sanches-Fernandes, G.M.M.; Louvado, A.; Gomes, N.C.M.; Costa, R. Bacteriome Structure, Function, and Probiotics in Fish Larviculture: The Good, the Bad, and the Gaps. Annu. Rev. Anim. Biosci. 2021, 9, 423–452. [Google Scholar] [CrossRef]
- Alvarez-Sieiro, P.; Montalbán-López, M.; Mu, D.; Kuipers, O.P. Bacteriocins of Lactic Acid Bacteria: Extending the Family. Appl. Microbiol. Biotechnol. 2016, 100, 2939–2951. [Google Scholar] [CrossRef]
- Imade, E.E.; Omonigho, S.E.; Babalola, O.O.; Enagbonma, B.J. Lactic Acid Bacterial Bacteriocins and Their Bioactive Properties against Food-Associated Antibiotic-Resistant Bacteria. Ann. Microbiol. 2021, 71, 44. [Google Scholar] [CrossRef]
- Rocchetti, M.T.; Russo, P.; Capozzi, V.; Drider, D.; Spano, G.; Fiocco, D. Bioprospecting Antimicrobials from Lactiplantibacillus Plantarum: Key Factors Underlying Its Probiotic Action. Int. J. Mol. Sci. 2021, 22, 12076. [Google Scholar] [CrossRef]
- Pérez-Sánchez, T.; Balcázar, J.L.; Merrifield, D.L.; Carnevali, O.; Gioacchini, G.; de Blas, I.; Ruiz-Zarzuela, I. Expression of Immune-Related Genes in Rainbow Trout (Oncorhynchus Mykiss) Induced by Probiotic Bacteria during Lactococcus Garvieae Infection. Fish Shellfish Immunol. 2011, 31, 196–201. [Google Scholar] [CrossRef]
- Pérez-Sánchez, T.; Mora-Sánchez, B.; Vargas, A.; Balcázar, J.L. Changes in Intestinal Microbiota and Disease Resistance Following Dietary Postbiotic Supplementation in Rainbow Trout (Oncorhynchus Mykiss). Microb. Pathog. 2020, 142, 104060. [Google Scholar] [CrossRef]
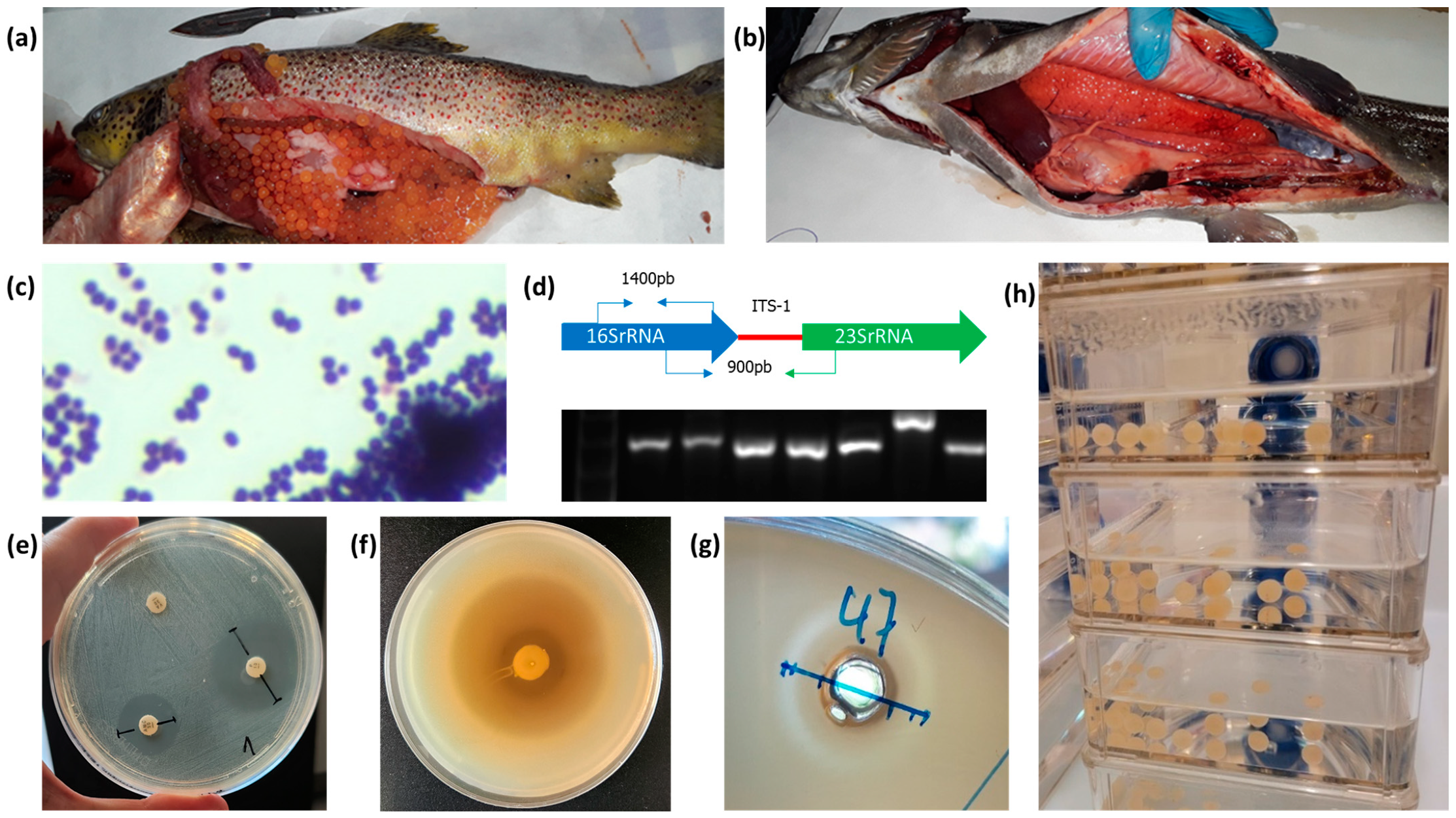
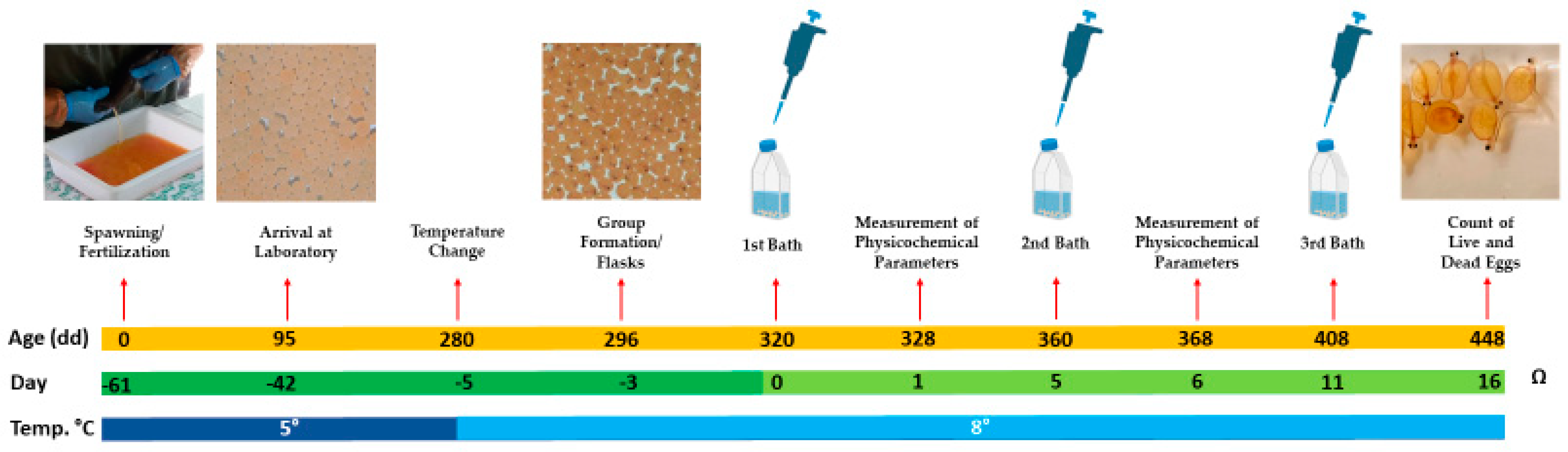


| Bacteria | Step | Temp. °C | Time and Growth Conditions |
|---|---|---|---|
| Isolation of LAB | Inoculation of samples | 20–25 | 24 h to 48 h in MRS broth |
| Isolation of colonies | 24 h to 48 h on MRS agar | ||
| Selection/sub-culture of colonies | 24 h to 48 h in MRS broth | ||
| Isolation of presumptive fish pathogenic bacteria | Inoculation of samples | 20–25 | 16 h to 48 h in TSB, AOE or MRS broth |
| Isolation of colonies | 16 h to 48 h on TSA, AOE agar or MRS agar | ||
| Selection/sub-culture of colonies | 16 h to 48 h in TSB, AOE or MRS broth | ||
| Lactiplantibacillus sp. | AST | 37 | 24 h in TSB, then 48 h on TSA |
| Agar plug diffusion method | 24 h in MRS broth, 24 h on buffered MRS agar | ||
| Agar well diffusion method | 24 h in MRS broth | ||
| Lactococcus lactis | AST | 37 | 48 h in TSB, then 48 h on TSA |
| Agar plug diffusion method | 48 h in MRS broth, 48 h on buffered MRS agar | ||
| Agar well diffusion method | 48 h in MRS broth | ||
| Leuconostoc mesenteroides | AST | 30 | 24 h in TSB, then 48 h on TSA |
| Agar plug diffusion method | 24 h in MRS broth, 24 h on buffered MRS agar | ||
| Agar well diffusion method | 24 h in TSB | ||
| Pediococcus acidilactici | AST | 30 | 24 h in TSB, then 48 h in TSA |
| Agar plug diffusion method | 24 h in MRS broth, 24 h on buffered MRS agar | ||
| Agar well diffusion method | 24 h in MRS broth | ||
| A. salmonicida subsp. salmonicida | Antagonistic assays | 25 | 24 h in TSB and after the mass inoculation, 24 h in TSA |
| Carnobacterium maltaromaticum | Antagonistic assays | 25 | 24 h in TSB and after the mass inoculation, 24 h in TSA |
| Yersinia ruckeri | Antagonistic assays | 25 | 24 h in TSB and after the mass inoculation, 24 h in TSA |
| Vagococcus salmoninarum | Antagonistic assays | 25 | 48 h in TSB and after the mass inoculation, 48 h in TSA |
| Lactococcus garvieae | Antagonistic assays | 37 | 24 h in TSB and after the mass inoculation, 24 h in TSA |
| Vibrio jasicida | Antagonistic assays | 30 | 24 h in marine broth and after the mass inoculation, 24 h in marine agar |
| Pathogen | Primer | Sequence 5′ to 3′ | Amplicon/ Reference |
|---|---|---|---|
| Flavobacterium columnare | 72 Seghou F | 5′-GAAGGAGCTTGTTCCTTT-3′ | 1260 pb [11] |
| 1260 Seghou R | 5′-GCCTACTTGCGTAGTG-3′ | ||
| Aeromonas salmonicida subsp. salmonicida | aopP Balcázar F | 5′-CGGAACGTAATCTGAATTGTTCTTTTC-3′ | 340 pb [3] |
| aopP Balcázar R | 5′-ATTGCTTATCGAGGCAGCCAAC-3′ | ||
| C. maltaromaticum | 16S Mohsina F | 5′-GAGGGTCATTGGAAACTGGA-3′ | 219 pb [5] |
| 16S Mohsina F | 5′-CGGAAACCCTCCAACACTTA-3′ | ||
| Vagococcus salmoninarum | sal Torres F | 5′-GACGCTTTCGGGTGTCACTA-3′ | 543 pb [9] |
| sal Torres R | 5′-CAGACCAGAGAGTCGCCTTC-3′ | ||
| Yersinia ruckeri | glnA Keeling F | 5′-TCCAGCACCAAATACGAAGG-3′ | 113 pb [4] |
| glnA Keeling R | 5′-ACATGGCAGAACGCAGATC-3′ | ||
| glnA Keeling P | HEX-5′-AAGGCGGTTACTTCCCGGTTCC-3′-BHQ1 | ||
| F. psychrophilum | sig Marancik F | 5′-GGTAGCGGAACCGGAAATG-3′ | 77 pb [8] |
| sig Marancik R | 5′-TTTCTGCCACCTAGCGAATACC-3′ | ||
| sig Marancik P | FAM-5′-CGCTTCCTGAGCCAGA-3′-BHQ1 | ||
| Lactococcus garvieae | ITS Chapela F | 5′-ACTTTATTCAGTTTTGAGGGGTCT-3′ | 190 pb [10] |
| ITS Chapela R | 5′-TTTAACGTCTTCGTTGACCAGA-3′ | ||
| ITS Chapela P | HEX-5′-AGAGAAGGGGCCTTAGCTC-3′-MGB | ||
| Renibacterium salmoninarum | RS 1238 Elliot F | 5′-GTGACCAACACCCAGATATCCA-3′ | 69 pb [32] |
| RS 1307 Elliot R | 5′-TCGCCAGACCACCATTTACC-3′ | ||
| RS 1262 Elliot P | FAM-5′-CACCAGATGGAGCAAC-3′-MGB | ||
| Elongation factor 1α salmonids (Housekeeping gene) | elf-1a GIM-2 F | 5′-GCCCCTCCAGGAYGTYTACAA-3′ | 146 pb [34] |
| elf-1a GIM-2 R | 5′-CCACACGGCCCACRGGTAC-3′ | ||
| elf-1a GIM-2 P | FAM-5′-ATCGGYGGTATTGGAAC-3′-MGB |
| Species/Strain | Genbank Accession No. | Genomic Nucleotide Position |
|---|---|---|
| Lactococcus lactis subsp. cremoris KW2 | CP004884.1 | 500179–502418 |
| Leuconostoc mesenteroides subsp. dextranicum strain DSM 20484 | CP012009.1 | 374968–377287 |
| Leuconostoc mesenteroides subsp. jonggajibkimchii strain DRC1506 | CP014611.1 | 22983–25302 |
| Leuconostoc mesenteroides subsp. mesenteroides FM06 | CP020731.1 | 718521–720840 |
| Leuconostoc mesenteroides subsp. suionicum strain LT-38 | AP017935.1 | 270279–272598 |
| Pediococcus acidilactici strain JQII-5 | CP023654.1 | 219495–221665 |
| Lactiplantibacillus plantarum subsp. plantarum strain E1 | CP031771.1 | 2889706–2891876 |
| Lactiplantibacillus pentosus strain ZFM222 | CP032654.1 | 1589825–1591995 |
| Lactococcus lactis subsp. lactis strain UC073 | CP068698.2 | 429176–431415 |
| Bacillus subtilis subsp. subtilis strain BGSC 10A5 | CP101936.1 | 184980–187588 |
| Staphylococcus aureus strain OPD001-1 | CP121234.1 | 635770–638640 |
| Host | Isolation Source | Species/Strain | pb. | Genbank Accession No. |
|---|---|---|---|---|
| O. mykiss | Intestinal mucus | P. acidilactici strain Om-Ci-Gu-1 | 2160 | OR734325 |
| O. mykiss | Intestinal mucus | P. acidilactici strain Om-Ci-Gu-5 | 1580 | OR734326 |
| O. mykiss | Intestinal mucus | Lactococcus lactis strain Om-Ci-Gu-12 | 1354 | OR734327 |
| O. mykiss | Intestinal mucus | Lactococcus lactis strain Om-Ci-Gu-18 | 2241 | OR734328 |
| O. mykiss | Intestinal mucus | Lactiplantibacillus sp. strain Om-Ci-Gu-25 | 2171 | OR734329 |
| O. mykiss | Intestinal mucus | Leuconostoc mesenteroides strain Om-Ci-Gu-28 | 1364 | OR734330 |
| O. mykiss | Intestinal mucus | C. divergens Om-V-Gu-34 | 2203 | OR753895 |
| O. mykiss | Mouth mucus | Lactiplantibacillus sp. strain Om-V-M-37 | 1376 | OR734332 |
| O. mykiss | Mouth mucus | C. maltaromaticum strain Om-Ca-M-31 | 2373 | OR753882 |
| O. mykiss | Mouth mucus | C. maltaromaticum strain Om-Ca-M-36c1 | 2365 | OR753883 |
| O. mykiss | Mouth mucus | C. maltaromaticum strain Om-Ca-M-36c2 | 2355 | OR753884 |
| O. mykiss | Gills mucus | Lactiplantibacillus sp. strain Om-Ci-Gi-13 | 1587 | OR734324 |
| S. trutta | Intestinal mucus | Lactiplantibacillus sp. strain St-RP-Gu-7 | 1560 | OR734338 |
| S. trutta | Intestinal mucus | P. acidilactici strain St-RT-Gu-10g | 2171 | OR734342 |
| S. trutta | Intestinal mucus | P. acidilactici strain St-RT-Gu-10p | 1580 | OR734343 |
| S. trutta | Intestinal mucus | C. maltaromaticum strain St-BC-Gu-33 | 2359 | OR753887 |
| S. trutta | Intestinal mucus | C. maltaromaticum strain St-PS-Gu-41 | 2332 | OR753889 |
| S. trutta | Mouth mucus | C. maltaromaticum strain St-Ba-M-35 | 2366 | OR753885 |
| S. trutta | Mouth mucus | C. maltaromaticum strain St-Ba-M-42 | 2371 | OR753886 |
| S. trutta | Gills mucus | Lactiplantibacillus sp. strain St-RP-Gi-4 | 1561 | OR734335 |
| S. trutta | Gills mucus | Lactiplantibacillus sp. strain St-RP-Gi-14 | 2165 | OR734336 |
| S. trutta | Gills mucus | Lactiplantibacillus sp. strain St-RP-Gi-19 | 2172 | OR734337 |
| S. trutta | Gills mucus | Lactiplantibacillus sp. strain St-RT-Gi-8 | 1571 | OR734339 |
| S. trutta | Gills mucus | Lactiplantibacillus sp. strain St-RT-Gi-15 | 2144 | OR734340 |
| S. trutta | Gills mucus | Lactiplantibacillus sp. strain St-RT-Gi-27 | 1569 | OR734341 |
| S. trutta | Gills mucus | C. maltaromaticum strain St-RP-Gi-2 | 2320 | OR753890 |
| S. trutta | Gills mucus | C. maltaromaticum strain St-RP-Gi-9 | 2323 | OR753891 |
| S. trutta | Gills mucus | C. maltaromaticum strain St-RP-Gi-24 | 2348 | OR753892 |
| S. trutta | Gills mucus | C. maltaromaticum strain St-RT-Gi-26p | 2336 | OR753896 |
| S. trutta | Skin mucus | Aerococcus sp. strain St-BC-SK-82 | 2230 | OR753888 |
| O. mykiss | Liver | Y. ruckeri strain Om-Ca-L-54 | 2497 | OR763346 |
| O. mykiss | Liver | V. salmoninarum strain Om-V-L-69 | 1419 | OR763343 |
| O. mykiss | Head kidney | Lactiplantibacillus sp. strain Om-V-HK-47 | 1600 | OR734331 |
| O. mykiss | Head kidney | Lactococcus garvieae strain Om-Pe-HK-61 | 2320 | OR763345 |
| O. mykiss | Swim bladder | Leuconostoc mesenteroides strain Om-V-SB-48 | 1620 | OR734333 |
| O. mykiss | Swim bladder | Leuconostoc mesenteroides strain Om-V-SB-49 | 2320 | OR734334 |
| D. labrax | Liver | V. jasicida strain Dl-Cu-L-65 | 1637 | OR763344 |
| S. trutta | Head kidney | C. maltaromaticum strain St-PS-HK-63 | 1609 | OR753893 |
| S. trutta | Head kidney | C. maltaromaticum strain St-PS-HK-64 | 1642 | OR753894 |
| Antimicrobial Agent | Lactococcus lactis Om-Ci-Gu-12 | Leuconostoc mesenteroides Om-Ci-Gu-28 | Leuconostoc mesenteroides Om-V-SB-48 | P. acidilactici Om-Ci-GU-1 | P. acidilactici Om-Ci-GU-5 | P. acidilactici St-RT-Gu-10p | Lactiplantibacillus sp. St-RP-Gi-4 | Lactiplantibacillus sp. St-RT-Gi-27 | Lactiplantibacillus sp. Om-V-HK-47 | Lactiplantibacillus sp. St-RP-Gu-7 | Lactiplantibacillus sp. Om-Ci-Gu-25 | Lactiplantibacillus sp. Om-V-M-37 | Lactiplantibacillus sp. St-RP-Gi-14 | Lactiplantibacillus sp. St-RT-Gi-8 | Lactiplantibacillus sp. Om-Ci-Gi-13 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Glycopeptides | Vancomycin | 0 R | 0 R | 0 R | 0 R | 0 R | 0 R | 0 R | 0 R | 0 R | 0 R | 0 R | 0 R | 0 R | 0 R | 0 R |
| Penicillins | Amoxicillin/Clavulanic | 20 | 20 | 18 | 20 | 20 | 21 | 22 | 26 | 23 | 22 | 24 | 36 | 21 | 25 | 29 |
| Penicillin G | 27 S | 23 S | 25 S | 27 S | 25 S | 26 S | 0 R | 0 R | 0 R | 0 R | 0 R | 26 S | 0 R | 0 R | 0 R | |
| Ampicillin | 12 R | 23 S | 15 R | 13 R | 12 R | 13 R | 12 R | 15 R | 16 R | 15 R | 12 R | 33 S | 23 S | 18 S | 17 S | |
| Oxacillin | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Aminoglycosides | Kanamycin | 0 | 11 | 20 | 11 | 11 | 11 | 17 | 16 | 16 | 18 | 16 | 24 | 17 | 19 | 18 |
| Neomycin | 18 | 16 | 25 | 16 | 16 | 17 | 20 | 22 | 19 | 21 | 23 | 27 | 20 | 23 | 22 | |
| Streptomycin | 9 | 8 | 19 | 9 | 10 | 9 | 19 | 23 | 23 | 22 | 25 | 26 | 15 | 24 | 23 | |
| Gentamicin | 18 | 14 | 24 | 15 | 16 | 14 | 21 | 19 | 21 | 20 | 22 | 27 | 18 | 23 | 21 | |
| Dihydrofolate reduc. | Trimethoprim | 0 | 0 | 0 | 0 | 0 | 0 | 23 | 16 | 14 | 22 | 25 | 20 | 0 | 16 | 16 |
| Macrolides | Erythromycin | 33 S | 40 S | 35 S | 44 S | 35 S | 44 S | 37 S | 36 S | 35 S | 43 S | 47 S | 35 S | 43 S | 32 S | 35 S |
| Cephalosporins | Cefuroxime | 17 | 17 | 16 | 18 | 20 | 19 | 33 | 31 | 21 | 22 | 29 | 28 | 26 | 20 | 25 |
| Ceftriaxone | 0 R | 0 | 0 | 0 | 0 | 0 | 38 | 35 | 23 | 26 | 35 | 36 | 33 | 20 | 26 | |
| Tetracyclines | Tetracycline | 20 S | 17I | 28 S | 20 S | 19 S | 21 S | 22 S | 21 S | 19 S | 22 S | 22 S | 23 S | 22 S | 18I | 19 S |
| Doxycycline | 21 S | 20 S | 30 S | 26 S | 25 S | 24 S | 27 S | 27 S | 22 S | 24 S | 27 S | 23 S | 25 S | 19 S | 21 S | |
| Oxytetracycline | 19 | 19 | 30 | 23 | 20 | 22 | 25 | 23 | 21 | 23 | 25 | 22 | 22 | 20 | 21 | |
| Lincosamides | Clindamycin | 33 | 32 | 32 | 35 | 32 | 34 | 26 | 17 | 12 | 14 | 17 | 19 | 12 | 13 | 13 |
| Quinolones | Nalidixic Acid | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Flumequine | 0 | 0 | 0 | 0 | 0 | 0 | 15 | 15 | 14 | 13 | 14 | 15 | 12 | 14 | 13 | |
| Ciprofloxacin | 0 R | 0 R | 19 I | 0 R | 0 R | 0 R | 14 R | 9 R | 9 R | 18 I | 21 S | 15 R | 16 I | 10 R | 12 R | |
| Nitrofurans | Nitrofurantoine | 23 S | 21 S | 0 R | 20 S | 22 S | 22 S | 31 S | 36 S | 18 S | 37 S | 31 S | 25 S | 33 S | 17 S | 19 S |
| Amphenicols | Florfenicol | 30 S | 33 S | 35 S | 32 S | 30 S | 28 S | 32 S | 34 S | 33 S | 36 S | 35 S | 30 S | 30 S | 33 S | 34 S |
| Chloramphenicol | 27 S | 25 S | 30 S | 25 S | 26 S | 30 S | 36 S | 32 S | 29 S | 33 S | 35 S | 28 S | 30 S | 25 S | 27 S | |
| Pathogen | Lactococcus lactis Om-Ci-Gu-12 | Leuconostoc mesenteroides Om-Ci-Gu-28 | Leuconostoc mesenteroides Om-V-SB-48 | P. acidilactici Om-Ci-GU-1 | P. acidilactici Om-Ci-GU-5 | P. acidilactici St-RT-Gu-10p | Lactiplantibacillus sp. St-RP-Gi-4 | Lactiplantibacillus sp. St-RT-Gi-27 | Lactiplantibacillus sp. Om-V-HK-47 | Lactiplantibacillus sp. St-RP-Gu-7 | Lactiplantibacillus sp. Om-Ci-Gu-25 | Lactiplantibacillus sp. Om-V-M-37 | Lactiplantibacillus sp. St-RP-Gi-14 | Lactiplantibacillus sp. St-RT-Gi-8 | Lactiplantibacillus sp. Om-Ci-Gi-13 | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C. maltaromaticum St-PS-HK-63 | 10 | 11 | 8 | 14 | 0 | 11 | 12 | 10 | 11 | 11 | 12 | 10 | 12 | 12 | 12 | |||
| V. salmoninarum Om-V-L-69 | 13 | 20 | 16 | 15 | 13 | 13 | 17 | 17 | 21 | 17 | 19 | 21 | 21 | 22 | 30 | |||
| Lactococcus garvieae Om-Pe-HK-61 | 10 | 11 | 5 | 7 | 6 | 8 | 7 | 10 | 0 | 0 | 10 | 7 | 11 | 8 | 13 | |||
| Y. ruckeri Om-Ca-L-54 | 12 | 0 | 0 | 9 | 14 | 13 | 8 | 11 | 15 | 14 | 11 | 10 | 9 | 11 | 15 | |||
| V. jasicida Dl-Cu-L-65 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||
| A. salmonicida subsp. salmonicida St-Mu-Sk | 32 | 16 | 0 | 12 | 19 | 27 | 29 | 20 | 26 | 12 | 30 | 50 | 32 | 18 | 32 | |||
| None (0 mm) | Slight (1 ≤ 10 mm) | Medium (11 ≤ 20 mm) | High (21–30 mm) | Very high (>30 mm) | ||||||||||||||
| Pathogen | Lactococcus lactis Om-Ci-Gu-12 | Leuconostoc mesenteroides Om-Ci-Gu-28 | Leuconostoc mesenteroides Om-V-SB-48 | P. acidilactici Om-Ci-GU-1 | P. acidilactici Om-Ci-GU-5 | P. acidilactici St-RT-Gu-10p | Lactiplantibacillus sp. St-RP-Gi-4 | Lactiplantibacillus sp. St-RT-Gi-27 | Lactiplantibacillus sp. Om-V-HK-47 | Lactiplantibacillus sp. St-RP-Gu-7 | Lactiplantibacillus sp. Om-Ci-Gu-25 | Lactiplantibacillus sp. Om-V-M-37 | Lactiplantibacillus sp. St-RP-Gi-14 | Lactiplantibacillus sp. St-RT-Gi-8 | Lactiplantibacillus sp. Om-Ci-Gi-13 | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C. maltaromaticum St-PS-HK-63 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 5 | 5 | 5 | 5 | 6 | 4 | 3 | 5 | |||
| V. salmoninarum Om-V-L-69 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 7 | 6 | 7 | 6 | 7 | 5 | 5 | 5 | |||
| Lactococcus garvieae Om-Pe-HK-61 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||
| Y. ruckeri Om-Ca-L-54 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||
| V. jasicida Dl-Cu-L-65 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||
| A. salmonicida subsp. salmonicida St-Mu-Sk | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||
| None (<1 mm) | Slight (1 ≤ 3 mm) | Medium (4 ≤ 6 mm) | High (7 ≤ 9 mm) | Very high (>10 mm) | ||||||||||||||
| Flask/Replicate | Assessment of Fish Eggs | Lactococcus lactis Om-Ci-Gu-12 | Leuconostoc mesenteroides Om-V-SB-48 | P. acidilactici St-RT-Gu-10p | Lactiplantibacillus sp. St-RP-Gi-4 | Lactiplantibacillus sp. Om-V-HK-47 | Lactiplantibacillus sp. Om-Ci-Gi-13 | Control |
|---|---|---|---|---|---|---|---|---|
| 1 | Dead | 5 | 1 | 1 | 6 | 7 | 8 | 1 |
| Living | 0 | 1 | 2 | 0 | 0 | 0 | 0 | |
| Hatched | 5 | 8 | 7 | 4 | 3 | 2 | 9 | |
| 2 | Dead | 5 | 0 | 0 | 7 | 6 | 8 | 2 |
| Living | 1 | 1 | 4 | 0 | 0 | 0 | 0 | |
| Hatched | 4 | 9 | 6 | 3 | 4 | 2 | 8 | |
| 3 | Dead | 4 | 1 | 0 | 4 | 9 | 4 | 0 |
| Living | 2 | 3 | 2 | 0 | 0 | 0 | 0 | |
| Hatched | 4 | 6 | 8 | 6 | 1 | 6 | 10 | |
| 4 | Dead | 1 | 2 | 4 | 9 | 4 | 7 | 5 |
| Living | 0 | 1 | 1 | 0 | 0 | 0 | 1 | |
| Hatched | 9 | 7 | 5 | 1 | 6 | 3 | 4 | |
| 5 | Dead | 2 | 8 | 2 | 9 | 7 | 3 | 1 |
| Living | 1 | 0 | 1 | 0 | 0 | 0 | 0 | |
| Hatched | 7 | 2 | 7 | 1 | 3 | 7 | 9 | |
| 6 | Dead | 1 | 5 | 3 | 7 | 8 | 8 | 2 |
| Living | 1 | 0 | 2 | 0 | 0 | 0 | 0 | |
| Hatched | 8 | 5 | 5 | 3 | 2 | 2 | 8 | |
| 7 | Dead | 5 | 6 | 5 | 9 | 5 | 8 | 1 |
| Living | 0 | 0 | 1 | 0 | 0 | 0 | 1 | |
| Hatched | 5 | 4 | 4 | 1 | 5 | 2 | 8 | |
| 8 | Dead | 0 | 3 | 2 | 6 | 6 | 7 | 4 |
| Living | 0 | 1 | 0 | 0 | 0 | 0 | 1 | |
| Hatched | 10 | 6 | 8 | 4 | 4 | 3 | 5 | |
| 9 | Dead | 2 | 8 | 4 | 4 | 9 | 10 | 3 |
| Living | 0 | 0 | 1 | 0 | 0 | 0 | 0 | |
| Hatched | 8 | 2 | 5 | 6 | 1 | 0 | 7 | |
| 10 | Dead | 3 | 8 | 5 | 7 | 8 | 8 | 2 |
| Living | 2 | 0 | 0 | 0 | 0 | 0 | 2 | |
| Hatched | 5 | 2 | 5 | 3 | 2 | 2 | 6 |
| Day of Assay | Parameter | Lactococcus lactis Om-Ci-Gu-12 | Leuconostoc mesenteroides Om-V-SB-48 | P. acidilactici St-RT-Gu-10p | Lactiplantibacillus sp. St-RP-Gi-4 | Lactiplantibacillus sp. Om-V-HK-47 | Lactiplantibacillus sp. Om-Ci-Gi-13 | Control |
|---|---|---|---|---|---|---|---|---|
| 24 h after 1st bath | O2% | 106.1 | 108.7 | 93.2 | 96.0 | 81.2 | 95.5 | 97.9 |
| O2 (mg/mL) | 10.4 | 10.2 | 8.7 | 8.9 | 7.6 | 9.0 | 9.4 | |
| pH | 7.6 | 7.5 | 7.5 | 7.6 | 7.4 | 7.6 | 7.4 | |
| Conductivity | 2.4 | 2.3 | 2.4 | 2.3 | 2.4 | 2.3 | 1.6 | |
| Turbidity (NTU) | 102 | 209 | 124 | 414 | 598 | 493 | 0.74 | |
| 24 h after 2nd bath | O2% | 84.8 | 87.4 | 86.1 | 73.4 | 83.0 | 72.2 | 93.8 |
| O2 mg/mL | 9.1 | 9.6 | 8.2 | 7.0 | 9.4 | 6.82 | 9.0 | |
| pH | 7.6 | 7.6 | 7.8 | 7.8 | 7.8 | 7.55 | 7.9 | |
| Conductivity | 2.5 | 2.6 | 2.6 | 2.7 | 2.6 | 2.78 | 1.7 | |
| Turbidity (NTU) | 209 | 289 | 272 | 586 | 468 | 571 | 0.77 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vargas-González, A.; Barajas, M.; Pérez-Sánchez, T. Isolation of Lactic Acid Bacteria (LAB) from Salmonids for Potential Use as Probiotics: In Vitro Assays and Toxicity Assessment of Salmo trutta Embryonated Eggs. Animals 2024, 14, 200. https://doi.org/10.3390/ani14020200
Vargas-González A, Barajas M, Pérez-Sánchez T. Isolation of Lactic Acid Bacteria (LAB) from Salmonids for Potential Use as Probiotics: In Vitro Assays and Toxicity Assessment of Salmo trutta Embryonated Eggs. Animals. 2024; 14(2):200. https://doi.org/10.3390/ani14020200
Chicago/Turabian StyleVargas-González, Augusto, Miguel Barajas, and Tania Pérez-Sánchez. 2024. "Isolation of Lactic Acid Bacteria (LAB) from Salmonids for Potential Use as Probiotics: In Vitro Assays and Toxicity Assessment of Salmo trutta Embryonated Eggs" Animals 14, no. 2: 200. https://doi.org/10.3390/ani14020200
APA StyleVargas-González, A., Barajas, M., & Pérez-Sánchez, T. (2024). Isolation of Lactic Acid Bacteria (LAB) from Salmonids for Potential Use as Probiotics: In Vitro Assays and Toxicity Assessment of Salmo trutta Embryonated Eggs. Animals, 14(2), 200. https://doi.org/10.3390/ani14020200






