Influence of Sires on Population Substructure in Dülmen Wild Horses
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Approval
2.2. Sample Collection
2.3. DNA Extraction and Microsatellite Analysis
2.4. Statistical Analysis
2.4.1. Genetic Diversity and Population Differentiation
2.4.2. Assessment of Population Structure and Admixture
3. Results
3.1. Characteristics and Genetic Diversity of the Microsatellite Markers
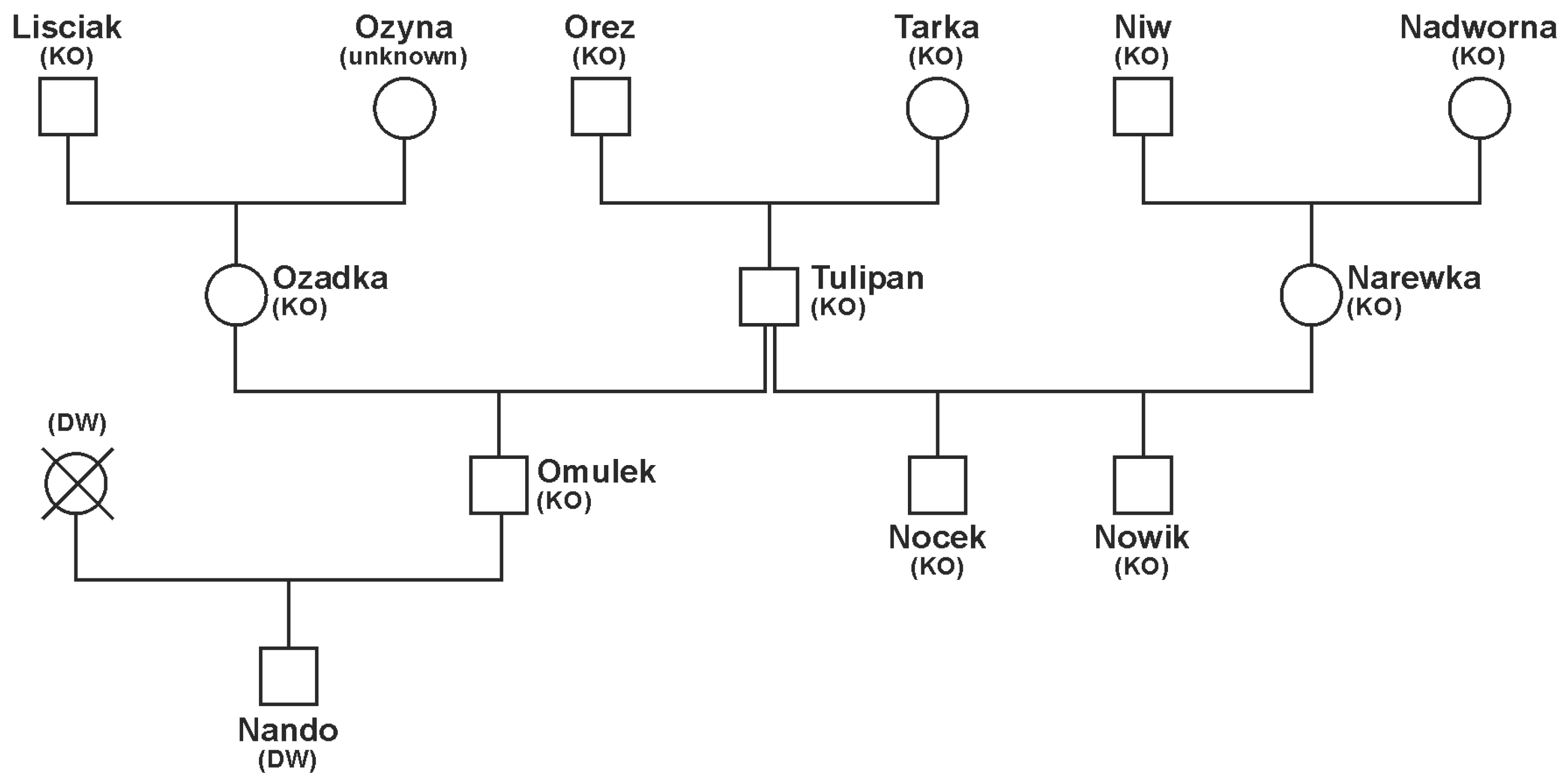
3.2. Pedigrees of Stallions and Distribution of Progeny by Stallions
3.3. Genetic Diversity of Paternal Half-Sib Groups
3.4. Genetic Diversity of Birth Cohorts
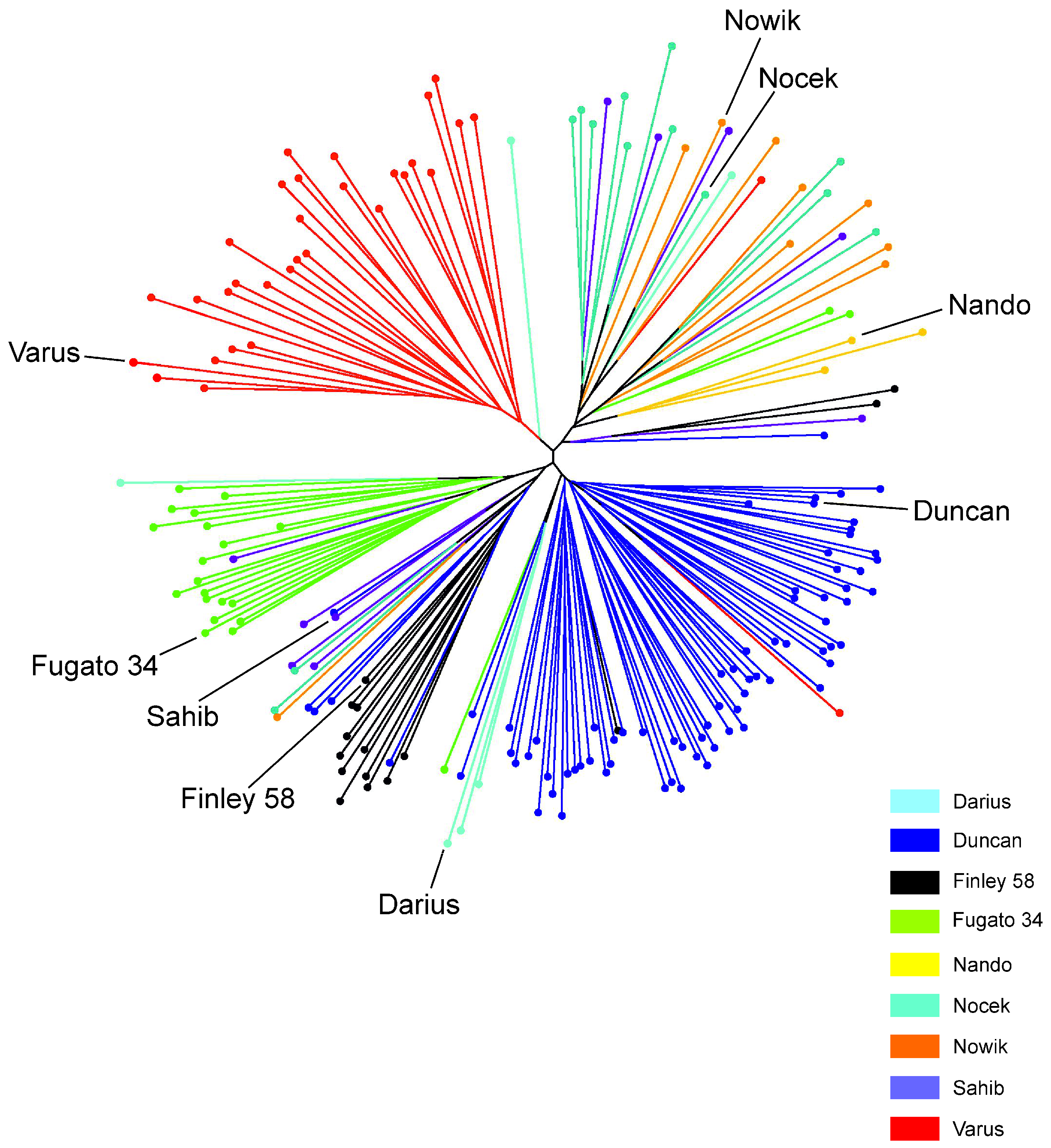
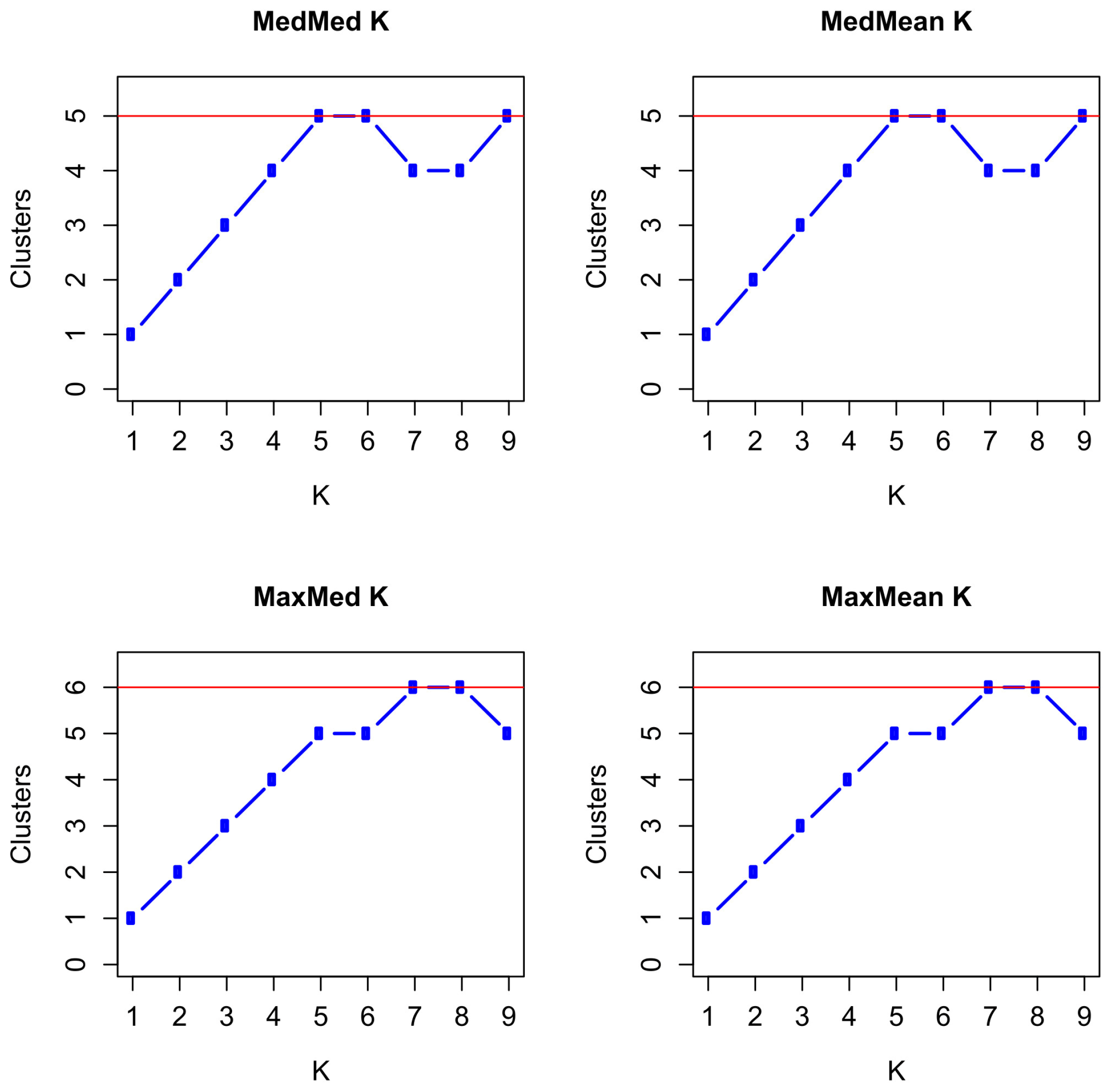
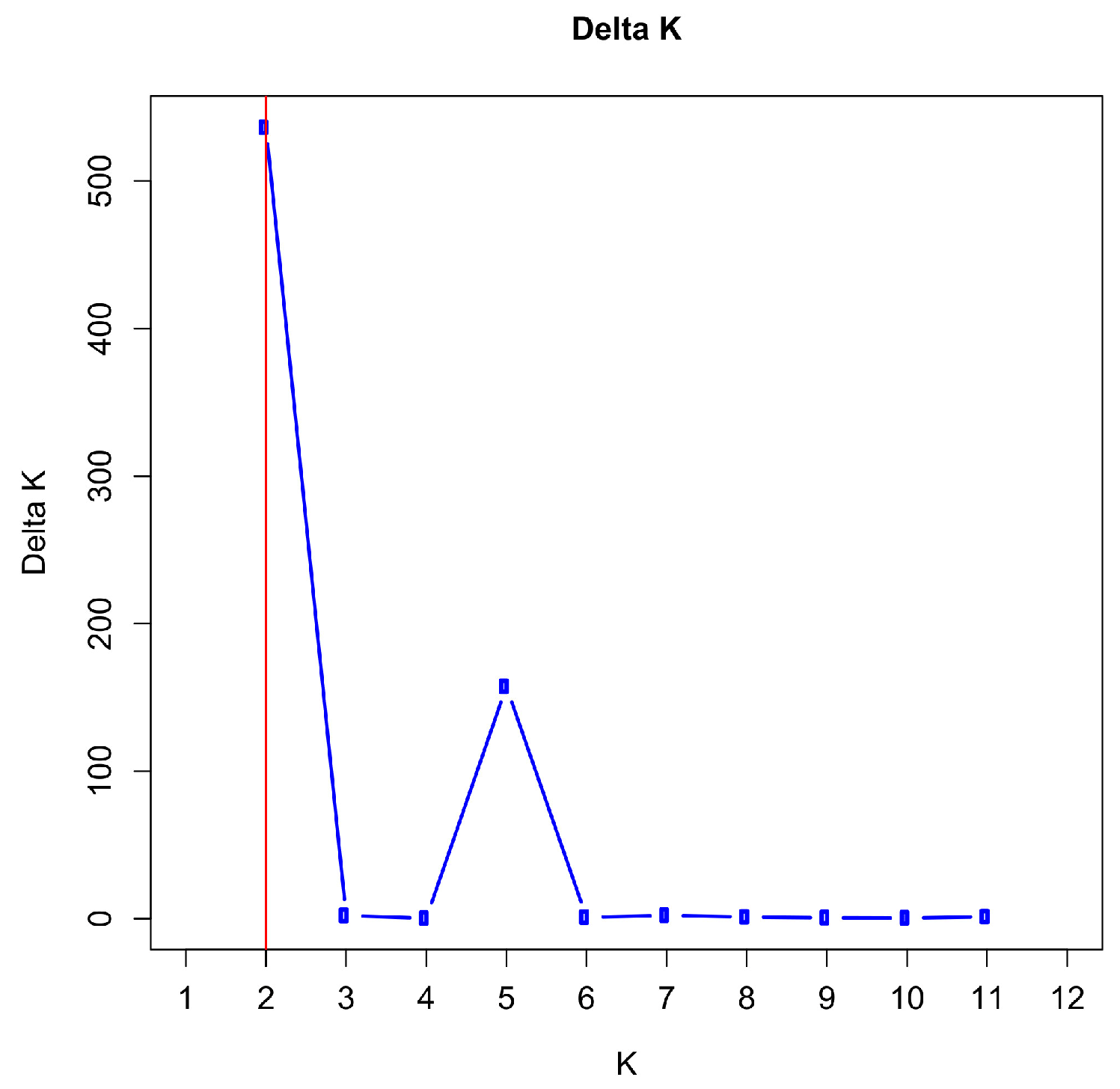
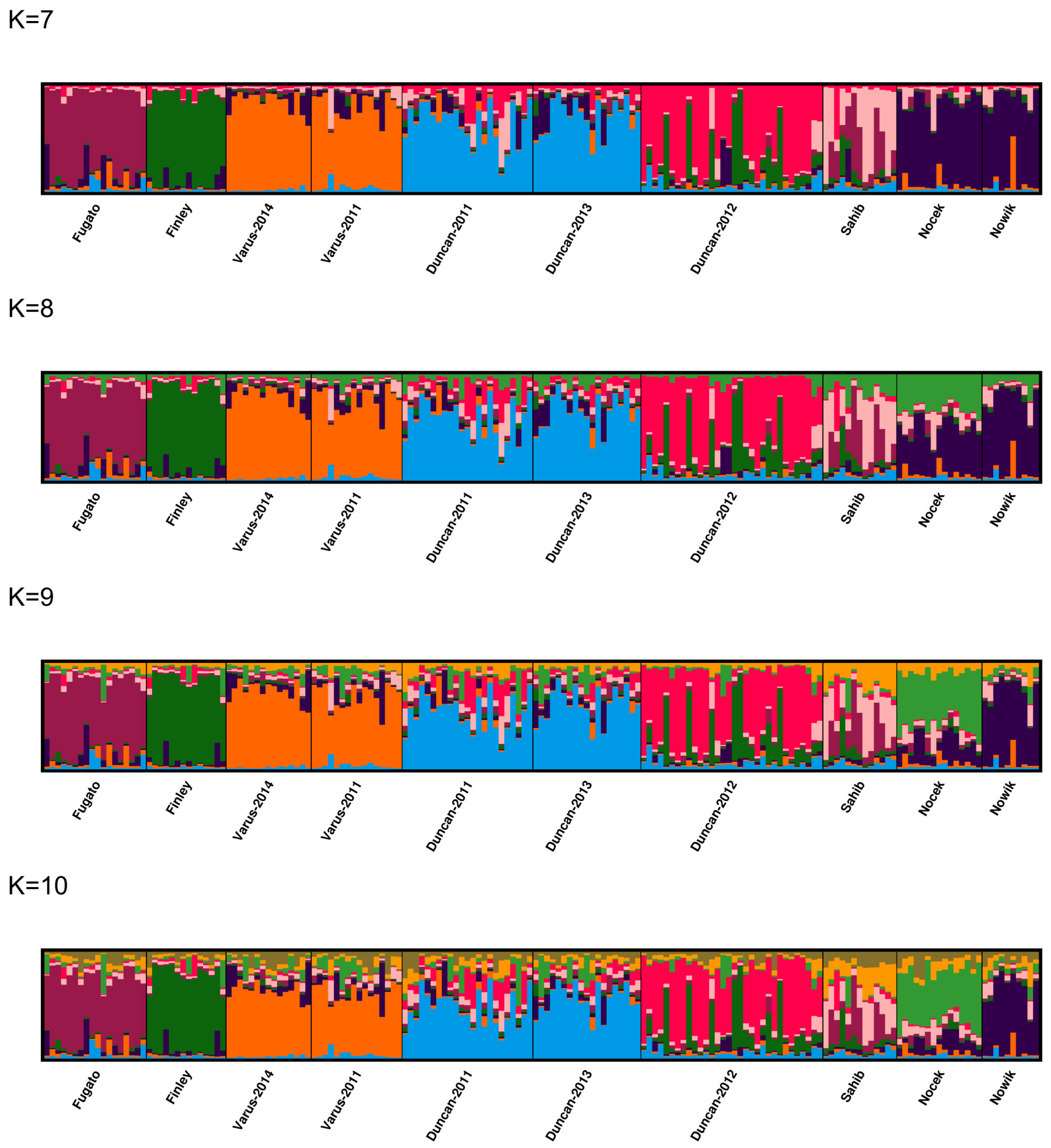
3.5. Population Structure and Admixture
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Opora, J. Die Wildbahngestüte Westfalens: Geschichte, Entwicklung und Zukunft. Ph.D. Thesis, Tierärztliche Hochschule Hannover, Hannover, Germany, 2006. [Google Scholar]
- Aberle, K.S.; Hamann, H.; Drögemuller, C.; Distl, O. Genetic diversity in German draught horse breeds compared with a group of primitive, riding and wild horses by means of microsatellite DNA markers. Anim. Genet. 2004, 35, 270–277. [Google Scholar] [CrossRef] [PubMed]
- Druml, T.; Curik, I.; Baumung, R.; Aberle, K.; Distl, O.; Sölkner, J. Individual-based assessment of population structure and admixture in Austrian, Croatian and German draught horses. Heredity 2007, 98, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Duderstadt, S.; Distl, O. Genetic Diversity and Population Structure of Dülmen Wild, Liebenthal and Polish Konik Horses in Comparison with Przewalski, Sorraia, German Draught and Riding Horses. Animals 2024, 14, 2221. [Google Scholar] [CrossRef] [PubMed]
- Fornal, A.; Kowalska, K.; Zabek, T.; Piestrzynska-Kajtoch, A.; Musiał, A.D.; Ropka-Molik, K. Genetic Diversity and Population Structure of Polish Konik Horse Based on Individuals from All the Male Founder Lines and Microsatellite Markers. Animals 2020, 10, 1569. [Google Scholar] [CrossRef]
- Fornal, A.; Kowalska, K.; Zabek, T.; Piestrzynska-Kajtoch, A.; Musiał, A.D.; Ropka-Molik, K. Genetic Variability and Population Structure of Polish Konik Horse Maternal Lines Based on Microsatellite Markers. Genes 2021, 12, 546. [Google Scholar] [CrossRef]
- Castaneda, C.; Juras, R.; Khanshour, A.; Randlaht, I.; Wallner, B.; Rigler, D.; Lindgren, G.; Raudsepp, T.; Cothran, E.G. Population Genetic Analysis of the Estonian Native Horse Suggests Diverse and Distinct Genetics, Ancient Origin and Contribution from Unique Patrilines. Genes 2019, 10, 629. [Google Scholar] [CrossRef]
- Mackowski, M.; Mucha, S.; Cholewinski, G.; Cieslak, J. Genetic diversity in Hucul and Polish primitive horse breeds. Arch. Anim. Breed. 2015, 58, 23–31. [Google Scholar] [CrossRef]
- Gralak, B.; Niemczewski, C.; Jaworski, Z. Genetic polymorphism of 12 micorsatellite markers in Polish Primitive Horse. Anim. Sci. Pap. Rep. 2001, 19, 277–283. [Google Scholar]
- Canon, J.; Checa, M.; Carleos, C.; Vega-Pla, J.; Vallejo, M.; Dunner, S. The genetic structure of Spanish Celtic horse breeds inferred from microsatellite data. Anim. Genet. 2000, 31, 39–48. [Google Scholar] [CrossRef]
- Plante, Y.; Vega-Pla, J.L.; Lucas, Z.; Colling, D.; De March, B.; Buchanan, F. Genetic diversity in a feral horse population from Sable Island, Canada. J. Hered. 2007, 98, 594–602. [Google Scholar] [CrossRef]
- Prystupa, J.M.; Juras, R.; Cothran, E.G.; Buchanan, F.C.; Plante, Y. Genetic diversity and admixture among Canadian Mountain and Moorland and Nordic pony populations. Animal 2012, 6, 19–30. [Google Scholar] [CrossRef] [PubMed]
- Solis, A.; Jugo, B.M.; Meriaux, J.C.; Iriondo, M.; Mazon, L.I.; Aguirre, A.I.; Vicario, A.; Estomba, A. Genetic diversity within and among four South European native horse breeds based on microsatellite DNA analysis: Implications for conservation. J. Hered. 2005, 96, 670–678. [Google Scholar] [CrossRef] [PubMed]
- Tozaki, T.; Takezaki, N.; Hasegawa, T.; Ishida, N.; Kurosawa, M.; Tomita, M.; Saitou, N.; Mukoyama, H. Microsatellite variation in Japanese and Asian horses and their phylogenetic relationship using a European horse outgroup. J. Hered. 2003, 94, 374–380. [Google Scholar] [CrossRef] [PubMed]
- Vega-Pla, J.L.; Calderón, J.; Rodríguez-Gallardo, P.; Martinez, A.; Rico, C. Saving feral horse populations: Does it really matter? A case study of wild horses from Doñana National Park in southern Spain. Anim. Genet. 2006, 37, 571–578. [Google Scholar] [CrossRef]
- Vostry, L.; Kracikova, O.; Hofmanova, B.; Czernekova, V.; Kott, T.; Pribyl, J. Intra-line and inter-line genetic diversity in sire lines of the Old Kladruber horse based on microsatellite analysis of DNA. Czech J. Anim. Sci. 2011, 56, 163–175. [Google Scholar] [CrossRef]
- Ząbek, T.; Nogai, A.; Radko, A.; Nogai, J.; Słota, E. Genetic variation of Polish endangered Biłgoraj horses and two common horse breeds in microsatellite loci. J. Appl. Genet. 2005, 46, 299–305. [Google Scholar]
- Dorji, J.; Tamang, S.; Tshewang, T.; Dorji, T.; Dorji, T.Y. Genetic diversity and population structure of three traditional horse breeds of Bhutan based on 29 DNA microsatellite markers. PLoS ONE 2018, 13, e0199376. [Google Scholar] [CrossRef] [PubMed]
- Jaworska, J.; Ropka-Molik, K.; Wocławek-Potocka, I.; Siemieniuch, M. Inter- and intrabreed diversity of the major histocompatibility complex (MHC) in primitive and draft horse breeds. PLoS ONE 2020, 15, e0228658. [Google Scholar] [CrossRef]
- Van Oosterhout, C.; Hutchinson, W.F.; Wills, D.P.; Shipley, P. MICRO-CHECKER: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Not. 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Kalinowski, S.T.; Taper, M.L.; Marshall, T.C. Revising how the computer program Cervus accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 2007, 16, 1099–1106. [Google Scholar] [CrossRef]
- Weir, B.S.; Cockerham, C.C. Estimating F-statistics for the analysis of population structure. Evolution 1984, 38, 1358–1370. [Google Scholar] [CrossRef] [PubMed]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Nei, M. Genetic distance between populations. Am. Nat. 1972, 106, 283–292. [Google Scholar] [CrossRef]
- Perrier, X.; Jacquemoud-Collet, J.P. Dissimilarity Analysis and Representation for Windows; CIRAD-BIOS UMR AGAP: Montpellier, France, 2014; DARwin 6.0.021. [Google Scholar]
- Do, C.; Waples, R.S.; Peel, D.; Macbeth, G.M.; Tillett, B.J.; Ovenden, J.R. NeEstimator v2: Re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol. Ecol. Resour. 2014, 14, 209–214. [Google Scholar] [CrossRef]
- Waples, R.S.; Do, C. Linkage disequilibrium estimates of contemporary Ne using highly variable genetic markers: A largely untapped resource for applied conservation and evolution. Ecol. Appl. 2010, 3, 244–262. [Google Scholar] [CrossRef]
- Simon, D.L.; Buchenauer, D. Genetic Diversity of European Livestock Breeds; Wageningen Pers: Wageningen, The Netherlands, 1993. [Google Scholar]
- FAO. Domestic Animal Diversity Information System (DAD-IS); FAO: Rome, Italy, 2024. [Google Scholar]
- Piry, S.; Luikart, G.; Cornuet, J.M. Computer note. BOTTLENECK: A computer program for detecting recent reductions in the effective size using allele frequency data. J. Hered. 1999, 90, 502–503. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar] [CrossRef]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Dominant markers and null alleles. Mol. Ecol. Notes 2007, 7, 574–578. [Google Scholar] [CrossRef]
- Hubisz, M.J.; Falush, D.; Stephens, M.; Pritchard, J.K. Inferring weak population structure with the assistance of sample group information. Mol. Ecol. Resour. 2009, 9, 1322–1332. [Google Scholar] [CrossRef]
- Waples, R.S.; Gaggiotti, O. What is a population? An empirical evaluation of some genetic methods for identifying the number of gene pools and their degree of connectivity. Mol. Ecol. 2006, 15, 1419–1439. [Google Scholar] [CrossRef] [PubMed]
- Janes, J.K.; Miller, J.M.; Dupuis, J.R.; Malenfant, R.M.; Gorrell, J.C.; Cullingham, C.I.; Andrew, R.L. The K = 2 conundrum. Mol. Ecol. 2017, 26, 3594–3602. [Google Scholar] [CrossRef] [PubMed]
- Cullingham, C.I.; Miller, J.M.; Peery, R.M.; Dupuis, J.R.; Malenfant, R.M.; Gorrell, J.C.; Janes, J.K. Confidently identifying the correct K value using the ΔK method: When does K = 2? Mol. Ecol. 2020, 29, 862–869. [Google Scholar] [CrossRef] [PubMed]
- Puechmaille, S.J. The program structure does not reliably recover the correct population structure when sampling is uneven: Subsampling and new estimators alleviate the problem. Mol. Ecol. Resour. 2016, 16, 608–627. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Li, Y.-L.; Liu, J.-X. StructureSelector: A web-based software to select and visualize the optimal number of clusters using multiple methods. Mol. Ecol. Resour. 2018, 18, 176–177. [Google Scholar] [CrossRef]
- Kopelman, N.M.; Mayzel, J.; Jakobsson, M.; Rosenberg, N.A.; Mayrose, I. CLUMPAK: A program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 2015, 15, 1179–1191. [Google Scholar] [CrossRef]
- Zeeb, K. Das Verhalten des Pferdes bei der Auseinandersetzung mit dem Menschen. Ph.D. Thesis, Ludwig-Maximilians-Universität München, Tierärztliche Fakultät, München, Germany, 1958. [Google Scholar]
- Zeeb, K. Wildpferde in Dülmen. Wildpferde in Dülmen. Pferde, wie sie wirklich leben, beobachtet in freier Wildbahn; Hallwag: Bern, Switzerland, 1965. [Google Scholar]
- Kaminski, S.; Hering, D.M.; Jaworski, Z.; Zabolewicz, T.; Rusc, A.; Kaminski, S.; Hering, D.M.; Jaworski, Z.; Zabolewicz, T.; Rusc, A. Assessment of genomic inbreeding in Polish Konik horses. Pol. J. Vet. Sci. 2017, 20, 603–605. [Google Scholar] [CrossRef] [PubMed]
- Cieslak, J.; Wodas, L.; Borowska, A.; Cothran, E.G.; Khanshour, A.M.; Mackowski, M. Characterization of the Polish Primitive Horse (Konik) maternal lines using mitochondrial D-loop sequence variation. PeerJ 2017, 5, e3714. [Google Scholar] [CrossRef]
- Musiał, A.D.; Radović, L.; Stefaniuk-Szmukier, M.; Bieniek, A.; Wallner, B.; Ropka-Molik, K. Mitochondrial DNA and Y chromosome reveal the genetic structure of the native Polish Konik horse population. PeerJ 2024, 12, e17549. [Google Scholar] [CrossRef]
- Howard, J.T.; Pryce, J.E.; Baes, C.; Maltecca, C. Invited review: Inbreeding in the genomics era: Inbreeding, inbreeding depression, and management of genomic variability. J. Dairy Sci. 2017, 100, 6009–6024. [Google Scholar] [CrossRef] [PubMed]




| Year of Siring | Birth Year | Year of Auction | Stallions | Mares | Foals Born Alive | Male Foals at Auctions |
|---|---|---|---|---|---|---|
| 2001 | 2002 | 2003 | 3 | 238 | 107 | 38 (36%) |
| 2010 | 2011 | 2012 | 3 | ~350 | 122 | 41 (34%) |
| 2011 | 2012 | 2013 | 2 | ~350 | 133 | 46 (35%) |
| 2012 | 2013 | 2014 | 3 | 406 | 65 | 27 (42%) |
| 2013 | 2014 | 2015 | 2 | 370 | 92 | 33 (36%) |
| Total | 9 | 519 | 185 (36%) |
| Stallion | 2002 | 2011 | 2012 | 2013 | 2014 | Total |
|---|---|---|---|---|---|---|
| Nocek | 15 | - | - | - | - | 15 |
| Nowik | 10 | - | - | - | - | 10 |
| Sahib | 13 | - | - | - | - | 13 |
| Varus | - | 16 | - | - | 15 | 31 |
| Duncan | - | 23 | 32 | 19 | - | 74 |
| Nando | - | 2 | - | - | - | 2 |
| Finley 58 | - | - | 14 | - | - | 14 |
| Darius | - | - | - | 5 | - | 5 |
| Fugato 34 | - | - | - | 3 | 18 | 21 |
| Total | 38 | 41 | 46 | 27 | 33 | 185 |
| Population | N | Ho | He | uHe | MNA | Ne | I | FIS |
|---|---|---|---|---|---|---|---|---|
| Sahib | 13 | 0.692 | 0.608 | 0.635 | 4.655 | 2.908 | 1.171 | −0.141 |
| Darius | 5 | 0.731 | 0.617 | 0.686 | 3.793 | 2.868 | 1.128 | −0.186 |
| Duncan | 74 | 0.682 | 0.613 | 0.617 | 5.897 | 2.939 | 1.218 | −0.118 |
| Finley 58 | 14 | 0.678 | 0.593 | 0.616 | 4.103 | 2.797 | 1.098 | −0.146 |
| Fugato 34 | 21 | 0.666 | 0.601 | 0.616 | 5.069 | 2.766 | 1.157 | −0.111 |
| Varus | 31 | 0.684 | 0.609 | 0.619 | 5.276 | 2.883 | 1.183 | −0.122 |
| Nocek | 15 | 0.704 | 0.602 | 0.627 | 4.621 | 2.904 | 1.163 | −0.162 |
| Nowik | 10 | 0.681 | 0.584 | 0.618 | 4.310 | 2.793 | 1.115 | −0.169 |
| Nando | 2 | 0.672 | 0.474 | 0.644 | 2.310 | 2.083 | 0.736 | −0.412 |
| Total (Mean) | 185 | 0.688 | 0.589 | 0.631 | 4.448 | 2.771 | 1.108 | −0.173 |
| Total (SE) | - | 0.014 | 0.010 | 0.011 | 0.102 | 0.057 | 0.023 | 0.016 |
| Birth Cohort | N | Ho | He | uHe | MNA | Ne | I | FIS | Neff | Neff-CI |
|---|---|---|---|---|---|---|---|---|---|---|
| 2002 | 38 | 0.694 | 0.650 | 0.660 | 5.690 | 3.448 | 1.321 | −0.074 | 67.2 | 53.7–88.0 |
| 2011 | 41 | 0.699 | 0.638 | 0.646 | 5.448 | 3.122 | 1.261 | −0.087 | 65.5 | 54.1–81.8 |
| 2012 | 46 | 0.681 | 0.620 | 0.627 | 5.276 | 2.998 | 1.203 | −0.102 | 95.7 | 74.8–129.9 |
| 2013 | 27 | 0.679 | 0.621 | 0.633 | 5.138 | 2.932 | 1.217 | −0.095 | 60.9 | 45.2–90.1 |
| 2014 | 33 | 0.660 | 0.644 | 0.654 | 5.207 | 3.074 | 1.249 | −0.024 | 44.8 | 37.1–55.6 |
| Total (Mean) | 185 | 0.683 | 0.635 | 0.644 | 5.352 | 3.115 | 1.250 | −0.076 | 92.6 | 86.7–99.1 |
| Total (SE) | - | 0.014 | 0.012 | 0.012 | 0.120 | 0.087 | 0.028 | 0.011 | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duderstadt, S.; Distl, O. Influence of Sires on Population Substructure in Dülmen Wild Horses. Animals 2024, 14, 2904. https://doi.org/10.3390/ani14192904
Duderstadt S, Distl O. Influence of Sires on Population Substructure in Dülmen Wild Horses. Animals. 2024; 14(19):2904. https://doi.org/10.3390/ani14192904
Chicago/Turabian StyleDuderstadt, Silke, and Ottmar Distl. 2024. "Influence of Sires on Population Substructure in Dülmen Wild Horses" Animals 14, no. 19: 2904. https://doi.org/10.3390/ani14192904
APA StyleDuderstadt, S., & Distl, O. (2024). Influence of Sires on Population Substructure in Dülmen Wild Horses. Animals, 14(19), 2904. https://doi.org/10.3390/ani14192904





