Additive and Dominance Genome-Wide Association Studies Reveal the Genetic Basis of Heterosis Related to Growth Traits of Duhua Hybrid Pigs
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Phenotype Data
2.3. Processing of Genotype Data
2.4. Calculation of Mid-Parent Heterosis
2.5. Estimation of Genetic Components
2.6. Estimation of Partial Genetic Values
2.7. Additive and Dominance Genome-Wide Association Studies
2.8. Identification and Functional Analysis of Candidate Genes
3. Results
3.1. Mid-Parent Heterosis in Two Traits, 100 AGE and 100 BF, of Duhua Pigs
3.2. Estimation of Genetic Components and Heritability
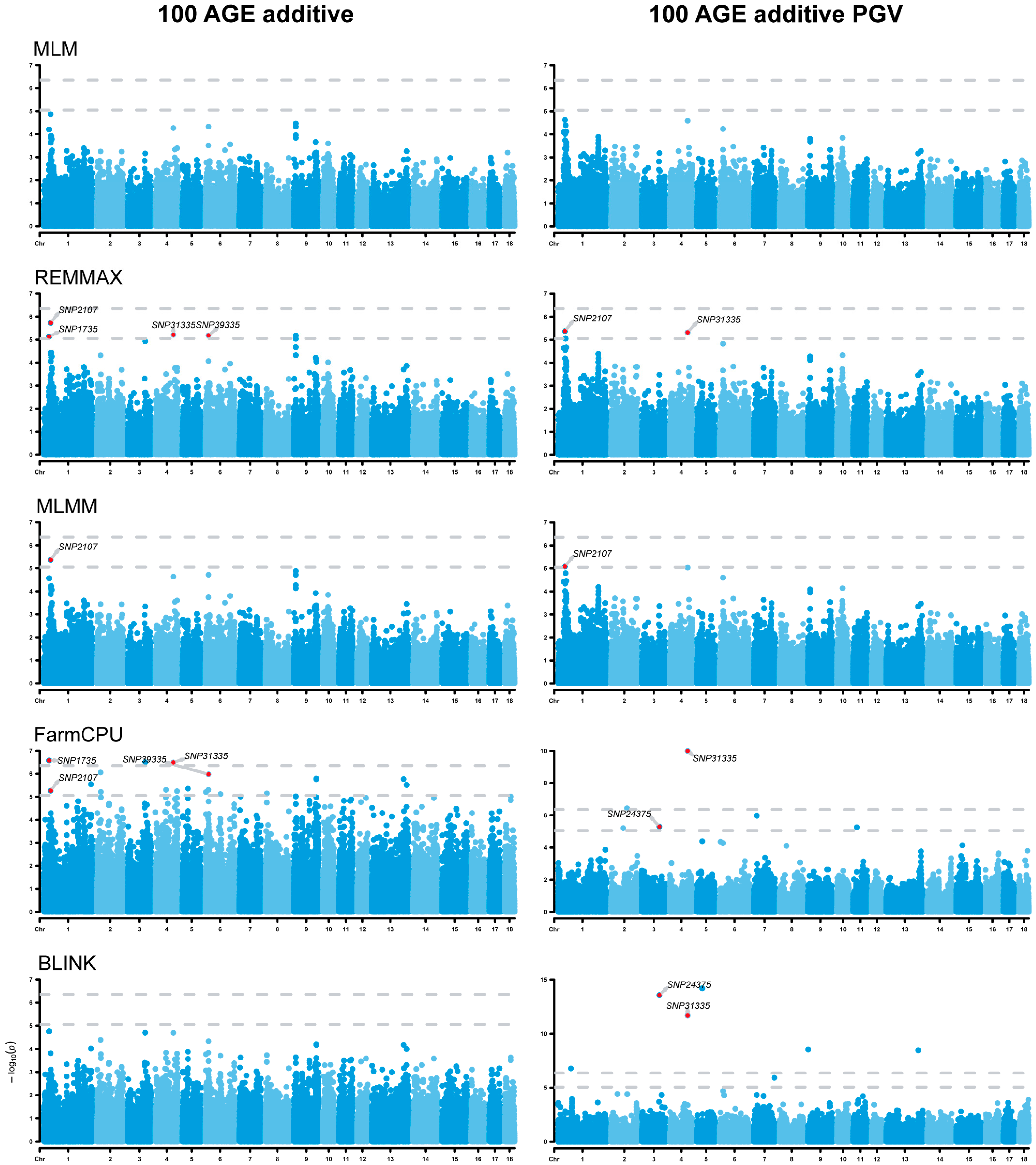
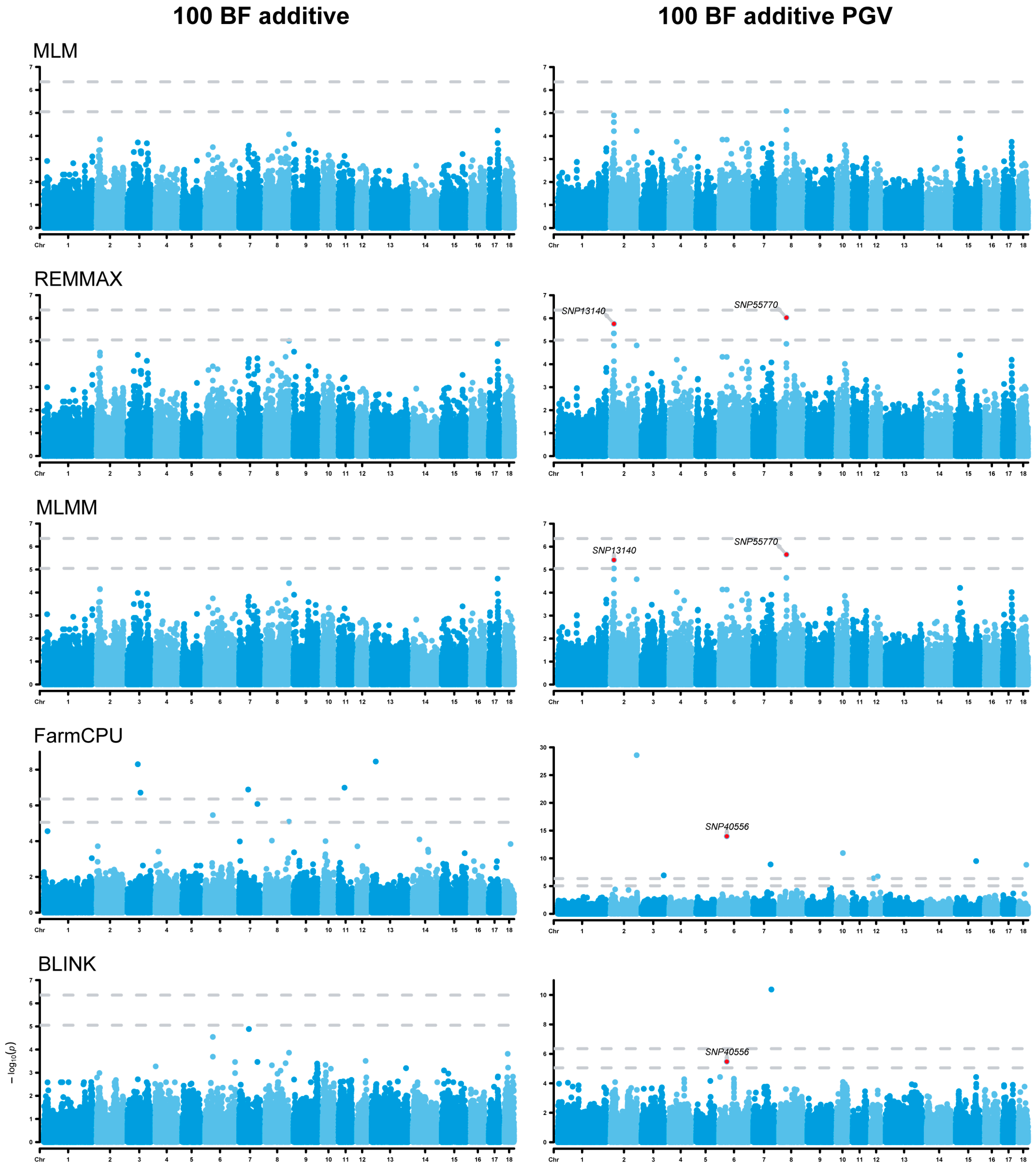
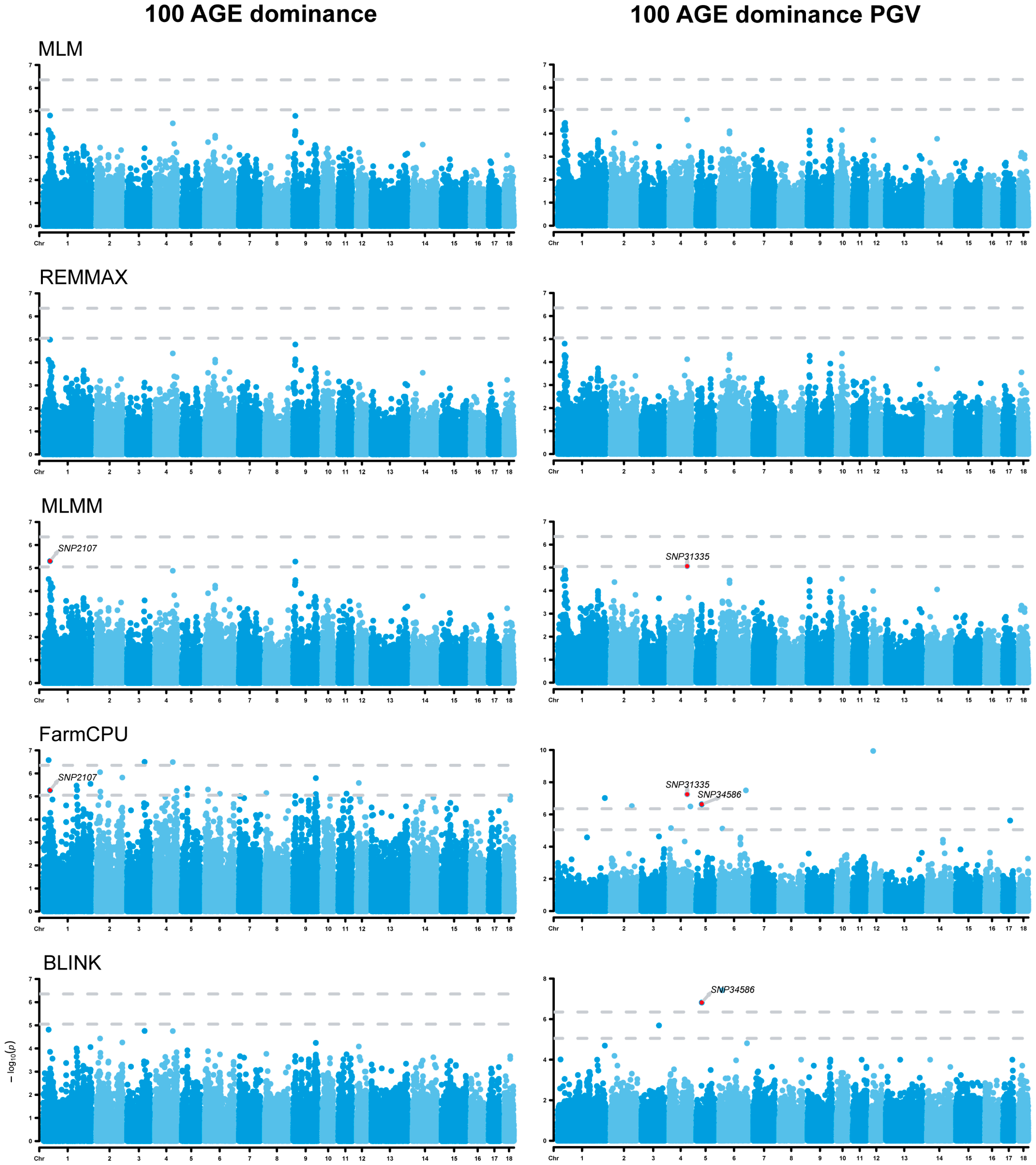
3.3. Genome-Wide Association Studies
3.4. E-GWAS on Additive and Dominance Simulations
3.5. Analysis of Additive Effects
3.6. Analysis of Dominance Effects
3.7. Functional Enrichment of Candidate Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shull, G.H. The Composition of a Field of Maize. J. Hered. 1908, os-4, 296–301. [Google Scholar] [CrossRef]
- Shull, G.H. What Is Heterosis. Genetics 1948, 33, 439–446. [Google Scholar] [CrossRef]
- Mohammadpanah, M.; Mehrgardi, A.A.; Gilbert, H.; Larzul, C.; Mercat, M.J.; Esmailizadeh, A.; Momen, M.; Tusell, L. Genic and non-genic SNP contributions to additive and dominance genetic effects in purebred and crossbred pig traits. Sci. Rep. 2022, 12, 3795. [Google Scholar] [CrossRef] [PubMed]
- Esfandyari, H.; Thekkoot, D.; Kemp, R.; Plastow, G.; Dekkers, J. Genetic parameters and purebred-crossbred genetic correlations for growth, meat quality, and carcass traits in pigs. J. Anim. Sci. 2020, 98, skaa379. [Google Scholar] [CrossRef] [PubMed]
- Steyn, Y.; Lourenco, D.A.; Chen, C.Y.; Valente, B.D.; Holl, J.; Herring, W.O.; Misztal, I. Optimal definition of contemporary groups for crossbred pigs in a joint purebred and crossbred genetic evaluation. J. Anim. Sci. 2021, 99, skaa396. [Google Scholar] [CrossRef]
- Fontanesi, L.; Schiavo, G.; Galimberti, G.; Calò, D.G.; Russo, V. A genomewide association study for average daily gain in Italian Large White pigs. J. Anim. Sci. 2014, 92, 1385–1394. [Google Scholar] [CrossRef]
- Ding, R.R.; Yang, M.; Wang, X.W.; Quan, J.P.; Zhuang, Z.W.; Zhou, S.P.; Li, S.Y.; Xu, Z.; Zheng, E.Q.; Cai, G.Y.; et al. Genetic Architecture of Feeding Behavior and Feed Efficiency in a Duroc Pig Population. Front. Genet. 2018, 9, 220. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.S.; Xu, J.Y.; Yin, L.L.; Yin, D.; Zhu, M.; Yu, M.; Li, X.Y.; Zhao, S.H.; Liu, X.L. Genome-Wide Association Study Reveals Candidate Genes for Growth Relevant Traits in Pigs. Front. Genet. 2019, 10, 302. [Google Scholar] [CrossRef]
- Cassady, J.P.; Young, L.D.; Leymaster, K.A. Heterosis and recombination effects on pig growth and carcass traits. J. Anim. Sci. 2002, 80, 2286–2302. [Google Scholar] [CrossRef]
- Visscher, P.; Pong-Wong, R.; Whittemore, C.; Haley, C. Impact of biotechnology on (cross)breeding programmes in pigs. Livest. Prod. Sci. 2000, 65, 57–70. [Google Scholar] [CrossRef]
- Fu, L.; Jiang, Y.; Wang, C.L.; Mei, M.R.; Zhou, Z.W.; Jiang, Y.F.; Song, H.L.; Ding, X.D. A Genome-Wide Association Study on Feed Efficiency Related Traits in Landrace Pigs. Front. Genet. 2020, 11, 692. [Google Scholar] [CrossRef]
- Wu, P.X.; Wang, K.; Yang, Q.; Zhou, J.; Liu, D.J.; Liu, Y.H.; Ma, J.D.; Tang, Q.; Jin, L.; Xiao, W.H.; et al. Whole-genome re-sequencing association study for direct genetic effects and social genetic effects of six growth traits in Large White pigs. Sci. Rep. 2019, 9, 9667. [Google Scholar] [CrossRef]
- Jiang, Y.; Tang, S.; Wang, C.; Wang, Y.; Qin, Y.; Wang, Y.; Zhang, J.; Song, H.; Mi, S.; Yu, F.; et al. A genome-wide association study of growth and fatness traits in two pig populations with different genetic backgrounds. J. Anim. Sci. 2018, 96, 806–816. [Google Scholar] [CrossRef]
- Cantor, R.M.; Lange, K.; Sinsheimer, J.S. Prioritizing GWAS Results. A Review of Statistical Methods and Recommendations for Their Application. Am. J. Hum. Genet. 2010, 86, 6–22. [Google Scholar] [CrossRef] [PubMed]
- Korte, A.; Farlow, A. The advantages and limitations of trait analysis with GWAS: A review. Plant Methods 2013, 9, 29. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.H.; Liu, S.; Li, W.N.; Mao, R.H.; Zhuo, Y.; Xing, W.K.; Liu, J.; Wang, C.; Zhou, L.; Lei, M.G.; et al. Genome-Wide Association Study Reveals Additive and Non-Additive Effects on Growth Traits in Duroc Pigs. Genes 2022, 13, 1454. [Google Scholar] [CrossRef]
- Li, X.; Zhang, Y.B.; Tang, Z.L.; Yu, M.; Liu, B.; Fan, B.; Xu, S.P.; Peng, Z.Z.; Zhao, S.H.; Zhu, M.J. Comparative Estimation on Three-Way Heterosis in Pigs Reveals Genetic Bias of the Widely Used Empirical Formula. J. Anim. Vet. Adv. 2009, 8, 1212–1218. [Google Scholar]
- Toro, M.A.; Varona, L. A note on mate allocation for dominance handling in genomic selection. Genet. Sel. Evol. 2010, 42, 33. [Google Scholar] [CrossRef]
- Vitezica, Z.G.; Varona, L.; Legarra, A. On the Additive and Dominant Variance and Covariance of Individuals Within the Genomic Selection Scope. Genetics 2013, 195, 1223. [Google Scholar] [CrossRef]
- Su, G.S.; Christensen, O.F.; Ostersen, T.; Henryon, M.; Lund, M.S. Estimating Additive and Non-Additive Genetic Variances and Predicting Genetic Merits Using Genome-Wide Dense Single Nucleotide Polymorphism Markers. PLoS ONE 2012, 7, e45293. [Google Scholar] [CrossRef]
- Lopes, M.S.; Bastiaansen, J.W.M.; Harlizius, B.; Knol, E.F.; Bovenhuis, H. A Genome-Wide Association Study Reveals Dominance Effects on Number of Teats in Pigs. PLoS ONE 2014, 9, e105867. [Google Scholar] [CrossRef] [PubMed]
- Garcia, A.A.F.; Wang, S.C.; Melchinger, A.E.; Zeng, Z.B. Quantitative Trait Loci Mapping and The Genetic Basis of Heterosis in Maize and Rice. Genetics 2008, 180, 1707–1724. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Long, H.; Feng, S.M.; Ma, T.T.; Wang, M.F.; Niu, L.Z.; Zhang, X.Y.; Wang, L.N.; Lei, Y.; Chen, Y.L.; et al. Trait correlated expression combined with eQTL and ASE analyses identified novel candidate genes affecting intramuscular fat. BMC Genom. 2021, 22, 805. [Google Scholar] [CrossRef] [PubMed]
- Terada, K.; Ohtani, T.; Ogawa, S.; Hirooka, H. Genetic parameters for carcass and meat quality traits in Jinhua, Duroc, and their crossbred pigs. J. Anim. Breed Genet. 2024, 141, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.S.; Jin, S.K.; Choi, Y.I.; Lee, J.J. Effects of Duroc Breeding Lines on Carcass Composition and Meat Quality. Korean J. Food Sci. Anim. Resour. 2015, 35, 80–85. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.S.; Wang, Z.G.; Wang, P.; Cheng, L.Y.; Li, J.H.; Luo, Y.F.; Yang, L.F.; Li, L.F.; Zeng, J.H.; Hu, B. Integrative analysis of proteomics and lipidomic profiles reveal the fat deposition and meat quality in Duroc x Guangdong small spotted pig. Front. Vet. Sci. 2024, 11, 1361441. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Browning, B.L.; Tian, X.W.; Zhou, Y.; Browning, S.R. Fast two-stage phasing of large-scale sequence data. Am. J. Hum. Genet. 2021, 108, 1880–1890. [Google Scholar] [CrossRef] [PubMed]
- Maize Hoeci: Mid-Parent, Better-Parent and Standard Heterosis of Experimental Crosses in Maize. Int. J. Trop. Agric. 2021, 39, 285–290.
- Pérez, P.; de los Campos, G. Genome-Wide Regression and Prediction with the BGLR Statistical Package. Genetics 2014, 198, 483–495. [Google Scholar] [CrossRef]
- Pérez-Rodríguez, P.; de los Campos, G. Multitrait Bayesian shrinkage and variable selection models with the BGLR-R package. Genetics 2022, 222, iyac112. [Google Scholar] [CrossRef] [PubMed]
- VanRaden, P.M. Efficient Methods to Compute Genomic Predictions. J. Dairy Sci. 2008, 91, 4414–4423. [Google Scholar] [CrossRef] [PubMed]
- Hayes, B.J.; Visscher, P.M.; Goddard, M.E. Increased accuracy of artificial selection by using the realized relationship matrix. Genet. Res. 2009, 91, 47–60. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.L.; Xu, F.J.; Qiao, J.K.; Che, Z.X.; Xiang, T.; Liu, X.L.; Li, X.Y.; Zhao, S.H.; Zhu, M.J. E-GWAS: An ensemble-like GWAS strategy that provides effective control over false positive rates without decreasing true positives. Genet. Sel. Evol. 2023, 55, 46. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.M.; Pressoir, G.; Briggs, W.H.; Bi, I.V.; Yamasaki, M.; Doebley, J.F.; McMullen, M.D.; Gaut, B.S.; Nielsen, D.M.; Holland, J.B.; et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 2006, 38, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Tang, H.; Liu, J.F.; Xu, S.Z.; Zhang, Q.; Ning, C. Rapid epistatic mixed-model association studies by controlling multiple polygenic effects. Bioinformatics 2020, 36, 4833–4837. [Google Scholar] [CrossRef] [PubMed]
- Segura, V.; Vilhjálmsson, B.J.; Platt, A.; Korte, A.; Seren, Ü.; Long, Q.; Nordborg, M. An efficient multi-locus mixed-model approach for genome-wide association studies in structured populations. Nat. Genet. 2012, 44, 825–830. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.L.; Huang, M.; Fan, B.; Buckler, E.S.; Zhang, Z.W. Iterative Usage of Fixed and Random Effect Models for Powerful and Efficient Genome-Wide Association Studies. PLoS Genet. 2016, 12, e1005767. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.L.; Zhang, H.H.; Tang, Z.S.; Xu, J.Y.; Yin, D.; Zhang, Z.W.; Yuan, X.H.; Zhu, M.J.; Zhao, S.H.; Li, X.Y.; et al. rMVP: A Memory-efficient, Visualization-enhanced, and Parallel-accelerated Tool for Genome-wide Association Study. Genom. Proteom. Bioinf. 2021, 19, 619–628. [Google Scholar] [CrossRef]
- Huang, M.; Liu, X.L.; Zhou, Y.; Summers, R.M.; Zhang, Z.W. BLINK: A package for the next level of genome-wide association studies with both individuals and markers in the millions. Gigascience 2019, 8, giy154. [Google Scholar] [CrossRef]
- Shaffer, J.P. Multiple Hypothesis-Testing. Annu. Rev. Psychol. 1995, 46, 561–584. [Google Scholar] [CrossRef]
- Cunningham, F.; Allen, J.E.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Austine-Orimoloye, O.; Azov, A.G.; Barnes, I.; Bennett, R.; et al. Ensembl 2022. Nucleic Acids. Res. 2022, 50, D988–D995. [Google Scholar] [CrossRef]
- Mao, X.Z.; Cai, T.; Olyarchuk, J.G.; Wei, L.P. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.M.; Mao, X.Z.; Cai, T.; Luo, J.C.; Wei, L.P. KOBAS server: A web-based platform for automated annotation and pathway identification. Nucleic Acids. Res. 2006, 34, W720–W724. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.B.; Zhang, Z.W. GAPIT Version 3: Boosting Power and Accuracy for Genomic Association and Prediction. Genom. Proteom. Bioinf. 2021, 19, 629–640. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, A.; Li, J.G.; Hu, W.C.; Yu, J.S.; Khan, S.U.; Khan, M.H.U.; Xie, G.S.; Wang, J.B.; Wang, L.Q. Uncovering genomic regions controlling plant architectural traits in hexaploid wheat using different GWAS models. Sci. Rep. 2021, 11, 6767. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zhong, H.; Meng, X.X.; Sun, T.; Li, Y.S.; Pinson, S.R.M.; Chang, S.K.C.; Peng, Z.H. Genome-wide association studies of ionomic and agronomic traits in USDA mini core collection of rice and comparative analyses of different mapping methods. BMC Plant Biol. 2020, 20, 441. [Google Scholar] [CrossRef] [PubMed]
- Nida, H.; Girma, G.; Mekonen, M.; Tirfessa, A.; Seyoum, A.; Bejiga, T.; Birhanu, C.; Dessalegn, K.; Senbetay, T.; Ayana, G.; et al. Genome-wide association analysis reveals seed protein loci as determinants of variations in grain mold resistance in sorghum. Theor. Appl. Genet. 2021, 134, 1167–1184. [Google Scholar] [CrossRef] [PubMed]
- Ning, C.; Kang, H.M.; Zhou, L.; Wang, D.; Wang, H.F.; Wang, A.G.; Fu, J.L.; Zhang, S.L.; Liu, J.F. Performance Gains in Genome-Wide Association Studies for Longitudinal Traits via Modeling Time-varied effects. Sci. Rep. 2017, 7, 590. [Google Scholar] [CrossRef]
- Sahana, G.; Guldbrandtsen, B.; Thomsen, B.; Holm, L.E.; Panitz, F.; Brondum, R.F.; Bendixen, C.; Lund, M.S. Genome-wide association study using high-density single nucleotide polymorphism arrays and whole-genome sequences for clinical mastitis traits in dairy cattle. J. Dairy Sci. 2014, 97, 7258–7275. [Google Scholar] [CrossRef]
- Fowler, K.E.; Pong-Wong, R.; Bauer, J.; Clemente, E.J.; Reitter, C.P.; Affara, N.A.; Waite, S.; Walling, G.A.; Griffin, D.K. Genome wide analysis reveals single nucleotide polymorphisms associated with fatness and putative novel copy number variants in three pig breeds. BMC Genom. 2013, 14, 784. [Google Scholar] [CrossRef] [PubMed]
- Long, Y.; Ruan, G.R.; Su, Y.; Xiao, S.J.; Zhang, Z.Y.; Ren, J.; Ding, N.S.; Huang, L.S. Genome-wide association study identifies QTLs for EBV of Backfat Thickness and average daily gain in Duroc pigs. Russ. J. Genet. 2014, 50, 1308–1315. [Google Scholar] [CrossRef]
- Rogers, S.; Macheda, M.L.; Docherty, S.E.; Carty, M.D.; Henderson, M.A.; Soeller, W.C.; Gibbs, E.M.; James, D.E.; Best, J.D. Identification of a novel glucose transporter-like protein-GLUT-12. Am. J. Physiol.-Endoc. Metab. 2002, 282, E733–E738. [Google Scholar] [CrossRef] [PubMed]
- White, M.A.; Tsouko, E.; Lin, C.C.; Rajapakshe, K.; Spencer, J.M.; Wilkenfeld, S.R.; Vakili, S.S.; Pulliam, T.L.; Awad, D.; Nikolos, F.; et al. GLUT12 promotes prostate cancer cell growth and is regulated by androgens and CaMKK2 signaling. Endocr.-Relat. Cancer 2018, 25, 453–469. [Google Scholar] [CrossRef] [PubMed]
- Grapes, L.; Rothschild, M.F. Investigation of a QTL region for loin eye area and fatness on pig Chromosome 1. Mamm. Genome 2006, 17, 657–668. [Google Scholar] [CrossRef]
- MacNeill, S.A. Structure and function of the GINS complex, a key component of the eukaryotic replisome. Biochem. J. 2010, 425, 489–500. [Google Scholar] [CrossRef]
- Das, S.K.; Ma, L.; Sharma, N.K. Adipose tissue gene expression and metabolic health of obese adults. Int. J. Obes. 2015, 39, 869–873. [Google Scholar] [CrossRef]
- Sajan, S.A.; Ganesh, J.; Shinde, D.N.; Powis, Z.; Scarano, M.I.; Stone, J.; Winter, S.; Tang, S. Biallelic disruption of is associated with a skeletal disorder characterised by rhizomelic shortening of extremities and dysmorphic features. J. Med. Genet. 2019, 56, 850–854. [Google Scholar] [CrossRef]
- Kinoshita, M.; Era, T.; Jakt, L.M.; Nishikawa, S.I. The novel protein kinase Vlk is essential for stromal function of mesenchymal cells. Development 2009, 136, 2069–2079. [Google Scholar] [CrossRef]
- Miao, Y.X.; Mei, Q.N.; Fu, C.K.; Liao, M.X.; Liu, Y.; Xu, X.W.; Li, X.Y.; Zhao, S.H.; Xiang, T. Genome-wide association and transcriptome studies identify candidate genes and pathways for feed conversion ratio in pigs. BMC Genom. 2021, 22, 294. [Google Scholar] [CrossRef]
- Ibáñez-Zamacona, M.E.; Poveda, A.; Rebato, E. Contribution of obesity associated genetic variants to anthropometric somatotype components. Anthr. Anz Ber Uber Biol-Anthr. Lit 2019, 76, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Wang, J.; Wang, K.; Wang, H.; Wu, Q.; Yang, C.; Yu, Y.; Ni, P.; Zhong, Y.; Song, Z.; et al. RNF217 regulates iron homeostasis through its E3 ubiquitin ligase activity by modulating ferroportin degradation. Blood 2021, 138, 689–705. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.Y.; Meng, X.R.; Hinney, A.; Song, J.Y.; Huang, T.; Ma, J.; Wang, H.J. Waist-hip ratio related genetic loci are associated with risk of impaired fasting glucose in Chinese children: A case control study. Nutr. Metab. 2018, 15, 34. [Google Scholar] [CrossRef] [PubMed]
- Niu, G.L.; Yang, Y.L.; Zhang, Y.Y.; Hua, C.J.; Wang, Z.S.; Tang, Z.L.; Li, K. Identifying suitable reference genes for gene expression analysis in developing skeletal muscle in pigs. PeerJ 2016, 4, e2428. [Google Scholar] [CrossRef]



| Trait | (SE) | (SE) | (SE) | (SE) | (SE) | |
|---|---|---|---|---|---|---|
| 100 AGE | 5.3080 (0.2004) | 5.6899 (0.2718) | 8.1597 (0.1523) | 0.3322 (0.0125) | 0.3561 (0.017) | 0.5173 |
| 100 BF | 3.8868 (0.1189) | 3.6087 (0.1518) | 6.4086 (0.1007) | 0.2523 (0.0077) | 0.2342 (0.0098) | 0.4814 |
| Trait | SNP | Chr | Pos (bp) | Model | Gene | Distance (bp) |
|---|---|---|---|---|---|---|
| 100 AGE | SNP1735 | 1 | 29,864,163 | REMMAX, FarmCPU | SLC2A12 | 0 |
| SNP2107 | 1 | 37,901,112 | REMMAX, MLMM, FarmCPU | RNF217 | 0 | |
| SNP31335 | 4 | 102,102,250 | REMMAX, FarmCPU | WARS2 | −219,180 | |
| SNP39335 | 6 | 20,128,919 | REMMAX, FarmCPU | GINS3 | +33,717 | |
| 100 AGE additive PGV | SNP2107 | 1 | 37,901,112 | REMMAX, MLMM | RNF217 | 0 |
| SNP24375 | 3 | 98,862,214 | FarmCPU, BLINK | PKDCC | +593,661 | |
| SNP31335 | 4 | 102,102,250 | REMMAX, FarmCPU, BLINK | WARS2 | −219,180 | |
| 100 BF additive PGV | SNP13140 | 2 | 19,545,736 | REMMAX, MLMM | API5 | −551,496 |
| SNP40556 | 6 | 44,729,418 | FarmCPU, BLINK | FAM187B | +12,795 | |
| SNP55770 | 8 | 42,734,254 | MLM, REMMAX, MLMM | MAP9 | +390 |
| Trait | SNP | Chr | Pos (bp) | Model | Gene | Distance (bp) |
|---|---|---|---|---|---|---|
| 100 AGE | SNP2107 | 1 | 37,901,112 | MLMM, FarmCPU | RNF217 | 0 |
| 100 AGE dominance PGV | SNP31335 | 4 | 102,102,250 | MLMM, FarmCPU | WARS2 | −219,180 |
| SNP34586 | 5 | 30,709,370 | FarmCPU, BLINK | GRIP1 | 0 | |
| 100 BF dominance PGV | SNP13140 | 2 | 19,545,736 | MLMM, FarmCPU, BLINK | API5 | −551,496 |
| SNP51848 | 7 | 102,484,784 | FarmCPU, BLINK | NRXN3 | −546,486 | |
| SNP55770 | 8 | 42,734,254 | MLMM, FarmCPU, BLINK | MAP9 | +390 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qiao, J.; Li, K.; Miao, N.; Xu, F.; Han, P.; Dai, X.; Abdelkarim, O.F.; Zhu, M.; Zhao, Y. Additive and Dominance Genome-Wide Association Studies Reveal the Genetic Basis of Heterosis Related to Growth Traits of Duhua Hybrid Pigs. Animals 2024, 14, 1944. https://doi.org/10.3390/ani14131944
Qiao J, Li K, Miao N, Xu F, Han P, Dai X, Abdelkarim OF, Zhu M, Zhao Y. Additive and Dominance Genome-Wide Association Studies Reveal the Genetic Basis of Heterosis Related to Growth Traits of Duhua Hybrid Pigs. Animals. 2024; 14(13):1944. https://doi.org/10.3390/ani14131944
Chicago/Turabian StyleQiao, Jiakun, Kebiao Li, Na Miao, Fangjun Xu, Pingping Han, Xiangyu Dai, Omnia Fathy Abdelkarim, Mengjin Zhu, and Yunxiang Zhao. 2024. "Additive and Dominance Genome-Wide Association Studies Reveal the Genetic Basis of Heterosis Related to Growth Traits of Duhua Hybrid Pigs" Animals 14, no. 13: 1944. https://doi.org/10.3390/ani14131944
APA StyleQiao, J., Li, K., Miao, N., Xu, F., Han, P., Dai, X., Abdelkarim, O. F., Zhu, M., & Zhao, Y. (2024). Additive and Dominance Genome-Wide Association Studies Reveal the Genetic Basis of Heterosis Related to Growth Traits of Duhua Hybrid Pigs. Animals, 14(13), 1944. https://doi.org/10.3390/ani14131944





