m6A Methylation Mediates the Function of the circRNA-08436/miR-195/ELOVL6 Axis in Regards to Lipid Metabolism in Dairy Goat Mammary Glands
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Experimental Animals and Preparation of Samples
2.3. Differential Expression Analysis of circRNAs
2.4. Cell Culture and Treatment
2.5. Determination of TAG and Cholesterol
2.6. Determination of Intracellular Fatty Acid Content
2.7. Oil Red O Staining
2.8. 5-Ethynyl-2′-Deoxyuridine (EdU) Experiment
2.9. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
2.10. Construction of Circ08436 Overexpression Vector
2.11. Western Blotting
2.12. Dual-Luciferase Activity Analysis
2.13. Statistical Analysis
3. Results
3.1. High-Throughput Sequencing of circRNAs from Mammary Gland Tissue of Dairy Goats
3.2. Differential Expression Analysis of mRNAs
3.3. Enrichment Analysis and Pathway Network Diagram of Differential Genes
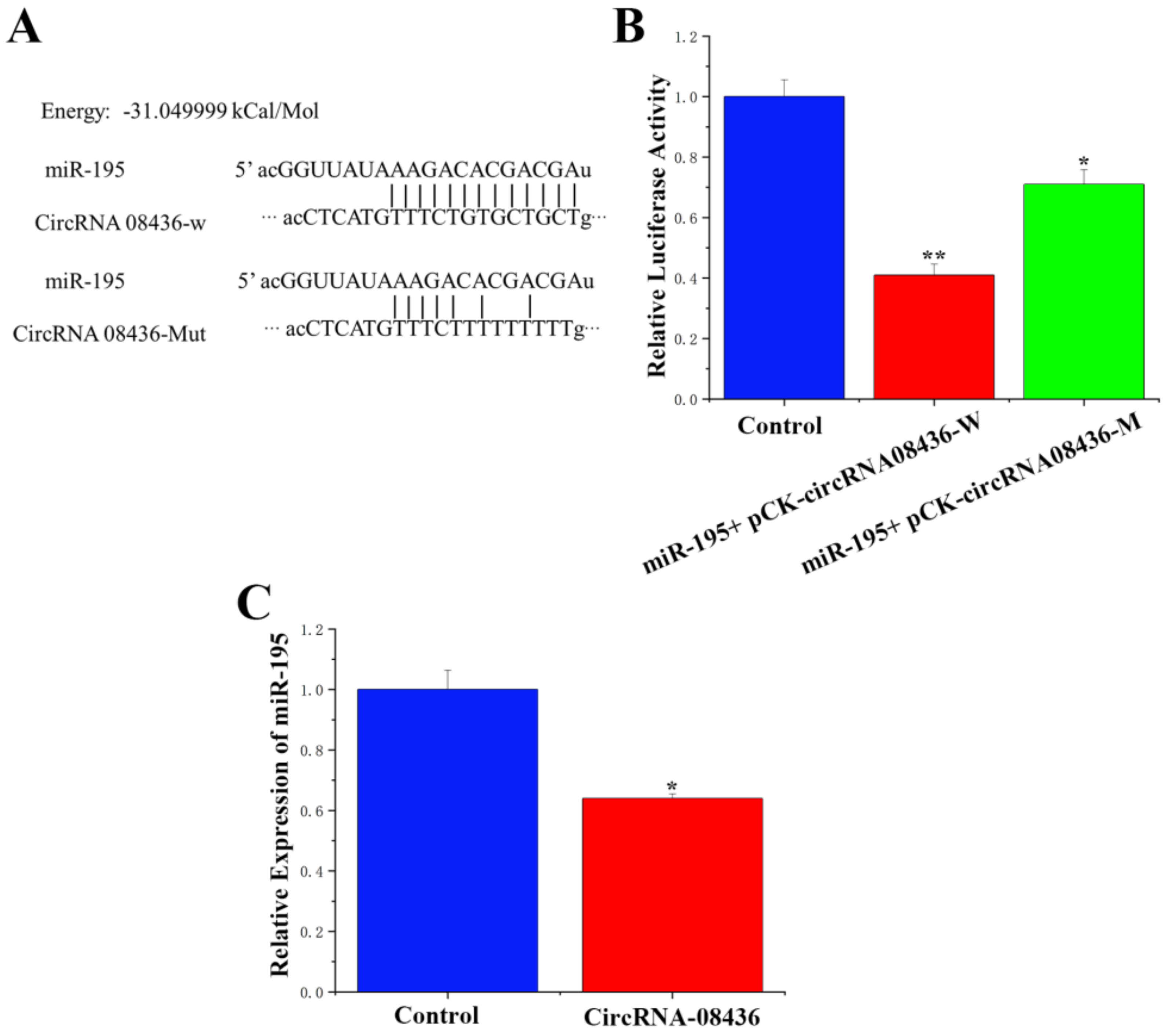
3.4. Adsorption and Binding of circRNA-08436 to miR-195
3.5. Specific Targeting of miR-195 on ELOVL6
3.6. Functional Verification of circRNA-08436 in GMECs
3.7. Functional Role of miR-195 and ELOVL6 in GMECs
3.8. Combination of circ08436 with miR-195 to Relieve the Inhibition of ELOVL6
3.9. Regulation of circRNA-08436 by m6A Methylation
4. Discussion
4.1. CircRNA-08436 Can Promote TAG Synthesis
4.2. CircRNA-08436 Can Competitively Bind to miR-195, Thereby Deregulating the Inhibitory Effect of miR-195 on the Target Gene ELOVL6
4.3. YTHDC2 May Play a Role in the Degradation of circRNA-08436 in the Cytoplasm
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nayik, G.A.; Jagdale, Y.D.; Gaikwad, S.A.; Devkatte, A.N.; Dar, A.H.; Dezmirean, D.S.; Bobis, O.; Ranjha, M.; Ansari, M.J.; Hemeg, H.A.; et al. Recent Insights Into Processing Approaches and Potential Health Benefits of Goat Milk and Its Products: A Review. Front. Nutr. 2021, 8, 789117. [Google Scholar] [CrossRef]
- Zine-eddine, Y.; Zinelabidine, L.H.; Kzaiber, F.; Oussama, A.; Boutoial, K. The use of paired comparison test on the goat’s milk adulteration detection (whole and semiskimmed). J. Sens. Stud. 2021, 36, e12690. [Google Scholar] [CrossRef]
- Sánchez-Macías, D.; Moreno-Indias, I.; Castro, N.; Morales-Delanuez, A.; Argüello, A. From goat colostrum to milk: Physical, chemical, and immune evolution from partum to 90 days postpartum. J. Dairy Sci. 2014, 97, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Gallier, S.; Tolenaars, L.; Prosser, C. Whole Goat Milk as a Source of Fat and Milk Fat Globule Membrane in Infant Formula. Nutrients 2020, 12, 3486. [Google Scholar] [CrossRef]
- Lu, Q.; Chen, Z.; Ji, D.; Mao, Y.; Jiang, Q.; Yang, Z.; Loor, J.J. Progress on the Regulation of Ruminant Milk Fat by Noncoding RNAs and ceRNAs. Front. Genet. 2021, 12, 733925. [Google Scholar] [CrossRef]
- Li, C.; Zhu, J.J.; Shi, H.B.; Luo, J.; Zhao, W.S.; Shi, H.P.; Xu, H.F.; Wang, H.; Loor, J.J. Comprehensive Transcriptome Profiling of Dairy Goat Mammary Gland Identifies Genes and Networks Crucial for Lactation and Fatty Acid Metabolism. Front. Genet. 2020, 11, 878. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, X.O.; Chen, T.; Xiang, J.F.; Yin, Q.F.; Xing, Y.H.; Zhu, S.; Yang, L.; Chen, L.L. Circular intronic long noncoding RNAs. Mol. Cell 2013, 51, 792–806. [Google Scholar] [CrossRef]
- Chen, Z.; Lu, Q.; Zhang, X.; Zhang, Z.; Cao, X.; Wang, K.; Lu, X.; Yang, Z.; Loor, J.J.; Jiao, P. Circ007071 Inhibits Unsaturated Fatty Acid Synthesis by Interacting with miR-103-5p to Enhance PPAR gamma Expression in the Dairy Goat Mammary Gland. J. Agric. Food Chem. 2022, 70, 13719–13729. [Google Scholar] [CrossRef]
- Chen, Z.; Zhou, J.; Wang, M.; Liu, J.; Zhang, L.; Loor, J.J.; Liang, Y.; Wu, H.; Yang, Z. Circ09863 Regulates Unsaturated Fatty Acid Metabolism by Adsorbing miR-27a-3p in Bovine Mammary Epithelial Cells. J. Agric. Food Chem. 2020, 68, 8589–8601. [Google Scholar] [CrossRef]
- He, L.; Li, H.; Wu, A.; Peng, Y.; Shu, G.; Yin, G. Functions of N6-methyladenosine and its role in cancer. Mol. Cancer 2019, 18, 176. [Google Scholar] [CrossRef]
- Wang, T.; Kong, S.; Tao, M.; Ju, S. The potential role of RNA N6-methyladenosine in Cancer progression. Mol. Cancer 2020, 19, 88. [Google Scholar] [CrossRef]
- Xin, Z.M.; Zhang, T.Y.; Lu, Q.Y.; Yang, Z.P.; Chen, Z. Progress of m(6)A Methylation in Lipid Metabolism in Humans and Animals. Agriculture 2022, 12, 1683. [Google Scholar] [CrossRef]
- Chiu, H.H.; Kuo, C.H. Gas chromatography-mass spectrometry-based analytical strategies for fatty acid analysis in biological samples. J. Food Drug Anal. 2020, 28, 60–73. [Google Scholar] [CrossRef]
- Chen, L.; Shan, G. Circular RNAs remain peculiarly unclear in biogenesis and function. Sci. China-Life Sci. 2015, 58, 616–618. [Google Scholar] [CrossRef]
- Kolakofsky, D. Isolation and characterization of Sendai virus DI-RNAs. Cell 1976, 8, 547–555. [Google Scholar] [CrossRef]
- Hsu, M.T.; Coca-Prados, M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 1979, 280, 339–340. [Google Scholar] [CrossRef]
- D’Ambrosi, S.; Visser, A.; Antunes-Ferreira, M.; Poutsma, A.; Giannoukakos, S.; Sol, N.; Sabrkhany, S.; Bahce, I.; Kuijpers, M.J.E.; Oude Egbrink, M.G.A.; et al. The Analysis of Platelet-Derived circRNA Repertoire as Potential Diagnostic Biomarker for Non-Small Cell Lung Cancer. Cancers 2021, 13, 4644. [Google Scholar] [CrossRef]
- Nan, X.-M.; Wang, J.-Q.; Chen, H.-Y.; Hu, H. MicroRNAs and Lipids Metabolism. Prog. Biochem. Biophys. 2013, 40, 118–129. [Google Scholar] [CrossRef]
- Zong, Y.R.; Wang, X.L.; Cui, B.; Xiong, X.W.; Wu, A.; Lin, C.R.; Zhang, Y.H. Decoding the regulatory roles of non-coding RNAs in cellular metabolism and disease. Mol. Ther. 2023, 31, 1562–1576. [Google Scholar] [CrossRef]
- Ma, B.H.; Liu, Z.Z.; Yan, W.; Wang, L.X.; He, H.B.; Zhang, A.J.; Li, Z.Y.; Zhao, Q.Z.; Liu, M.M.; Guan, S.Y.; et al. Circular RNAs acting as ceRNAs mediated by miRNAs may be involved in the synthesis of soybean fatty acids. Funct. Integr. Genom. 2021, 21, 435–450. [Google Scholar] [CrossRef]
- Tahri-Joutey, M.; Andreoletti, P.; Surapureddi, S.; Nasser, B.; Cherkaoui-Malki, M.; Latruffe, N. Mechanisms Mediating the Regulation of Peroxisomal Fatty Acid Beta-Oxidation by PPARα. Int. J. Mol. Sci. 2021, 22, 8969. [Google Scholar] [CrossRef]
- Lappas, M. Effect of pre-existing maternal obesity, gestational diabetes and adipokines on the expression of genes involved in lipid metabolism in adipose tissue. Metab. Clin. Exp. 2014, 63, 250–262. [Google Scholar] [CrossRef]
- Kazmierczak, D.; Hydbring, P. Construction of a Full-Length 3’ UTR Reporter System for Identification of Cell-Cycle Regulating MicroRNAs. In Cell Cycle Oscillators, 2 Edition: Methods and Protocols; Coutts, A.S., Weston, L., Eds.; Humana: New York, NY, USA, 2021; Volume 2329, pp. 81–94. [Google Scholar]
- Matsuzaka, T.; Shimano, H. Role of Fatty Acid Elongase Elovl6 in the Regulation of Fatty Acid Quality and Lifestyle-related Diseases. Yakugaku Zasshi-J. Pharm. Soc. Jpn. 2022, 142, 473–476. [Google Scholar] [CrossRef]
- Matsuzaka, T.; Shimano, H. Elovl6: A new player in fatty acid metabolism and insulin sensitivity. J. Mol. Med. 2009, 87, 379–384. [Google Scholar] [CrossRef] [PubMed]
- Junjvlieke, Z.; Khan, R.; Mei, C.; Cheng, G.; Wang, S.; Raza, S.H.A.; Hong, J.; Wang, X.; Yang, W.; Zan, L. Effect of ELOVL6 on the lipid metabolism of bovine adipocytes. Genomics 2020, 112, 2282–2290. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaka, T. Role of fatty acid elongase Elovl6 in the regulation of energy metabolism and pathophysiological significance in diabetes. Diabetol. Int. 2021, 12, 68–73. [Google Scholar] [CrossRef]
- Shimano, H. Novel qualitative aspects of tissue fatty acids related to metabolic regulation: Lessons from Elovl6 knockout. Prog. Lipid Res. 2012, 51, 267–271. [Google Scholar] [CrossRef]
- Yoshida, K.; Morishima, Y.; Ano, S.; Sakurai, H.; Kuramoto, K.; Tsunoda, Y.; Yazaki, K.; Nakajima, M.; Sherpa, M.T.; Matsuyama, M.; et al. ELOVL6 deficiency aggravates allergic airway inflammation through the ceramide-S1P pathway in mice. J. Allergy Clin. Immunol. 2023, 151, 1067–1080.e9. [Google Scholar] [CrossRef]
- He, Q.; Luo, J.; Wu, J.; Li, Z.; Yao, W.; Zang, S.; Niu, H. ELOVL6 promoter binding sites directly targeted by sterol regulatory element binding protein 1 in fatty acid synthesis of goat mammary epithelial cells. J. Dairy Sci. 2021, 104, 6253–6266. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Li, J.; Yang, H.; Yang, X.; Zhang, Y.; Yu, X.; Li, Y.; Chen, G.; Yang, Z. The potential role of m6A reader YTHDF1 as diagnostic biomarker and the signaling pathways in tumorigenesis and metastasis in pan-cancer. Cell Death Discov. 2023, 9, 34. [Google Scholar] [CrossRef] [PubMed]
- Murakami, S.; Jaffrey, S.R. Hidden codes in mRNA: Control of gene expression by m(6)A. Mol. Cell 2022, 82, 2236–2251. [Google Scholar] [CrossRef]
- Yan, H.Q.; Zhang, L.Q.; Cui, X.B.; Zheng, S.N.; Li, R.B. Roles and mechanisms of the m(6)A reader YTHDC1 in biological processes and diseases. Cell Death Discov. 2022, 8, 237. [Google Scholar] [CrossRef]
- Shi, R.K.; Ying, S.L.; Li, Y.D.; Zhu, L.Y.; Wang, X.; Jin, H.C. Linking the YTH domain to cancer: The importance of YTH family proteins in epigenetics. Cell Death Dis. 2021, 12, 346. [Google Scholar] [CrossRef]
- Roundtree, I.A.; Luo, G.Z.; Zhang, Z.; Wang, X.; Zhou, T.; Cui, Y.; Sha, J.; Huang, X.; Guerrero, L.; Xie, P.; et al. YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. eLife 2017, 6, e31311. [Google Scholar] [CrossRef]
- Chen, R.X.; Chen, X.; Xia, L.P.; Zhang, J.X.; Pan, Z.Z.; Ma, X.D.; Han, K.; Chen, J.W.; Judde, J.G.; Deas, O.; et al. N(6)-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat. Commun. 2019, 10, 4695. [Google Scholar] [CrossRef]
- Zhou, C.; Molinie, B.; Daneshvar, K.; Pondick, J.V.; Wang, J.; Van Wittenberghe, N.; Xing, Y.; Giallourakis, C.C.; Mullen, A.C. Genome-Wide Maps of m6A circRNAs Identify Widespread and Cell-Type-Specific Methylation Patterns that Are Distinct from mRNAs. Cell Rep. 2017, 20, 2262–2276. [Google Scholar] [CrossRef]
- Zaccara, S.; Jaffrey, S.R. A Unified Model for the Function of YTHDF Proteins in Regulating m(6)A-Modified mRNA. Cell 2020, 181, 1582–1595.e18. [Google Scholar] [CrossRef]












| Fatty Acid | Control | circ08436 |
|---|---|---|
| C16:0 (%) | 21.41 ± 0.14 | 41.12 ± 0.56 ** |
| C16:1 (%) | 10.12 ± 0.57 | 2.10 ± 0.34 ** |
| C18:0 (%) | 10.57 ± 0.34 | 18.41 ± 0.25 * |
| C18:1 (%) | 48.14 ± 0.35 | 34.83 ± 0.41 ** |
| C18:2 (%) | 9.76 ± 0.24 | 3.54 ± 0.24 * |
| SFA (%) | 31.98 | 59.53 |
| UFA (%) | 68.02 | 40.47 |
| UFA/SFA | 2.13 | 0.68 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Wu, Y.; Yang, S.; Gao, R.; Lv, X.; Yang, Z.; Jiao, P.; Zhang, N.; Loor, J.J.; Chen, Z. m6A Methylation Mediates the Function of the circRNA-08436/miR-195/ELOVL6 Axis in Regards to Lipid Metabolism in Dairy Goat Mammary Glands. Animals 2024, 14, 1715. https://doi.org/10.3390/ani14121715
Wang Y, Wu Y, Yang S, Gao R, Lv X, Yang Z, Jiao P, Zhang N, Loor JJ, Chen Z. m6A Methylation Mediates the Function of the circRNA-08436/miR-195/ELOVL6 Axis in Regards to Lipid Metabolism in Dairy Goat Mammary Glands. Animals. 2024; 14(12):1715. https://doi.org/10.3390/ani14121715
Chicago/Turabian StyleWang, Yu, Yanni Wu, Sitian Yang, Rui Gao, Xiaoyang Lv, Zhangping Yang, Peixin Jiao, Ning Zhang, Juan J. Loor, and Zhi Chen. 2024. "m6A Methylation Mediates the Function of the circRNA-08436/miR-195/ELOVL6 Axis in Regards to Lipid Metabolism in Dairy Goat Mammary Glands" Animals 14, no. 12: 1715. https://doi.org/10.3390/ani14121715
APA StyleWang, Y., Wu, Y., Yang, S., Gao, R., Lv, X., Yang, Z., Jiao, P., Zhang, N., Loor, J. J., & Chen, Z. (2024). m6A Methylation Mediates the Function of the circRNA-08436/miR-195/ELOVL6 Axis in Regards to Lipid Metabolism in Dairy Goat Mammary Glands. Animals, 14(12), 1715. https://doi.org/10.3390/ani14121715







