Exploring Variability: Inflammation Mediator Levels across Tissues and Time in Poultry Experimentally Infected by the G1a and G6 Genogroups of Infectious Bursal Disease Virus (IBDV)
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples
2.2. RNA Extraction and Reverse Transcription
2.3. Relative Quantification Assay Validation
2.4. Relative Quantification
2.5. Statistical Analysis
3. Results
3.1. Clinical Pathological and Virological Outcomes
3.2. IBDV Detection
3.3. Lesions
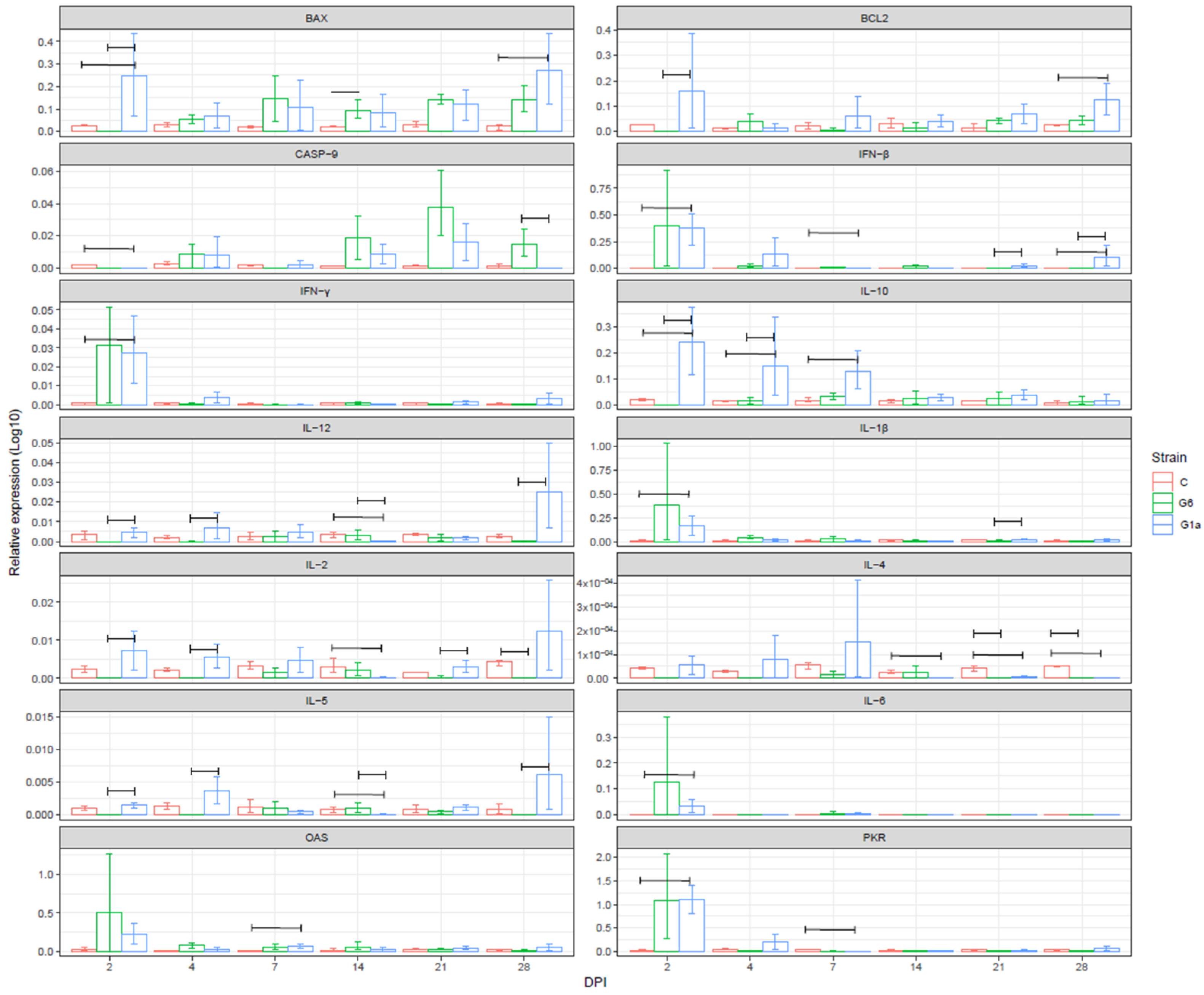
3.4. Gene Expression
3.4.1. Bursa of Fabricius
3.4.2. Thymus
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Franciosini, M.P.; Davidson, I. A Walk through Gumboro Disease. Poultry 2022, 1, 229–243. [Google Scholar] [CrossRef]
- Orakpoghenor, O.; Oladele, S.B.; Abdu, P.A. Infectious Bursal Disease: Transmission, Pathogenesis, Pathology and Control—An Overview. Worlds Poult. Sci. J. 2020, 76, 292–303. [Google Scholar] [CrossRef]
- Islam, M.R.; Nooruzzaman, M.; Rahman, T.; Mumu, T.T.; Rahman, M.M.; Chowdhury, E.H.; Eterradossi, N.; Müller, H. A Unified Genotypic Classification of Infectious Bursal Disease Virus Based on Both Genome Segments. Avian Pathol. 2021, 50, 190–206. [Google Scholar] [CrossRef] [PubMed]
- Rautenschlein, S.; Alkie, T.N. Infectious Bursal Disease Virus in Poultry: Current Status and Future Prospects. Vet. Med. Res. Rep. 2016, 7, 9. [Google Scholar] [CrossRef] [PubMed]
- Duffy, S. Why Are RNA Virus Mutation Rates so Damn High? PLoS Biol. 2018, 16, e3000003. [Google Scholar] [CrossRef] [PubMed]
- de Fraga, A.P.; Gräf, T.; Coltro, V.P.; Ikuta, N.; Fonseca, A.S.K.; Majó, N.; Lunge, V.R. Phylodynamic Analyses of Brazilian Antigenic Variants of Infectious Bursal Disease Virus. Infect. Genet. Evol. 2019, 73, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Cosgrove, A.S. An Apparently New Disease of Chickens: Avian Nephrosis. Avian Dis. 1962, 6, 385. [Google Scholar] [CrossRef]
- Lasher, H.N.; Davis, V.S. History of Infectious Bursal Disease in the U.S.A.—The First Two Decades. Avian Dis. 1997, 41, 11–19. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, X.; Gao, Y.; Qi, X. The Over-40-Years-Epidemic of Infectious Bursal Disease Virus in China. Viruses 2022, 14, 2253. [Google Scholar] [CrossRef]
- Michel, L.O.; Jackwood, D.J. Classification of Infectious Bursal Disease Virus into Genogroups. Arch. Virol. 2017, 162, 3661–3670. [Google Scholar] [CrossRef]
- Wang, Y.L.; Fan, L.J.; Jiang, N.; Gao, L.; Li, K.; Gao, Y.L.; Liu, C.J.; Cui, H.Y.; Pan, Q.; Zhang, Y.P.; et al. An Improved Scheme for Infectious Bursal Disease Virus Genotype Classification Based on Both Genome-Segments A and B. J. Integr. Agric. 2021, 20, 1372–1381. [Google Scholar] [CrossRef]
- Lupini, C.; Giovanardi, D.; Pesente, P.; Bonci, M.; Felice, V.; Rossi, G.; Morandini, E.; Cecchinato, M.; Catelli, E. A Molecular Epidemiology Study Based on VP2 Gene Sequences Reveals That a New Genotype of Infectious Bursal Disease Virus Is Dominantly Prevalent in Italy. Avian Pathol. 2016, 45, 458–464. [Google Scholar] [CrossRef]
- Felice, V.; Franzo, G.; Catelli, E.; Di Francesco, A.; Bonci, M.; Cecchinato, M.; Mescolini, G.; Giovanardi, D.; Pesente, P.; Lupini, C. Genome Sequence Analysis of a Distinctive Italian Infectious Bursal Disease Virus. Poult. Sci. 2017, 96, 4370–4377. [Google Scholar] [CrossRef] [PubMed]
- Lupini, C.; Felice, V.; Silveira, F.; Mescolini, G.; Berto, G.; Listorti, V.; Cecchinato, M.; Catelli, E. Comparative In Vivo Pathogenicity Study of an ITA Genotype Isolate (G6) of Infectious Bursal Disease Virus. Transbound. Emerg. Dis. 2020, 67, 1025–1031. [Google Scholar] [CrossRef]
- Legnardi, M.; Poletto, F.; Alam, S.; Cherfane, A.; Le-Tallec, B.; Franzo, G.; Tucciarone, C.M.; Lupini, C.; Pasotto, D.; Cecchinato, M. Molecular Epidemiology of Infectious Bursal Disease Virus in the Near East and Persian Gulf Regions. Avian Pathol. 2024, 53, 56–67. [Google Scholar] [CrossRef]
- Mahgoub, H.A. An Overview of Infectious Bursal Disease. Arch. Virol. 2012, 157, 2047–2057. [Google Scholar] [CrossRef] [PubMed]
- Umar, S.; Munir, M.T.; Ahsan, U.; Raza, I.; Chowdhury, M.R.; Ahmed, Z.; Shah, M.A.A. Immunosuppressive Interactions of Viral Diseases in Poultry. Worlds Poult. Sci. J. 2016, 73, 121–135. [Google Scholar] [CrossRef]
- Sharma, J.M.; Kim, I.J.; Rautenschlein, S.; Yeh, H.Y. Infectious Bursal Disease Virus of Chickens: Pathogenesis and Immunosuppression. Dev. Comp. Immunol. 2000, 24, 223–235. [Google Scholar] [CrossRef]
- Rautenschlein, S.; Yeh, H.Y.; Sharma, J.M. Comparative Immunopathogenesis of Mild, Intermediate, and Virulent Strains of Classic Infectious Bursal Disease Virus. Avian Dis. 2003, 47, 66–78. [Google Scholar] [CrossRef]
- Silveira, F.; Felice, V.; Franzo, G.; Mescolini, G.; Catelli, E.; Cecchinato, M.; Berto, G.; Listorti, V.; Lupini, C. Inoculation of Specific Pathogen-Free Chickens with an Infectious Bursal Disease Virus of the ITA Genotype (G6) Leads to a High and Persistent Viral Load in Lymphoid Tissues and to a Delayed Antiviral Response. Vet. Microbiol. 2019, 235, 136–142. [Google Scholar] [CrossRef]
- Lam, K.M. Morphological Evidence of Apoptosis in Chickens Infected with Infectious Bursal Disease Virus. J. Comp. Pathol. 1997, 116, 367–377. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Lecompte, J.C.; Niño-Fong, R.; Lopez, A.; Markham, R.J.F.; Kibenge, F.S.B. Infectious Bursal Disease Virus (IBDV) Induces Apoptosis in Chicken B Cells. Comp. Immunol. Microbiol. Infect. Dis. 2005, 28, 321–337. [Google Scholar] [CrossRef] [PubMed]
- Rauf, A.; Khatri, M.; Murgia, M.V.; Jung, K.; Saif, Y.M. Differential Modulation of Cytokine, Chemokine and Toll like Receptor Expression in Chickens Infected with Classical and Variant Infectious Bursal Disease Virus. Vet. Res. 2011, 42, 85. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Y.; Wang, Y.; Guo, Y.; Xiong, Y.; Chen, M.; Zhao, A.; Liu, H. T Cell Subset Profile and Inflammatory Cytokine Properties in the Gut-Associated Lymphoid Tissues of Chickens during Infectious Bursal Disease Virus (IBDV) Infection. Arch. Virol. 2020, 165, 2249–2258. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Chen, J.; Xiang, Y.; Chen, Y.; Shen, W.; Wang, W.; Li, Y.; Wei, P.; He, X. Differential Modulation of Innate Antiviral Profiles in the Intestinal Lamina Propria Cells of Chickens Infected with Infectious Bursal Disease Viruses of Different Virulence. Viruses 2022, 14, 393. [Google Scholar] [CrossRef] [PubMed]
- Eldaghayes, I.; Rothwell, L.; Williams, A.; Withers, D.; Balu, S.; Davison, F.; Kaiser, P. Infectious Bursal Disease Virus: Strains That Differ in Virulence Differentially Modulate the Innate Immune Response to Infection in the Chicken Bursa. Viral Immunol. 2006, 19, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Liu, W.; Zhang, J.; Liu, Z.; Wang, M.; Wang, L.; Zhou, H.; Jiang, Y.; Cui, W.; Qiao, X.; et al. Very Virulent Infectious Bursal Disease Virus-Induced Immune Injury Is Involved in Inflammation, Apoptosis, and Inflammatory Cytokines Imbalance in the Bursa of Fabricius. Dev. Comp. Immunol. 2021, 114, 103839. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Zheng, S.J. Infectious Bursal Disease Virus-Host Interactions: Multifunctional Viral Proteins That Perform Multiple and Differing Jobs. Int. J. Mol. Sci. 2017, 18, 161. [Google Scholar] [CrossRef] [PubMed]
- Ingrao, F.; Rauw, F.; Lambrecht, B.; Van den Berg, T. Infectious Bursal Disease: A Complex Host-Pathogen Interaction. Dev. Comp. Immunol. 2013, 41, 429–438. [Google Scholar] [CrossRef]
- Dobner, M.; Auerbach, M.; Mundt, E.; Icken, W.; Rautenschlein, S. Genotype-Associated Differences in Bursal Recovery after Infectious Bursal Disease Virus (IBDV) Inoculation. Vet. Immunol. Immunopathol. 2020, 220, 109993. [Google Scholar] [CrossRef]
- Jackwood, D.J.; Schat, K.A.; Michel, L.O.; de Wit, S. A Proposed Nomenclature for Infectious Bursal Disease Virus Isolates. Avian Pathol. 2018, 47, 576–584. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, M.; Han, H.; Yuan, J.; Li, Z. Comparison of the Expression of Cytokine Genes in the Bursal Tissues of the Chickens Following Challenge with Infectious Bursal Disease Viruses of Varying Virulence. Virol. J. 2010, 7, 364. [Google Scholar] [CrossRef]
- Ginestet, C. Ggplot2: Elegant Graphics for Data Analysis. J. R. Stat. Soc. Ser. A Stat. Soc. 2011, 174, 245–246. [Google Scholar] [CrossRef]
- Kassambara, A. Ggpubr: ‘ggplot2’ based Publication Ready Plots. R Package Version 2018, 2. Available online: https://rpkgs.datanovia.com/ggpubr/ (accessed on 12 March 2024).
- Wu, Z.; Hu, T.; Rothwell, L.; Vervelde, L.; Kaiser, P.; Boulton, K.; Nolan, M.J.; Tomley, F.M.; Blake, D.P.; Hume, D.A. Analysis of the Function of IL-10 in Chickens Using Specific Neutralising Antibodies and a Sensitive Capture ELISA. Dev. Comp. Immunol. 2016, 63, 206. [Google Scholar] [CrossRef]

| Primer | Sequence | Reference |
|---|---|---|
| ACTIN-F | TGCTGTGTTCCCATCTATCG | [27] |
| ACTIN-R | TTGGTGACAATACCGTGTTCA | [27] |
| GADPH-F | TGCTGCCCAGAACATCATCC | [27] |
| GADPH-R | ACGGCAGGTCAGGTCAACAA | [27] |
| BAX-F | GTGATGGCATGGGACATAGCTC | [27] |
| BAX-R | TGGCGTAGACCTTGCGGATAA | [27] |
| BCL2-F | ATCGTCGCCTTCTTCGAGTT | [27] |
| BCL2-R | ATCCCATCCTCCGTTGTCCT | [27] |
| CASP-9-F | CCGAAGGAGCAAGCACG | [27] |
| CASP-9-R | AGGTTGGACTGGGATGGAC | [27] |
| IFN-B-F | TTCTCCTGCAACCATCTTC | [25] |
| IFN-B-R | GAGGTGGAGCCGTATTCT | [25] |
| IFN-G-F | ATCATACTGAGCCAGATTGTTTCG | [27] |
| IFN-G-R | TCTTTCACCTTCTTCACGCCAT | [27] |
| IL-10-F | AGCTGAGGGTGAAGTTTGAGGAA | [27] |
| IL-10-R | CAGGACCTCATCTGTGTAGAAGCG | [27] |
| IL-12-F | AGGTGGGTCTGGCTTT | [27] |
| IL-12-R | TTCTGAGACTGGTGGCTTCACTTCC | [27] |
| IL-1B-F | GCTCTACATGTCGTGTGTGATGAG | [27] |
| IL-1B-R | TGTCGATGTCCCGCATGA | [27] |
| IL-2-F | TTCTGGGACCACTGTATGCTCTT | [32] |
| IL-2-R | TACCGACAAAGTGAGAATCAATCAG | [32] |
| IL-4-F | GCTCTTATGCAAAGCCTCCACAA | [27] |
| IL-4-R | TGCTGCTGGCATTCAGGAGC | [27] |
| IL-5-F | GGAACGGCACTGTTGAAAAATAA | [32] |
| IL-5-R | TTCTCCCTCTCCTGTCAGTTGTG | [32] |
| IL-6-F | GCTCGCCGGCTTCGA | [27] |
| IL-6-R | GGTAGGTCTGAAAGGCGAACAG | [27] |
| OAS-F | CACGGCCTCTTCTACGACA | [25] |
| OAS-R | TGGGCCATACGGTGTAGACT | [25] |
| PKR-F | CCTCTGCTGGCCTTACTGTCA | [25] |
| PKR-R | AAGAGAGGCAGAAGGAATAATTTGCC | [25] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Franzo, G.; Dotto, G.; Lupini, C.; Legnardi, M.; Tucciarone, C.M.; Poletto, F.; Catelli, E.; Graziosi, G.; Cecchinato, M.; Pasotto, D. Exploring Variability: Inflammation Mediator Levels across Tissues and Time in Poultry Experimentally Infected by the G1a and G6 Genogroups of Infectious Bursal Disease Virus (IBDV). Animals 2024, 14, 1619. https://doi.org/10.3390/ani14111619
Franzo G, Dotto G, Lupini C, Legnardi M, Tucciarone CM, Poletto F, Catelli E, Graziosi G, Cecchinato M, Pasotto D. Exploring Variability: Inflammation Mediator Levels across Tissues and Time in Poultry Experimentally Infected by the G1a and G6 Genogroups of Infectious Bursal Disease Virus (IBDV). Animals. 2024; 14(11):1619. https://doi.org/10.3390/ani14111619
Chicago/Turabian StyleFranzo, Giovanni, Giorgia Dotto, Caterina Lupini, Matteo Legnardi, Claudia Maria Tucciarone, Francesca Poletto, Elena Catelli, Giulia Graziosi, Mattia Cecchinato, and Daniela Pasotto. 2024. "Exploring Variability: Inflammation Mediator Levels across Tissues and Time in Poultry Experimentally Infected by the G1a and G6 Genogroups of Infectious Bursal Disease Virus (IBDV)" Animals 14, no. 11: 1619. https://doi.org/10.3390/ani14111619
APA StyleFranzo, G., Dotto, G., Lupini, C., Legnardi, M., Tucciarone, C. M., Poletto, F., Catelli, E., Graziosi, G., Cecchinato, M., & Pasotto, D. (2024). Exploring Variability: Inflammation Mediator Levels across Tissues and Time in Poultry Experimentally Infected by the G1a and G6 Genogroups of Infectious Bursal Disease Virus (IBDV). Animals, 14(11), 1619. https://doi.org/10.3390/ani14111619









