A Functional Single-Nucleotide Polymorphism Upstream of the Collagen Type III Gene Is Associated with Catastrophic Fracture Risk in Thoroughbred Horses
Abstract
Simple Summary
Abstract
1. Introduction
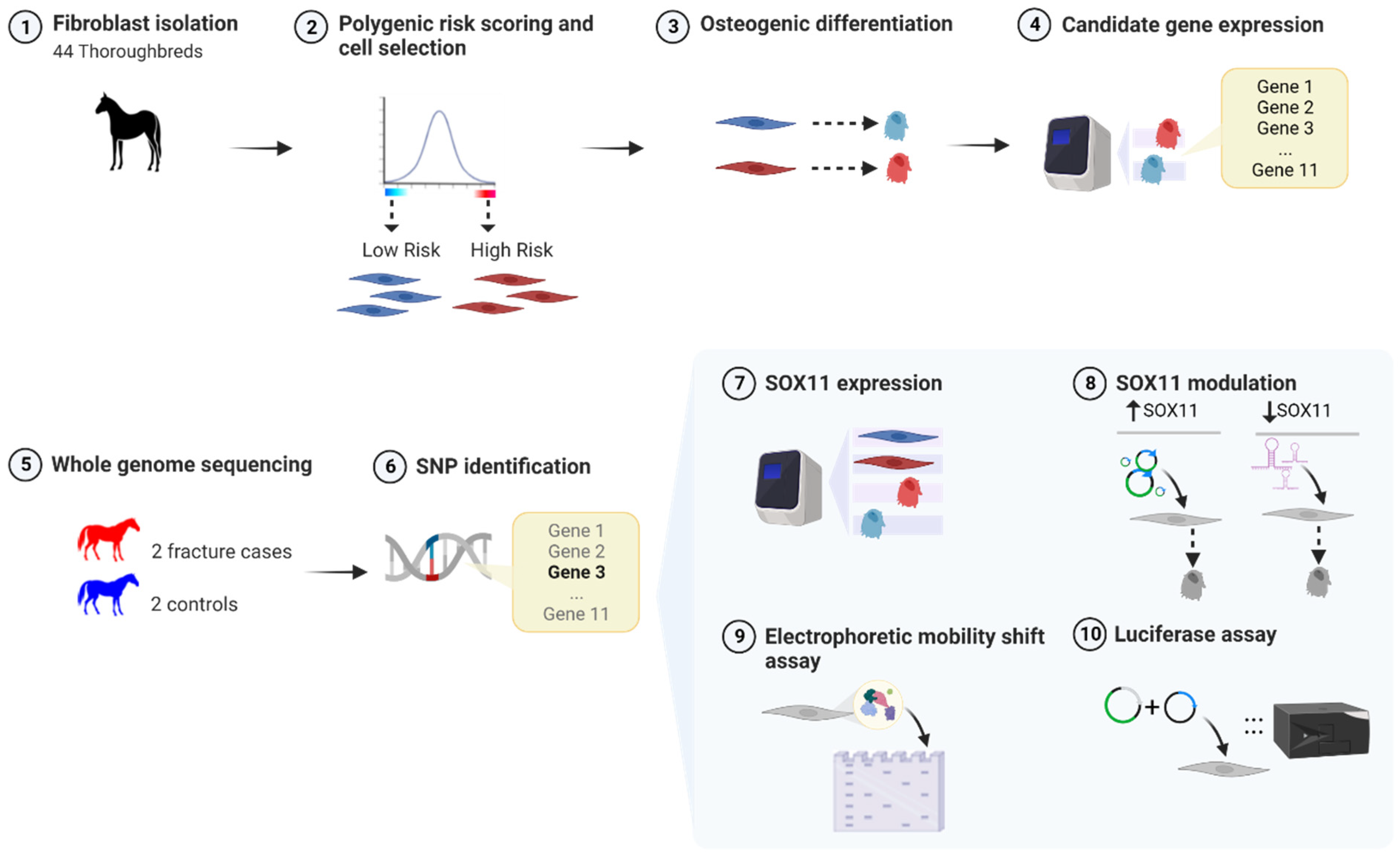
2. Materials and Methods
2.1. Skin Fibroblast Cell Isolation and Culture
2.2. Polygenic Risk Scoring of Thoroughbred Skin Fibroblasts
2.3. Osteoblast Differentiation
2.4. Histological Staining
2.5. RNA Extraction, cDNA Synthesis, and Quantitative PCR
2.6. Immunocytochemistry
2.7. Whole-Genome Sequencing
2.8. Genotyping for the COL3A1 SNP
2.9. Overexpression of SOX11
2.10. SOX11 Knockdown
2.11. ELISA
2.12. Electrophoretic Mobility Shift Assay (EMSA)
2.13. Construction of the Luciferase Reporter Plasmids
2.14. Luciferase Assays
2.15. Statistical Analysis
3. Results
3.1. Application of a Polygenic Risk Score to Select Thoroughbred Cells for Use
3.2. Skin Fibroblasts Can Differentiate into Osteoblast-like Cells In Vitro
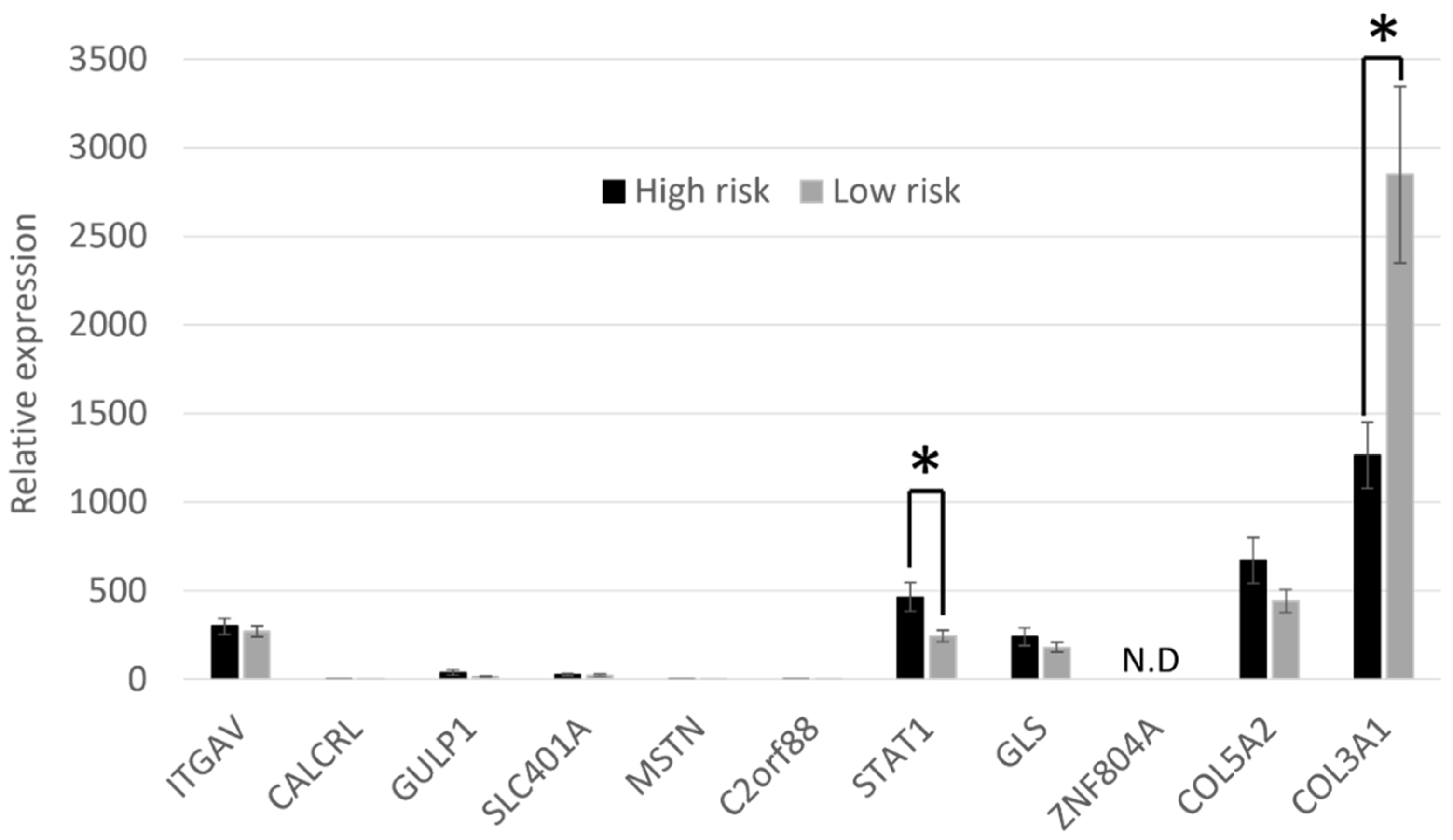
3.3. Candidate Gene Expression Analysis Reveals a Significant Difference in STAT1 and COL3A1 Gene Expression in Skin-Derived Osteoblasts from High- and Low-Risk Thoroughbred Horses
3.4. WGS Revealed a SNP in the Region Upstream of COL3A1 That Is Predicted to Result in the Loss of a SOX11 Binding Site and Is Significantly Associated with Fracture Risk
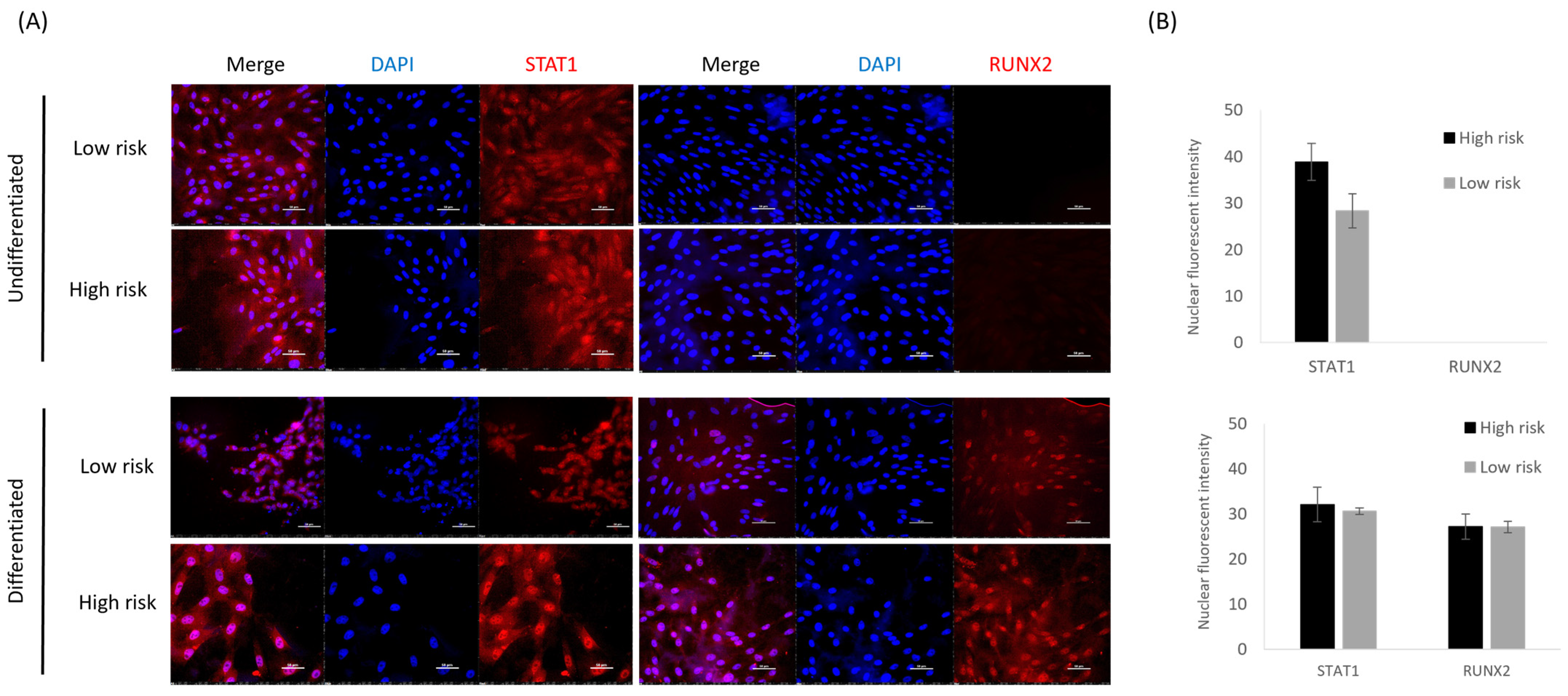
3.5. SOX11 Is Expressed in Undifferentiated and Osteoblast-Differentiated Skin Fibroblasts but Is Not Differentially Expressed between High- and Low-Risk Horses
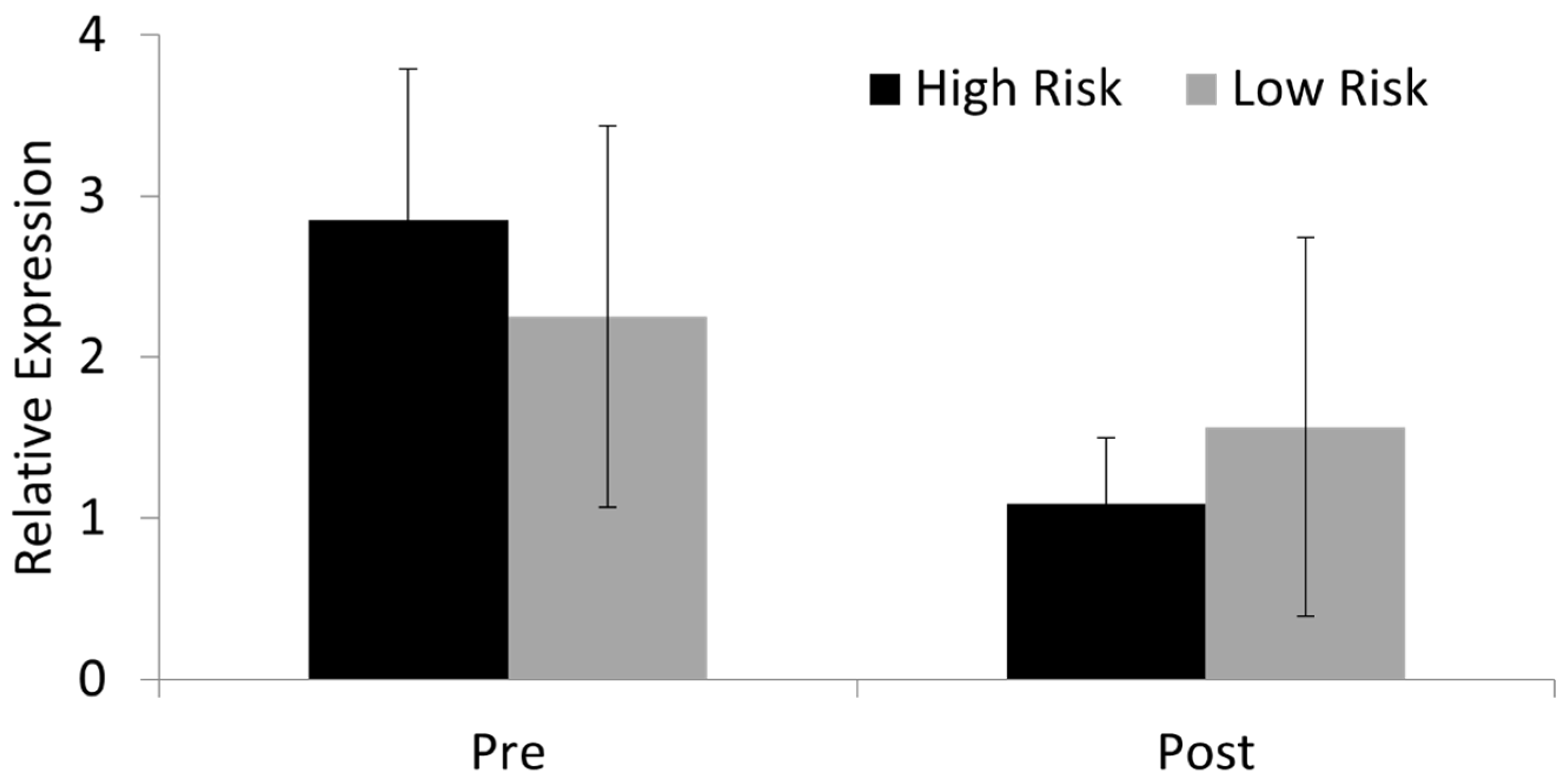
3.6. SOX11 Modulation in Skin Fibroblasts Results in Significant Changes in the Expression of COL3A1
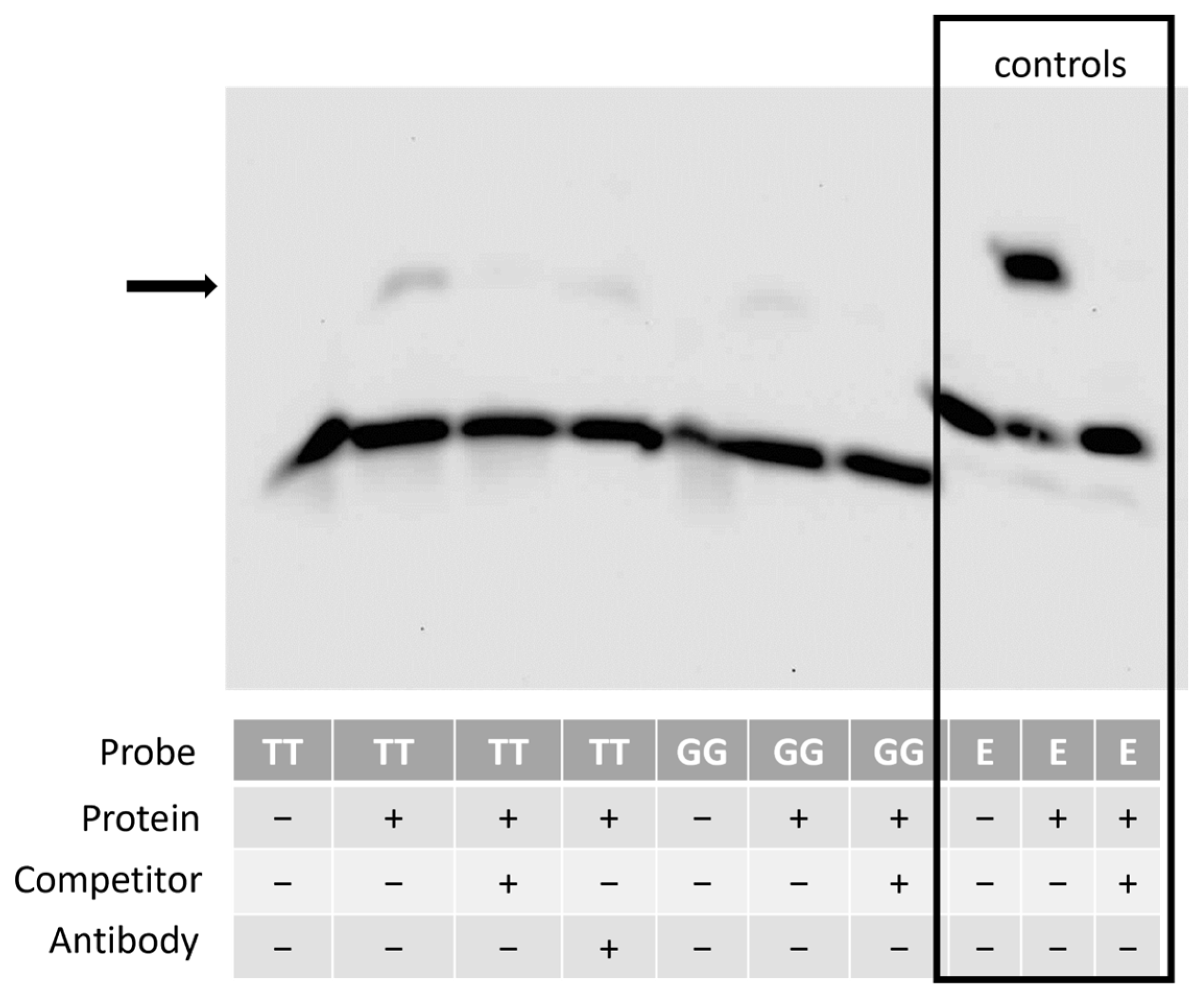
3.7. The Region Containing the SNP Binds to Nuclear Proteins from Equine Cells
3.8. The Region Containing the SNP Has Promoter Activity, and the SNP Affects Reporter Gene Expression in a Genetic-Background-Dependent Manner
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Parkin, T.D.; Clegg, P.D.; French, N.P.; Proudman, C.J.; Riggs, C.M.; Singer, E.R.; Webbon, P.M.; Morgan, K.L. Risk of fatal distal limb fractures among Thoroughbreds involved in the five types of racing in the United Kingdom. Vet. Rec. 2004, 154, 493–497. [Google Scholar] [CrossRef] [PubMed]
- McKee, S.L. An update on racing fatalities in the UK. Equine Vet. Ed. 1995, 7, 202–204. [Google Scholar] [CrossRef]
- Georgopoulos, S.P.; Parkin, T.D. Risk factors for equine fractures in Thoroughbred flat racing in North America. Prev. Vet. Med. 2017, 139, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Parkin, T.D.; Clegg, P.D.; French, N.P.; Proudman, C.J.; Riggs, C.M.; Singer, E.R.; Webbon, P.M.; Morgan, K.L. Race- and course-level risk factors for fatal distal limb fracture in racing Thoroughbreds. Equine Vet. J. 2004, 36, 521–526. [Google Scholar] [CrossRef] [PubMed]
- Verheyen, K.; Price, J.; Lanyon, L.; Wood, J.L.N. Exercise distance and speed affect the risk of fracture in racehorses. Bone 2006, 39, 1322–1330. [Google Scholar] [CrossRef] [PubMed]
- Anthenill, L.A.; Stover, S.M.; Gardner, I.A.; Hill, A.E. Risk factors for proximal sesamoid bone fractures associated with exercise history and horseshoe characteristics in Thoroughbred racehorses. AJVR 2007, 68, 760–771. [Google Scholar] [CrossRef] [PubMed]
- Kristoffersen, M.; Parkin, T.D.; Singer, E.R. Catastrophic biaxial proximal sesamoid bone fractures in UK Thoroughbred races (1994–2004): Horse characteristics and racing history. Equine Vet. J. 2010, 45, 420–424. [Google Scholar] [CrossRef]
- Welsh, C.E.; Lewis, T.W.; Blott, S.C.; Mellor, D.J.; Stirk, A.J.; Parkin, T.D. Estimates of genetic parameters of distal limb fracture and superficial digital flexor tendon injury in UK Thoroughbred racehorses. Vet. J. 2014, 200, 253–256. [Google Scholar] [CrossRef][Green Version]
- Blott, S.C.; Swinburne, J.E.; Sibbons, C.; Fox-Clipsham, L.Y.; Helwegen, M.; Hillyer, L.; Parkin, T.D.; Newton, J.R.; Vaudin, M. A genome-wide association study demonstrates significant genetic variation for fracture risk in Thoroughbred racehorses. BMC Genom. 2014, 15, 147. [Google Scholar] [CrossRef]
- Blott, S.C.; Swinburne, J.E. Predictive Method for Bone Fracture Risk in Horses and Humans. 2015. Available online: https://patentscope.wipo.int/search/en/detail.jsf?docId=WO2015019097 (accessed on 4 May 2023).
- Tozaki, T.; Kusano, K.; Ishikawa, Y.; Kushiro, A.; Nomura, M.; Kikuchi, M.; Kakoi, H.; Hirota, K.; Miyake, T.; Hill, E.W.; et al. A candidate-SNP retrospective cohort study for fracture risk in Japanese Thoroughbred racehorses. Anim. Genet. 2020, 51, 43–50. [Google Scholar] [CrossRef]
- Bulathsinhala, L.; Hughes, J.M.; McKinnon, C.J.; Kardouni, J.R.; Guerriere, K.I.; Popp, K.L.; Matheny, R.W., Jr.; Bouxsein, M.L. Risk of Stress Fracture Varies by Race/Ethnic Origin in a Cohort Study of 1.3 Million US Army Soldiers. J. Bone Miner. Res. 2017, 32, 1546–1553. [Google Scholar] [CrossRef] [PubMed]
- Korvala, J.; Hartikka, H.; Pihlajamaki, H.; Solovieva, S.; Ruohola, J.P.; Sahi, T.; Barral, S.; Ott, J.; Ala-Kokko, L.; Mannikko, M. Genetic predisposition for femoral neck stress fractures in military conscripts. BMC Genet. 2010, 11, 95. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Chang, Q.; Huang, T.; Huang, C. Prospective cohort study of the risk factors for stress fractures in Chinese male infantry recruits. J. Int. Med. Res. 2016, 44, 787–795. [Google Scholar] [CrossRef] [PubMed]
- Varley, I.; Greeves, J.P.; Sale, C.; Friedman, E.; Moran, D.S.; Yanovich, R.; Wilson, P.J.; Gartland, A.; Hughes, D.C.; Stellingwerff, T.; et al. Functional polymorphisms in the P2X7 receptor gene are associated with stress fracture injury. Purinergic Signal 2016, 12, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Warden, S.J.; Burr, D.B.; Brukner, P.D. Stress fractures: Pathophysiology, epidemiology, and risk factors. Curr. Osteoporos. Rep. 2006, 4, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Warden, S.J.; Creaby, M.W.; Bryant, A.L.; Crossley, K.M. Stress fracture risk factors in female football players and their clinical implications. Br. J. Sports Med. 2007, 41 (Suppl. 1), i38–i43. [Google Scholar] [CrossRef] [PubMed]
- Long, P.; Samnakay, P.; Jenner, P.; Rose, S. A yeast two-hybrid screen reveals that osteopontin associates with MAP1A and MAP1B in addition to other proteins linked to microtubule stability, apoptosis and protein degradation in the human brain. Eur. J. Neurosci. 2012, 36, 2733–2742. [Google Scholar] [CrossRef]
- Logan, N.J.; Camman, M.; Williams, G.; Higgins, C.A. Demethylation of ITGAV accelerates osteogenic differentiation in a blast-induced heterotopic ossification in vitro cell culture model. Bone 2018, 117, 149–160. [Google Scholar] [CrossRef]
- Wang, L.; Shi, X.; Zhao, R.; Halloran, B.P.; Clark, D.J.; Jacobs, C.R.; Kingery, W.S. Calcitonin-gene-related peptide stimulates stromal cell osteogenic differentiation and inhibits RANKL induced NF-kappaB activation, osteoclastogenesis and bone resorption. Bone 2010, 46, 1369–1379. [Google Scholar] [CrossRef]
- Park, G.; Park, S.-Y.; Lee, E.-H.; Lee, Y.-J.; Kim, S.-Y.; Kim, S.-H.; Kim, Y.-H.; Kim, I.-S.; Kim, J.-E. High trabecular bone mass induced by reduced function of osteoclasts in GULP1-deficient mice. In Proceedings of the European Calcified Tissue Society Conference ECTS, Rome, Italy, 14–17 May 2016. [Google Scholar]
- Volk, S.W.; Shah, S.R.; Cohen, A.J.; Wang, Y.; Brisson, B.K.; Vogel, L.K.; Hankenson, K.D.; Adams, S.L. Type III collagen regulates osteoblastogenesis and the quantity of trabecular bone. Calcif. Tissue Int. 2014, 94, 621–631. [Google Scholar] [CrossRef]
- Hong, D.; Chen, H.X.; Yu, H.Q.; Liang, Y.; Wang, C.; Lian, Q.Q.; Deng, H.T.; Ge, R.S. Morphological and proteomic analysis of early stage of osteoblast differentiation in osteoblastic progenitor cells. Exp. Cell Res. 2010, 316, 2291–2300. [Google Scholar] [CrossRef]
- Pereira, M.; Ko, J.-H.; Logan, J.; Protheroe, H.; Kim, K.-B.; Tan, A.L.M.; Croucher, P.I.; Park, K.-S.; Rotival, M.; Petretto, E.; et al. A trans-eQTL network regulates osteoclast multinucleation and bone mass. eLife 2020, 9, e55549. [Google Scholar] [CrossRef]
- Wu, L.-F.; Zhu, D.-C.; Wang, B.-H.; Lu, Y.-H.; He, P.; Zhang, Y.-H.; Gao, H.-Q.; Zhu, X.-W.; Xia, W.; Zhu, H.; et al. Relative abundance of mature myostatin rather than total myostatin is negatively associated with bone mineral density in Chinese. J. Cell. Mol. Med. 2018, 22, 1329–1336. [Google Scholar] [CrossRef]
- Bialek, P.; Parkington, J.; Li, X.; Gavin, D.; Wallace, C.; Zhang, J.; Root, A.; Yan, G.; Warner, L.; Seeherman, H.J.; et al. A myostatin and activin decoy receptor enhances bone formation in mice. Bone 2014, 60, 162–171. [Google Scholar] [CrossRef]
- Burger, M.G.; Steinitz, A.; Geurts, J.; Pippenger, B.E.; Schaefer, D.J.; Martin, I.; Barbero, A.; Pelttari, K. Ascorbic Acid Attenuates Senescence of Human Osteoarthritic Osteoblasts. Int. J. Mol. Sci. 2017, 18, 2517. [Google Scholar] [CrossRef]
- Kim, S.; Koga, T.; Isobe, M.; Kern, B.E.; Yokochi, T.; Chin, Y.E.; Karsenty, G.; Taniguchi, T.; Takayanagi, H. Stat1 functions as a cytoplasmic attenuator of Runx2 in the transcriptional program of osteoblast differentiation. Genes Dev. 2003, 17, 1979–1991. [Google Scholar] [CrossRef]
- Tajima, K.; Takaishi, H.; Takito, J.; Tohmonda, T.; Yoda, M.; Ota, N.; Kosaki, N.; Matsumoto, M.; Ikegami, H.; Nakamura, T.; et al. Inhibition of STAT1 accelerates bone fracture healing. J. Orthop. Res. 2010, 28, 937–941. [Google Scholar] [CrossRef]
- Xiao, L.; Naganawa, T.; Obugunde, E.; Gronowicz, G.; Ornitz, D.M.; Coffin, J.D.; Hurley, M.M. Stat1 controls postnatal bone formation by regulating fibroblast growth factor signaling in osteoblasts. J. Biol. Chem. 2004, 279, 27743–27752. [Google Scholar] [CrossRef]
- Yu, Y.; Newman, H.; Shen, L.; Sharma, D.; Hu, G.; Mirando, A.J.; Zhang, H.; Knudsen, E.; Zhang, G.-F.; Hilton, M.J.; et al. Glutamine Metabolism Regulates Proliferation and Lineage Allocation in Skeletal Stem Cells. Cell Metab. 2019, 29, 966–978.e964. [Google Scholar] [CrossRef]
- Groza, T.; Gomez, F.L.; Mashhadi, H.H.; Muñoz-Fuentes, V.; Gunes, O.; Wilson, R.; Cacheiro, P.; Frost, A.; Keskivali-Bond, P.; Vardal, B.; et al. The International Mouse Phenotyping Consortium: Comprehensive knockout phenotyping underpinning the study of human disease. Nucleic Acids Res. 2022, 51, D1038–D1045. [Google Scholar] [CrossRef]
- Flint, J.; Mackay, T.F. Genetic architecture of quantitative traits in mice, flies, and humans. Genome Res. 2009, 19, 723–733. [Google Scholar] [CrossRef]
- Li, Z.-k.; Zhou, Q. Cellular models for disease exploring and drug screening. Protein Cell 2010, 1, 355–362. [Google Scholar] [CrossRef][Green Version]
- Park, I.-H.; Arora, N.; Huo, H.; Maherali, N.; Ahfeldt, T.; Shimamura, A.; Lensch, M.W.; Cowan, C.; Hochedlinger, K.; Daley, G.Q. Disease-specific induced pluripotent stem (iPS) cells. Cell 2008, 134, 877–886. [Google Scholar] [CrossRef]
- Barral, S.; Kurian, M.A. Utility of Induced Pluripotent Stem Cells for the Study and Treatment of Genetic Diseases: Focus on Childhood Neurological Disorders. Front. Mol. Neurosci. 2016, 9, 78. [Google Scholar] [CrossRef]
- Chamberlain, S.J. Disease modelling using human iPSCs. Hum. Mol. Genet. 2016, 25, R173–R181. [Google Scholar] [CrossRef]
- Dobrindt, K.; Zhang, H.; Das, D.; Abdollahi, S.; Prorok, T.; Ghosh, S.; Weintraub, S.; Genovese, G.; Powell, S.K.; Lund, A.; et al. Publicly Available hiPSC Lines with Extreme Polygenic Risk Scores for Modeling Schizophrenia. Complex. Psychiatry 2021, 6, 68–82. [Google Scholar] [CrossRef]
- Coleman, J.R.I. Feasibility and application of polygenic score analysis to the morphology of human-induced pluripotent stem cells. Mol. Genet. Genom. 2022, 297, 1111–1122. [Google Scholar] [CrossRef]
- Choi, S.W.; Mak, T.S.-H.; O’Reilly, P.F. Tutorial: A guide to performing polygenic risk score analyses. Nat. Protoc. 2020, 15, 2759–2772. [Google Scholar] [CrossRef]
- Chung, W. Statistical models and computational tools for predicting complex traits and diseases. Genom. Inform. 2021, 19, e36. [Google Scholar] [CrossRef]
- Yang, J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real time quantitative PCR and the 2-deltadeltaCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Van der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; Del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From FastQ data to high confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinform. 2013, 43, 11.10.11–11.10.33. [Google Scholar] [CrossRef]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.S.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef]
- Gupta, S.; Stamatoyannopoulos, J.A.; Bailey, T.L.; Noble, W.S. Quantifying similarity between motifs. Genome Biol. 2007, 8, R24. [Google Scholar] [CrossRef]
- Jagannathan, V.; Gerber, V.; Rieder, S.; Tetens, J.; Thaller, G.; Drögemüller, C.; Leeb, T. Comprehensive characterization of horse genome variation by whole-genome sequencing of 88 horses. Anim. Genet. 2019, 50, 74–77. [Google Scholar] [CrossRef]
- Tozaki, T.; Ohnuma, A.; Kikuchi, M.; Ishige, T.; Kakoi, H.; Hirota, K.-I.; Kusano, K.; Nagata, S.-I. Rare and common variant discovery by whole-genome sequencing of 101 Thoroughbred racehorses. Sci. Rep. 2021, 11, 16057. [Google Scholar] [CrossRef]
- Durward-Akhurst, S.A.; Schaefer, R.J.; Grantham, B.; Carey, W.K.; Mickelson, J.R.; McCue, M.E. Genetic Variation and the Distribution of Variant Types in the Horse. Front. Genet. 2021, 12, 758366. [Google Scholar] [CrossRef]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef]
- Luo, Y.; Hitz, B.C.; Gabdank, I.; Hilton, J.A.; Kagda, M.S.; Lam, B.; Myers, Z.; Sud, P.; Jou, J.; Lin, K.; et al. New developments on the Encyclopedia of DNA Elements (ENCODE) data portal. Nucleic Acids Res. 2020, 48, D882–D889. [Google Scholar] [CrossRef]
- Liew, C.-G.; Draper, J.S.; Walsh, J.; Moore, H.; Andrews, P.W. Transient and Stable Transgene Expression in Human Embryonic Stem Cells. Stem Cells 2007, 25, 1521–1528. [Google Scholar] [CrossRef]
- Palomino Lago, E.; Jelbert, E.R.; Baird, A.; Lam, P.Y.; Guest, D.J. Equine induced pluripotent stem cells are responsive to inflammatory cytokines before and after differentiation into musculoskeletal cell types. In Vitr. Cell. Dev. Biol. Anim. 2023, 59, 514–527. [Google Scholar] [CrossRef]
- Smith, E.J.; Beaumont, R.E.; McClellan, A.; Sze, C.; Palomino Lago, E.; Hazelgrove, L.; Dudhia, J.; Smith, R.K.W.; Guest, D.J. Tumour necrosis factor alpha, interleukin 1 beta and interferon gamma have detrimental effects on equine tenocytes that cannot be rescued by IL-1RA or mesenchymal stromal cell-derived factors. Cell Tissue Res. 2023, 391, 523–544. [Google Scholar] [CrossRef]
- Cooper, G.M.; Stone, E.A.; Asimenos, G.; Green, E.D.; Batzoglou, S.; Sidow, A. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005, 15, 901–913. [Google Scholar] [CrossRef]
- Haseeb, A.; Lefebvre, V. The SOXE transcription factors-SOX8, SOX9 and SOX10-share a bi-partite transactivation mechanism. Nucleic Acids Res. 2019, 47, 6917–6931. [Google Scholar] [CrossRef]
- Haseeb, A.; Kc, R.; Angelozzi, M.; de Charleroy, C.; Rux, D.; Tower, R.J.; Yao, L.; Pellegrino da Silva, R.; Pacifici, M.; Qin, L.; et al. SOX9 keeps growth plates and articular cartilage healthy by inhibiting chondrocyte dedifferentiation/osteoblastic redifferentiation. Proc. Natl. Acad. Sci. USA 2021, 118, e2019152118. [Google Scholar] [CrossRef]
- Dy, P.; Penzo-Méndez, A.; Wang, H.; Pedraza, C.E.; Macklin, W.B.; Lefebvre, V. The three SoxC proteins--Sox4, Sox11 and Sox12--exhibit overlapping expression patterns and molecular properties. Nucleic Acids Res. 2008, 36, 3101–3117. [Google Scholar] [CrossRef]
- Gadi, J.; Jung, S.H.; Lee, M.J.; Jami, A.; Ruthala, K.; Kim, K.M.; Cho, N.H.; Jung, H.S.; Kim, C.H.; Lim, S.K. The transcription factor protein Sox11 enhances early osteoblast differentiation by facilitating proliferation and the survival of mesenchymal and osteoblast progenitors. J. Biol. Chem. 2013, 288, 25400–25413. [Google Scholar] [CrossRef] [PubMed]
- Kassem, M.; Ankersen, L.; Eriksen, E.F.; Clark, B.F.C.; Rattan, S.I.S. Demonstration of cellular aging and senescence in serially passaged long-term cultures of human trabecular osteoblasts. Osteoporos. Int. 1997, 7, 514–524. [Google Scholar] [CrossRef]
- Lorenz, K.; Sicker, M.; Schmelzer, E.; Rupf, T.; Salvetter, J.; Schulz-Siegmund, M.; Bader, A. Multilineage differentiation potential of human dermal skin-derived fibroblasts. Exp. Dermatol. 2008, 17, 925–932. [Google Scholar] [CrossRef]
- Halcsik, E.; Forni, M.F.; Fujita, A.; Verano-Braga, T.; Jensen, O.N.; Sogayar, M.C. New insights in osteogenic differentiation revealed by mass spectrometric assessment of phosphorylated substrates in murine skin mesenchymal cells. BMC Cell Biol. 2013, 14, 47. [Google Scholar] [CrossRef]
- Glynn, E.R.; Londono, A.S.; Zinn, S.A.; Hoagland, T.A.; Govoni, K.E. Culture conditions for equine bone marrow mesenchymal stem cells and expression of key transcription factors during their differentiation into osteoblasts. J. Anim. Sci. Biotechnol. 2013, 4, 40. [Google Scholar] [CrossRef]
- Nagy, K.; Sung, H.-K.; Zhang, P.; Laflamme, S.; Vincent, P.; Agha-Mohammadi, S.; Woltjen, K.; Monetti, C.; Michael, I.P.; Smith, L.C.; et al. Induced pluripotent stem cell lines derived from equine fibroblasts. Stem Cell Rev. Rep. 2011, 7, 693–702. [Google Scholar] [CrossRef]
- Bavin, E.P.; Smith, O.; Baird, A.E.; Smith, L.C.; Guest, D.J. Equine Induced Pluripotent Stem Cells have a Reduced Tendon Differentiation Capacity Compared to Embryonic Stem Cells. Front. Vet. Sci. 2015, 2, 55. [Google Scholar] [CrossRef]
- Sharma, R.; Livesey, M.R.; Wyllie, D.J.; Proudfoot, C.; Whitelaw, B.; Hay, D.C.; Donadeu, X. Generation of functional neurons from feeder-free, keratinocyte-derived equine induced pluripotent stem cells. Stem Cells Dev. 2014, 23, 1524–1534. [Google Scholar] [CrossRef]
- Sun, T.C.; Riggs, C.M.; Cogger, N.; Wright, J.; Al-Alawneh, J.I. Noncatastrophic and catastrophic fractures in racing Thoroughbreds at the Hong Kong Jockey Club. Equine Vet. J. 2019, 51, 77–82. [Google Scholar] [CrossRef]
- Baird, A.; Lindsay, T.; Everett, A.; Iyemere, V.; Paterson, Y.Z.; McClellan, A.; Henson, F.M.D.; Guest, D.J. Osteoblast differentiation of equine induced pluripotent stem cells. Biol. Open 2018, 7, bio033514. [Google Scholar] [CrossRef]
- Guest, D.J.; Ousey, J.C.; Smith, M.R.W. Defining the expression of marker genes in equine mesenchymal stromal cells. Stem Cells Cloning Adv. Appl. 2008, 1, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Burk, J.; Ribitsch, I.; Gittel, C.; Juelke, H.; Kasper, C.; Staszyk, C.; Brehm, W. Growth and differentiation characteristics of equine mesenchymal stromal cells derived from different sources. Vet. J. 2013, 195, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Radcliffe, C.H.; Flaminio, J.B.F.; Fortier, L.A. Temporal analysis of equine bone marrow aspirate during establishment of putative mesenchymal progenitor cell populations. Stem Cells Dev. 2010, 19, 269–282. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.-H.; Golub, E.E.; Forbes, E.; Tokuoka, T.; Shapiro, I.M. Mechanism of action of β-glycerophosphate on bone cell mineralization. Calcif. Tissue Int. 1992, 51, 305–311. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Toyosawa, S.; Furuichi, T.; Kanatani, N.; Yoshida, C.; Liu, Y.; Himeno, M.; Narai, S.; Yamaguchi, A.; Komori, T. Overexpression of Cbfa1 in osteoblasts inhibits osteoblast maturation and causes osteopenia with multiple fractures. J. Cell Biol. 2001, 155, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Yen, J.L.; Lin, S.P.; Chen, M.R.; Niu, D.M. Clinical features of Ehlers-Danlos syndrome. J. Formos. Med. Assoc. Taiwan Yi Zhi 2006, 105, 475–480. [Google Scholar] [CrossRef] [PubMed]
- Kingsley, N.B.; Hamilton, N.A.; Lindgren, G.; Orlando, L.; Bailey, E.; Brooks, S.; McCue, M.; Kalbfleisch, T.S.; MacLeod, J.N.; Petersen, J.L.; et al. “Adopt-a-Tissue” Initiative Advances Efforts to Identify Tissue-Specific Histone Marks in the Mare. Front. Genet. 2021, 12, 649959. [Google Scholar] [CrossRef]
- Huber, C.D.; Kim, B.Y.; Lohmueller, K.E. Population genetic models of GERP scores suggest pervasive turnover of constrained sites across mammalian evolution. PLoS Genet. 2020, 16, e1008827. [Google Scholar] [CrossRef]
- Beisser, A.L.; McClure, S.; Wang, C.; Soring, K.; Garrison, R.; Peckham, B. Evaluation of catastrophic musculoskeletal injuries in Thoroughbreds and Quarter Horses at three Midwestern racetracks. J. Am. Vet. Med. Assoc. 2011, 239, 1236–1241. [Google Scholar] [CrossRef]
- Kido, T.; Sikora-Wohlfeld, W.; Kawashima, M.; Kikuchi, S.; Kamatani, N.; Patwardhan, A.; Chen, R.; Sirota, M.; Kodama, K.; Hadley, D.; et al. Are minor alleles more likely to be risk alleles? BMC Med. Genom. 2018, 11, 3. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Cox, N.J. The allelic architecture of human disease genes: Common disease-common variant...or not? Hum. Mol. Genet. 2002, 11, 2417–2423. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K. Are rare variants responsible for susceptibility to complex diseases? Am. J. Hum. Genet. 2001, 69, 124–137. [Google Scholar] [CrossRef] [PubMed]
- Gordon, A.R.; Outram, S.V.; Keramatipour, M.; Goddard, C.A.; Colledge, W.H.; Metcalfe, J.C.; Hager-Theodorides, A.L.; Crompton, T.; Kemp, P.R. Splenomegaly and modified erythropoiesis in KLF13-/-mice. J. Biol. Chem. 2008, 283, 11897–11904. [Google Scholar] [CrossRef] [PubMed]
- Yao, W.; Jiao, Y.; Zhou, Y.; Luo, X. KLF13 suppresses the proliferation and growth of colorectal cancer cells through transcriptionally inhibiting HMGCS1-mediated cholesterol biosynthesis. Cell Biosci. 2020, 10, 76. [Google Scholar] [CrossRef]
- Lavallée, G.; Andelfinger, G.; Nadeau, M.; Lefebvre, C.; Nemer, G.; Horb, M.E.; Nemer, M. The Kruppel-like transcription factor KLF13 is a novel regulator of heart development. EMBO J. 2006, 25, 5201–5213. [Google Scholar] [CrossRef] [PubMed]
- Leclerc, N.; Luppen, C.A.; Ho, V.V.; Nagpal, S.; Hacia, J.G.; Smith, E.; Frenkel, B. Gene expression profiling of glucocorticoid-inhibited osteoblasts. J. Mol. Endocrinol. 2004, 33, 175–193. [Google Scholar] [CrossRef] [PubMed]
- Martin, K.M.; Metcalfe, J.C.; Kemp, P.R. Expression of Klf9 and Klf13 in mouse development. Mech. Dev. 2001, 103, 149–151. [Google Scholar] [CrossRef]
- Xu, L.; Huang, S.; Hou, Y.; Liu, Y.; Ni, M.; Meng, F.; Wang, K.; Rui, Y.; Jiang, X.; Li, G. Sox11-modified mesenchymal stem cells (MSCs) accelerate bone fracture healing: Sox11 regulates differentiation and migration of MSCs. FASEB J. 2015, 29, 1143–1152. [Google Scholar] [CrossRef]
- Vogel, C.; Marcotte, E.M. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 2012, 13, 227–232. [Google Scholar] [CrossRef]
- Qin, J.Y.; Zhang, L.; Clift, K.L.; Hulur, I.; Xiang, A.P.; Ren, B.-Z.; Lahn, B.T. Systematic Comparison of Constitutive Promoters and the Doxycycline-Inducible Promoter. PLoS ONE 2010, 5, e10611. [Google Scholar] [CrossRef]
- Wiebe, M.S.; Nowling, T.K.; Rizzino, A. Identification of novel domains within Sox-2 and Sox-11 involved in autoinhibition of DNA binding and partnership specificity. J. Biol. Chem. 2003, 278, 17901–17911. [Google Scholar] [CrossRef] [PubMed]
- Kan, A.; Ikeda, T.; Fukai, A.; Nakagawa, T.; Nakamura, K.; Chung, U.-I.; Kawaguchi, H.; Tabin, C.J. SOX11 contributes to the regulation of GDF5 in joint maintenance. BMC Dev. Biol. 2013, 13, 4. [Google Scholar] [CrossRef] [PubMed]
- Bragdon, B.C.; Bahney, C.S. Origin of Reparative Stem Cells in Fracture Healing. Curr. Osteoporos. Rep. 2018, 16, 490–503. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Palomino Lago, E.; Baird, A.; Blott, S.C.; McPhail, R.E.; Ross, A.C.; Durward-Akhurst, S.A.; Guest, D.J. A Functional Single-Nucleotide Polymorphism Upstream of the Collagen Type III Gene Is Associated with Catastrophic Fracture Risk in Thoroughbred Horses. Animals 2024, 14, 116. https://doi.org/10.3390/ani14010116
Palomino Lago E, Baird A, Blott SC, McPhail RE, Ross AC, Durward-Akhurst SA, Guest DJ. A Functional Single-Nucleotide Polymorphism Upstream of the Collagen Type III Gene Is Associated with Catastrophic Fracture Risk in Thoroughbred Horses. Animals. 2024; 14(1):116. https://doi.org/10.3390/ani14010116
Chicago/Turabian StylePalomino Lago, Esther, Arabella Baird, Sarah C. Blott, Rhona E. McPhail, Amy C. Ross, Sian A. Durward-Akhurst, and Deborah J. Guest. 2024. "A Functional Single-Nucleotide Polymorphism Upstream of the Collagen Type III Gene Is Associated with Catastrophic Fracture Risk in Thoroughbred Horses" Animals 14, no. 1: 116. https://doi.org/10.3390/ani14010116
APA StylePalomino Lago, E., Baird, A., Blott, S. C., McPhail, R. E., Ross, A. C., Durward-Akhurst, S. A., & Guest, D. J. (2024). A Functional Single-Nucleotide Polymorphism Upstream of the Collagen Type III Gene Is Associated with Catastrophic Fracture Risk in Thoroughbred Horses. Animals, 14(1), 116. https://doi.org/10.3390/ani14010116







