In Silico Analysis of miRNA-Mediated Genes in the Regulation of Dog Testes Development from Immature to Adult Form
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. miRNA Data Profiling
2.2. Conserved Nucleotide Sequences
2.3. Identification of Target Genes of Differentially Expressed miRNAs
2.4. Gene Ontology Enrichment and KEGG Analysis
2.5. Identification and Analysis of Hub Gene
2.6. Real-Time Polymerase Chain Reaction for Determination of mRNA Expression of Hub Genes
2.7. Protein Immunoblots
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- DeFalco, T.; Capel, B. Gonad morphogenesis in vertebrates: Divergent means to a convergent end. Annu. Rev. Cell Dev. Biol. 2009, 25, 457–482. [Google Scholar] [CrossRef]
- Piprek, R.P. Molecular and cellular machinery of gonadal differentiation in mammals. Int. J. Dev. Biol. 2010, 54, 779–786. [Google Scholar] [CrossRef]
- Carletti, M.Z.; Christenson, L.K. MicroRNA in the ovary and female reproductive tract. J. Anim. Sci. 2009, 87 (Suppl. S14), E29–E38. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Kokkinaki, M.; Pant, D.; Gallicano, G.I.; Dym, M. Small RNA molecules in the regulation of spermatogenesis. Reproduction 2009, 137, 901–911. [Google Scholar] [CrossRef]
- Kotaja, N. MicroRNAs and spermatogenesis. Fertil. Steril. 2014, 101, 1552–1562. [Google Scholar] [CrossRef]
- Hu, J.; Sun, F.; Handel, M.A. Nuclear localization of EIF4G3 suggests a role for the XY body in translational regulation during spermatogenesis in mice. Biol. Reprod. 2018, 98, 102–114. [Google Scholar] [CrossRef] [PubMed]
- Yan, N.; Lu, Y.; Sun, H.; Tao, D.; Zhang, S.; Liu, W.; Ma, Y. Microarray for microRNA profiling in mouse testis tissues. Reproduction 2007, 134, 73–79. [Google Scholar] [CrossRef]
- Luo, L.; Ye, L.; Liu, G.; Shao, G.; Zheng, R.; Ren, Z.; Zuo, B.; Xu, D.; Lei, M.; Jiang, S.; et al. Microarray-based approach identifies differentially expressed microRNAs in porcine sexually immature and mature testes. PLoS ONE 2010, 5, e11744. [Google Scholar] [CrossRef] [PubMed]
- Lian, C.; Sun, B.; Niu, S.; Yang, R.; Liu, B.; Lu, C.; Meng, J.; Qiu, Z.; Zhang, L.; Zhao, Z. A comparative profile of the microRNA transcriptome in immature and mature porcine testes using Solexa deep sequencing. FEBS J. 2012, 279, 964–975. [Google Scholar] [CrossRef]
- Romero, Y.; Meikar, O.; Papaioannou, M.D.; Conne, B.; Grey, C.; Weier, M.; Pralong, F.; De Massy, B.; Kaessmann, H.; Vassalli, J.D.; et al. Dicer1 depletion in male germ cells leads to infertility due to cumulative meiotic and spermiogenic defects. PLoS ONE 2011, 6, e25241. [Google Scholar] [CrossRef]
- Liu, D.; Li, L.; Fu, H.; Li, S.; Li, J. Inactivation of Dicer1 has a severe cumulative impact on the formation of mature germ cells in mouse testes. Biochem. Biophys. Res. Commun. 2012, 422, 114–120. [Google Scholar] [CrossRef] [PubMed]
- Korhonen, H.M.; Meikar, O.; Yadav, R.P.; Papaioannou, M.D.; Romero, Y.; Da Ros, M.; Herrera, P.L.; Toppari, J.; Nef, S.; Kotaja, N. Dicer is required for haploid male germ cell differentiation in mice. PLoS ONE 2011, 6, e24821. [Google Scholar] [CrossRef] [PubMed]
- Korhonen, H. The Essential Role of DICER in Spermatogenesis and male Fertility. Ph.D. Thesis, University of Turku, Turku, Finland, 2016. [Google Scholar]
- Fu, M.; Xu, K.-H.; Xu, W.-M. Research advances of Dicer in regulating reproductive function. Hereditas 2016, 38, 612–622. [Google Scholar] [CrossRef] [PubMed]
- Yadav, R.P.; Kotaja, N. Small RNAs in spermatogenesis. Mol. Cell. Endocrinol. 2014, 382, 498–508. [Google Scholar] [CrossRef]
- Chen, X.; Li, X.; Guo, J.; Zhang, P.; Zeng, W. The roles of microRNAs in regulation of mammalian spermatogenesis. J. Anim. Sci. Biotechnol. 2017, 8, 35. [Google Scholar] [CrossRef] [PubMed]
- Ro, S.; Park, C.; Sanders, K.M.; McCarrey, J.R.; Yan, W. Cloning and expression profiling of testis-expressed microRNAs. Dev. Biol. 2007, 311, 592–602. [Google Scholar] [CrossRef]
- Tay, Y.; Zhang, J.; Thomson, A.M.; Lim, B.; Rigoutsos, I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature 2008, 455, 1124–1128. [Google Scholar] [CrossRef]
- Lytle, J.R.; Yario, T.A.; Steitz, J.A. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc. Natl. Acad. Sci. USA 2007, 104, 9667–9672. [Google Scholar] [CrossRef]
- Kasimanickam, V.R.; Kasimanickam, R.K. Differential expression of microRNAs in sexually immature and mature canine testes. Theriogenology 2015, 83, 394–398. [Google Scholar] [CrossRef]
- Kasimanickam, V.R.; Kasimanickam, R.K.; Dernell, W.S. Dysregulated microRNA clusters in response to retinoic acid and CYP26B1 inhibitor induced testicular function in dogs. PLoS ONE 2014, 9, e99433. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Kozomara, A.; Griffiths-Jones, S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014, 42, D68–D73. [Google Scholar] [CrossRef]
- Chang, L.; Zhou, G.; Soufan, O.; Xia, J. miRNet 2.0: Network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res. 2020, 48, W244–W251. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Xia, J. MicroRNA Regulatory Network Analysis Using miRNet 2.0. Methods Mol. Biol. 2023, 2594, 185–204. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef] [PubMed]
- Gustavsen, J.A.; Pai, S.; Isserlin, R.; Demchak, B.; Pico, A.R. RCy3: Network biology using Cytoscape from within R. F1000Research 2019, 8, 1774. [Google Scholar] [CrossRef]
- Chin, C.H.; Chen, S.H.; Wu, H.H.; Ho, C.W.; Ko, M.T.; Lin, C.Y. CytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Sys. Biol. 2014, 8 (Suppl. S4), S11. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef]
- Kasimanickam, V.R. Expression of retinoic acid-metabolizing enzymes, ALDH1A1, ALDH1A2, ALDH1A3, CYP26A1, CYP26B1 and CYP26C1 in canine testis during post-natal development. Reprod. Domest. Anim. 2016, 51, 901–909. [Google Scholar] [CrossRef]
- Hayashi, K.; de Sousa Lopes, S.M.C.; Kaneda, M.; Tang, F.; Hajkova, P.; Lao, K.; O’Carroll, D.; Das, P.P.; Tarakhovsky, A.; Miska, E.A.; et al. MicroRNA biogenesis is required for mouse primordial germ cell development and spermatogenesis. PLoS ONE 2008, 3, e1738. [Google Scholar] [CrossRef]
- Comazzetto, S.; Giacomo, M.D.; Rasmussen, K.D.; Much, C.; Azzi, C.; Perlas, E.; Morgan, M.; O’Carroll, D. Oligoasthenoter-atozoospermia and Infertility in Mice Deficient for miR-34b/c and miR-449 Loci. PLoS Genet. 2014, 10, e1004597. [Google Scholar] [CrossRef]
- Yuan, S.; Tang, C.; Zhang, Y.; Wu, J.; Bao, J.; Zheng, H.; Xu, C.; Yan, W. mir-34b/c and mir-449a/b/c are required for spermatogenesis, but not for the first cleavage division in mice. Biol. Open 2015, 4, 212–223. [Google Scholar] [CrossRef] [PubMed]
- Abu-Halima, M.; Backes, C.; Leidinger, P.; Keller, A.; Lubbad, A.M.; Hammadeh, M.; Meese, E. MicroRNA expression profiles in human testicular tissues of infertile men with different histopathologic patterns. Fertil. Steril. 2014, 101, 78–86. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, X.; Mata, A.; Bassas, L.; Larriba, S. Altered miRNA Signature of Developing Germ-cells in Infertile Patients Relates to the Severity of Spermatogenic Failure and Persists in Spermatozoa. Sci. Rep. 2015, 5, 17991. [Google Scholar] [CrossRef]
- Smorag, L.; Zheng, Y.; Nolte, J.; Zechner, U.; Engel, W.; Pantakani, D.V. MicroRNA signature in various cell types of mouse spermatogenesis: Evidence for stage-specifically expressed miRNA-221, -203 and -34b-5p mediated spermatogenesis regulation. Biol. Cell 2012, 104, 677–692. [Google Scholar] [CrossRef] [PubMed]
- Bouhallier, F.; Allioli, N.; Lavial, F.; Chalmel, F.; Perrard, M.H.; Durand, P.; Samarut, J.; Pain, B.; Rouault, J.P. Role of miR-34c microRNA in the late steps of spermatogenesis. RNA 2010, 16, 720–731. [Google Scholar] [CrossRef]
- Abu-Halima, M.; Hammadeh, M.; Schmitt, J.; Leidinger, P.; Keller, A.; Meese, E.; Backes, C. Altered microRNA expression profiles of human spermatozoa in patients with different spermatogenic impairments. Fertil. Steril. 2013, 99, 12491255. [Google Scholar] [CrossRef]
- Zhang, S.; Yu, M.; Liu, C.; Wang, L.; Hu, Y.; Bai, Y.; Hua, J. MIR-34c regulates mouse embryonic stem cells differentiation into male germ-like cells through RARg. Cell Biochem. Funct. 2012, 30, 623–632. [Google Scholar] [CrossRef]
- Cha, Y.H.; Kim, N.H.; Park, C.; Lee, I.; Kim, H.S.; Yook, J.I. MiRNA-34 intrinsically links p53 tumor suppressor and Wnt signaling. Cell Cycle 2012, 11, 1273–1281. [Google Scholar] [CrossRef]
- Lombardi, A.P.; Royer, C.; Pisolato, R.; Cavalcanti, F.N.; Lucas, T.F.; Lazari, M.F.; Porto, C.S. Physiopathological aspects of the Wnt/b-catenin signaling pathway in the male reproductive system. Spermatogenesis 2013, 3, e23181. [Google Scholar] [CrossRef]
- Tanwar, P.S.; Tomoko, K.T.; Lihua, Z.; Poonam, R.; Taketo, M.M.; Jose, T. Constitutive WNT/beta-catenin signaling in murine sertoli cells disrupts their differentiation and ability to support spermatogenesis. Biol. Reprod. 2010, 82, 422. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.D.; Neerja, W.; Neetu, K.; Kanchan, S.; Shankar, P.B.; Majumdar, S.S. Dickkopf homolog 3 (dkk3) plays a crucial role upstream of WNT/β-catenin signaling for sertoli cell mediated regulation of spermatogenesis. PLoS ONE 2013, 8, e63603. [Google Scholar] [CrossRef]
- Rappaport, M.S.; Smith, E.P. Insulin-like growth factor I inhibits aromatization induced by follicle stimulating hormone in rat Sertoli cell culture. Biol. Reprod. 1996, 54, 446–452. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.A.; Lilianne, N.; Lauren, P.; Spicer, L.J.; Davis, J.S. Follicle-stimulating hormone amplifies insulin-like growth factor I-mediated activation of AKT/protein kinase b signaling in immature rat Sertoli cells. Endocrinology 2002, 143, 2259–2267. [Google Scholar] [CrossRef] [PubMed]
- Borland, K.; Mita, M.; Oppenheimer, C.L.; Blinderman, L.A.; Massague, J.; Hall, P.F.; Czech, M.P. The actions of insulin-like growth factors I and II on cultured Sertoli cells. Endocrinology 1984, 114, 240. [Google Scholar] [CrossRef] [PubMed]
- Mullaney, B.P.; Skinner, M.K. Transforming growth factor-beta (beta 1, beta 2, and beta 3) gene expression and action during pubertal development of the seminiferous tubule: Potential role at the onset of spermatogenesis. Mol. Endocrinol. 1993, 7, 67–76. [Google Scholar] [CrossRef]
- Catherine, I.; Loveland, K.L. Smads and cell fate: Distinct roles in specification, development, and tumorigenesis in the testis. IUBMB Life 2013, 65, 85–97. [Google Scholar] [CrossRef]
- Capra, E.; Turri, F.; Lazzari, B.; Cremonesi, P.; Gliozzi, T.M.; Fojadelli, I.; Stella, A.; Pizzi, F. Small RNA sequencing of cryopreserved semen from single bull revealed altered miRNAs and piRNAs expression between High- and Low-motile sperm populations. BMC Genom. 2017, 18, 14. [Google Scholar] [CrossRef]
- Fang, X.; Qin, L.; Yu, H.; Jiang, P.; Xia, L.; Gao, Z.; Yang, R.; Zhao, Y.; Yu, X.; Zhao, Z. Comprehensive Analysis of miRNAs and Target mRNAs between Immature and Mature Testis Tissue in Chinese Red Steppes Cattle. Animals 2021, 11, 3024. [Google Scholar] [CrossRef]
- Fischer, M. Census and evaluation of p53 target genes. Oncogene 2017, 36, 3943–3956. [Google Scholar] [CrossRef]
- Torgovnick, A.; Heger, J.M.; Liaki, V.; Isensee, J.; Schmitt, A.; Knittel, G.; Riabinska, A.; Beleggia, F.; Laurien, L.; Leeser, U.; et al. The Cdkn1aSUPER Mouse as a Tool to Study p53-Mediated Tumor Suppression. Cell Rep. 2018, 25, 1027–1039.e6. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Huang, Y.; Su, Z.; Zhu, Q.; Ge, Y.; Wang, G.; Wang, C.Q.; Mukai, M.; Holsberger, D.R.; Cooke, P.S.; et al. Deficiency of CDKN1A or both CDKN1A and CDKN1B affects the pubertal development of mouse Leydig cells. Biol. Reprod. 2015, 92, 77. [Google Scholar] [CrossRef] [PubMed]
- Hynes, N.E.; Horsch, K.; Olayioye, M.A.; Badache, A. The ErbB receptor tyrosine family as signal integrators. Endocr. Relat. Cancer 2001, 8, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Abé, K.; Eto, K.; Abé, S. Epidermal growth factor mediates spermatogonial proliferation in newt testis. Reprod. Biol. Endocrinol. 2008, 6, 7. [Google Scholar] [CrossRef]
- Chen, J.; Zeng, F.; Forrester, S.J.; Eguchi, S.; Zhang, M.Z.; Harris, R.C. Expression and Function of the Epidermal Growth Factor Receptor in Physiology and Disease. Physiol. Rev. 2016, 96, 1025–1069. [Google Scholar] [CrossRef]
- Castellano, E.; Santos, E. Functional specificity of ras isoforms: So similar but so different. Genes Cancer 2011, 2, 216–231. [Google Scholar] [CrossRef]
- Richards, J.S.; Fan, H.Y.; Liu, Z.; Tsoi, M.; Laguë, M.N.; Boyer, A.; Boerboom, D. Either Kras activation or Pten loss similarly enhance the dominant-stable CTNNB1-induced genetic program to promote granulosa cell tumor development in the ovary and testis. Oncogene 2012, 31, 1504–1520. [Google Scholar] [CrossRef]
- Schneider, M.R.; Yarden, Y. The EGFR-HER2 module: A stem cell approach to understanding a prime target and driver of solid tumors. Oncogene 2016, 35, 2949–2960. [Google Scholar] [CrossRef]
- Marte, B.M.; Downward, J. PKB/Akt: Connecting phosphoinositide 3-kinase to cell survival and beyond. Trends Biochem. Sci. 1997, 22, 355–358. [Google Scholar] [CrossRef] [PubMed]
- Deng, C.Y.; Lv, M.; Luo, B.H.; Zhao, S.Z.; Mo, Z.C.; Xie, Y.J. The role of the PI3K/AKT/mTOR signalling pathway in male reproduction. Curr. Mol. Med. 2021, 21, 539–548. [Google Scholar] [CrossRef]
- McDonald, C.A.; Millena, A.C.; Reddy, S.; Finlay, S.; Vizcarra, J.; Khan, S.A.; Davis, J.S. Follicle-Stimulating Hormone-Induced Aromatase in Immature Rat Sertoli Cells Requires an Active Phosphatidylinositol 3-Kinase Pathway and Is Inhibited via the Mitogen-Activated Protein Kinase Signaling Pathway. Mol. Endocrinol. 2006, 20, 608–618. [Google Scholar] [CrossRef]
- Kim, H.H.; Sierke, S.L.; Koland, J.G. Epidermal growth factor-dependent association of phosphatidylinositol 3-kinase with the erbB3 gene product. J. Biol. Chem. 1994, 269, 24747–24755. [Google Scholar] [CrossRef]
- Ueki, K.; Fruman, D.A.; Brachmann, S.M.; Tseng, Y.H.; Cantley, L.C.; Kahn, C.R. Molecular balance between the regulatory and catalytic subunits of phosphoinositide 3-kinase regulates cell signaling and survival. Mol. Cell Biol. 2002, 22, 965–977. [Google Scholar] [CrossRef] [PubMed]
- Bott, R.C.; McFee, R.M.; Clopton, D.T.; Toombs, C.; Cupp, A.S. Vascular endothelial growth factor and kinase domain Region receptor are involved in both seminiferous cord formation and vascular development during testis morphogenesis in the rat. Biol. Reprod. 2005, 75, 56–67. [Google Scholar] [CrossRef] [PubMed]
- Meroni, S.B.; Galardo, M.N.; Rindone, G.; Gorga, A.; Riera, M.F.; Cigorraga, S.B. Molecular mechanisms and signaling pathways involved in Sertoli cell proliferation. Front. Endocrinol. 2019, 10, 224. [Google Scholar] [CrossRef] [PubMed]
- Abel, M.H.; Baker, P.J.; Charlton, H.M.; Monteiro, A.; Verhoeven, G.; De Gendt, K.; Guillou, F.; O’Shaughnessy, P.J. Spermatogenesis and Sertoli cell activity in mice lacking Sertoli cell receptors for follicle-stimulating hormone and androgen. Endocrinology 2008, 149, 3279–3285. [Google Scholar] [CrossRef] [PubMed]
- Pitetti, J.L.; Calvel, P.; Zimmermann, C.; Conne, B.; Papaioannou, M.D.; Aubry, F.; Cederroth, C.R.; Urner, F.; Fumel, B.; Crausaz, M.; et al. An essential role for insulin and IGF1 receptors in regulating Sertoli cell proliferation, testis size, and FSH action in mice. Mol. Endocrinol. 2013, 27, 814–827. [Google Scholar] [CrossRef]
- Lin, T. Insulin like growth factor-1 regulation of Leydig cells. In The Leydig Cell; Payne, A.H., Hardy, M.P., Russell, L.D., Eds.; Cache River Press: Vienna, IL, USA, 1996; pp. 477–491. [Google Scholar]
- Nascimento, A.R.; Pimenta, M.T.; Lucas, T.F.; Royer, C.; Porto, C.S.; Lazari, M.F. Intracellular signaling pathways involved in the relaxin-induced proliferation of rat Sertoli cells. Eur. J. Pharmacol. 2012, 691, 28–291. [Google Scholar] [CrossRef]
- Rouiller-Fabre, V.; Lecref, L.; Gautier, C.; Saez, J.M.; Habert, R. Expression and effect of insulin-like growth factor I on rat fetal Leydig cell function and differentiation. Endocrinology 1998, 139, 2926–2934. [Google Scholar] [CrossRef]
- Abney, T.O.; Myers, R.B. 17 beta-estradiol inhibition of Leydig cell regeneration in the ethane dimethylsulfonate-treated mature rat. J. Androl. 1991, 12, 295–304. [Google Scholar]
- Meehan, T.; Schlatt, S.; O’Bryan, M.K.; de Kretser, D.M.; Loveland, K.L. Regulation of germ cell and Sertoli cell development by activin, follistatin, and FSH. Dev. Biol. 2000, 220, 225–237. [Google Scholar] [CrossRef] [PubMed]
- Buzzard, J.J.; Farnworth, P.G.; De Kretser, D.M.; O’Connor, A.E.; Wreford, N.G.; Morrison, J.R. Proliferative phase Sertoli cells display a developmentally regulated response to activin in vitro. Endocrinology 2003, 144, 474–483. [Google Scholar] [CrossRef] [PubMed]
- Zavadil, J.; Cermak, L.; Soto-Nieves, N.; Böttinger, E.P. Integration of TGF-beta/Smad and Jagged1/Notch signalling in epithelial-to-mesenchymal transition. EMBO J. 2004, 23, 1155–1165. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Liu, M.; Gonzalez-Perez, R.R. Role of Notch and its oncogenic signaling crosstalk in breast cancer. Biochim. Biophys. Acta 2011, 1815, 197–213. [Google Scholar] [CrossRef]
- Ishitani, T.; Hirao, T.; Suzuki, M.; Isoda, M.; Ishitani, S.; Harigaya, K.; Kitagawa, M.; Matsumoto, K.; Itoh, M. Nemo-like kinase suppresses Notch signalling by interfering with formation of the Notch active transcriptional complex. Nat. Cell Biol. 2010, 12, 278–285. [Google Scholar] [CrossRef]
- Petersen, C.; Boitani, C.; Fröysa, B.; Söder, O. Interleukin-1 is a potent growth factor for immature rat Sertoli cells. Mol. Cell Endocrinol. 2002, 186, 37–47. [Google Scholar] [CrossRef]
- Mauduit, C.; Besset, V.; Caussanel, V.; Benahmed, M. Tumor necrosis factor alpha receptor p55 is under hormonal (follicle-stimulating hormone) control in testicular Sertoli cells. Biochem. Biophys. Res. Commun. 1996, 224, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Maxwell, P.J.; Neisen, J.; Messenger, J.; Waugh, D.J. Tumor-derived CXCL8 signaling augments stroma-derived CCL2-promoted proliferation and CXCL12-mediated invasion of PTEN-deficient prostate cancer cells. Oncotarget 2014, 5, 4895–4908. [Google Scholar] [CrossRef]
- Lucas, T.F.G.; Lazari, M.F.M.; Porto, C.S. Differential role of the estrogen receptors ESR1 and ESR2 on the regulation of proteins involved with proliferation and differentiation of Sertoli cells from 15-day-old rats. Mol. Cell Endocrinol. 2014, 382, 84–96. [Google Scholar] [CrossRef]
- Cavaco, J.E.; Laurentino, S.S.; Barros, A.; Sousa, M.; Socorro, S. Estrogen receptors alpha and beta in human testis: Both isoforms are expressed. Syst. Biol. Reprod. Med. 2009, 55, 137–144. [Google Scholar] [CrossRef]
- Yang, W.R.; Zhu, F.W.; Zhang, J.J.; Wang, Y.; Zhang, J.H.; Lu, C.; Wang, X.Z. PI3K/Akt activated by GPR30 and Src regulates 17beta-estradiol-induced cultured immature boar Sertoli cells proliferation. Reprod. Sci. 2017, 24, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Hilbold, E.; Distl, O.; Hoedemaker, M.; Wilkening, S.; Behr, R.; Rajkovic, A.; Langeheine, M.; Rode, K.; Jung, K.; Metzger, J.; et al. Loss of Cx43 in murine Sertoli cells leads to altered prepubertal Sertoli cell maturation and impairment of the mitosis-meiosis switch. Cells 2020, 9, 676. [Google Scholar] [CrossRef] [PubMed]
- Holsberger, D.R.; Buchold, G.M.; Leal, M.C.; Kiesewetter, S.E.; O’Brien, D.A.; Hess, R.A.; França, L.R.; Kiyokawa, H.; Cooke, P.S. Cell-cycle inhibitors p27Kip1 and p21Cip1 regulate murine Sertoli cell proliferation. Biol. Reprod. 2005, 72, 1429–1436. [Google Scholar] [CrossRef] [PubMed]
- Valinezhad Orang, A.; Safaralizadeh, R.; Kazemzadeh-Bavili, M. Mechanisms of miRNA-mediated gene regulation from common Downregulation to mRNA-specific upregulation. Int. J. Genom. 2014, 2014, 970607. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Wong, S.C.; Chan, L.W.; Cho, W.C.; Yip, S.P.; Yung, B.Y. Multiple regression analysis of mRNA-miRNA associations in colorectal cancer pathway. Biomed. Res. Int. 2014, 2014, 676724. [Google Scholar] [CrossRef]
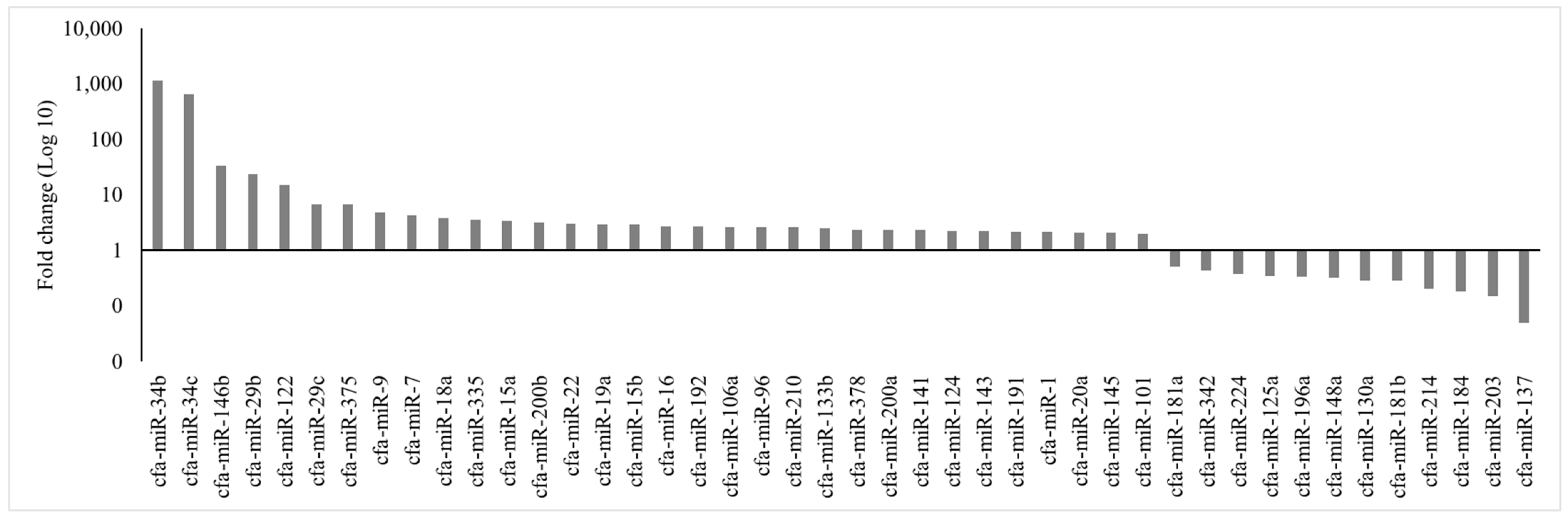

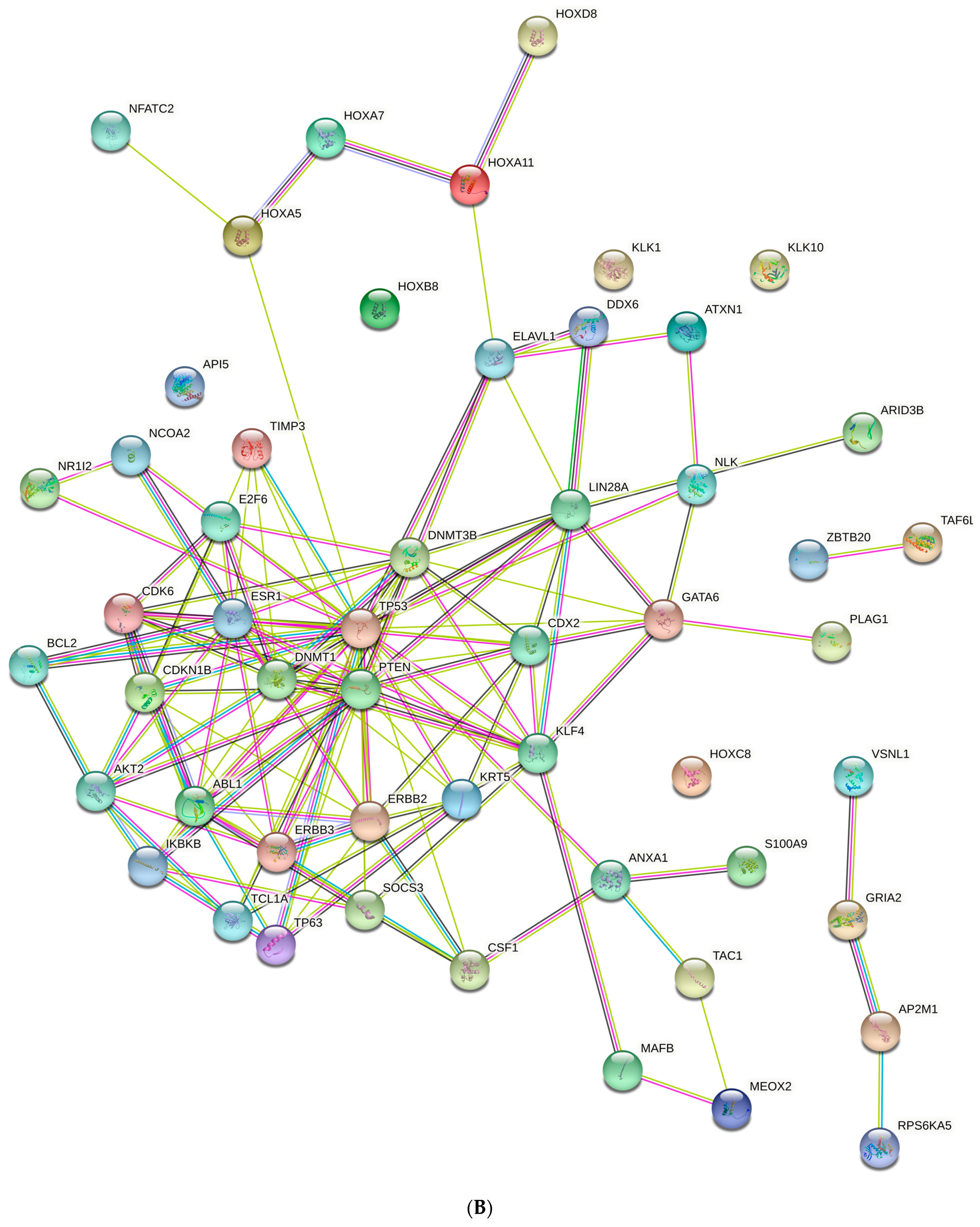
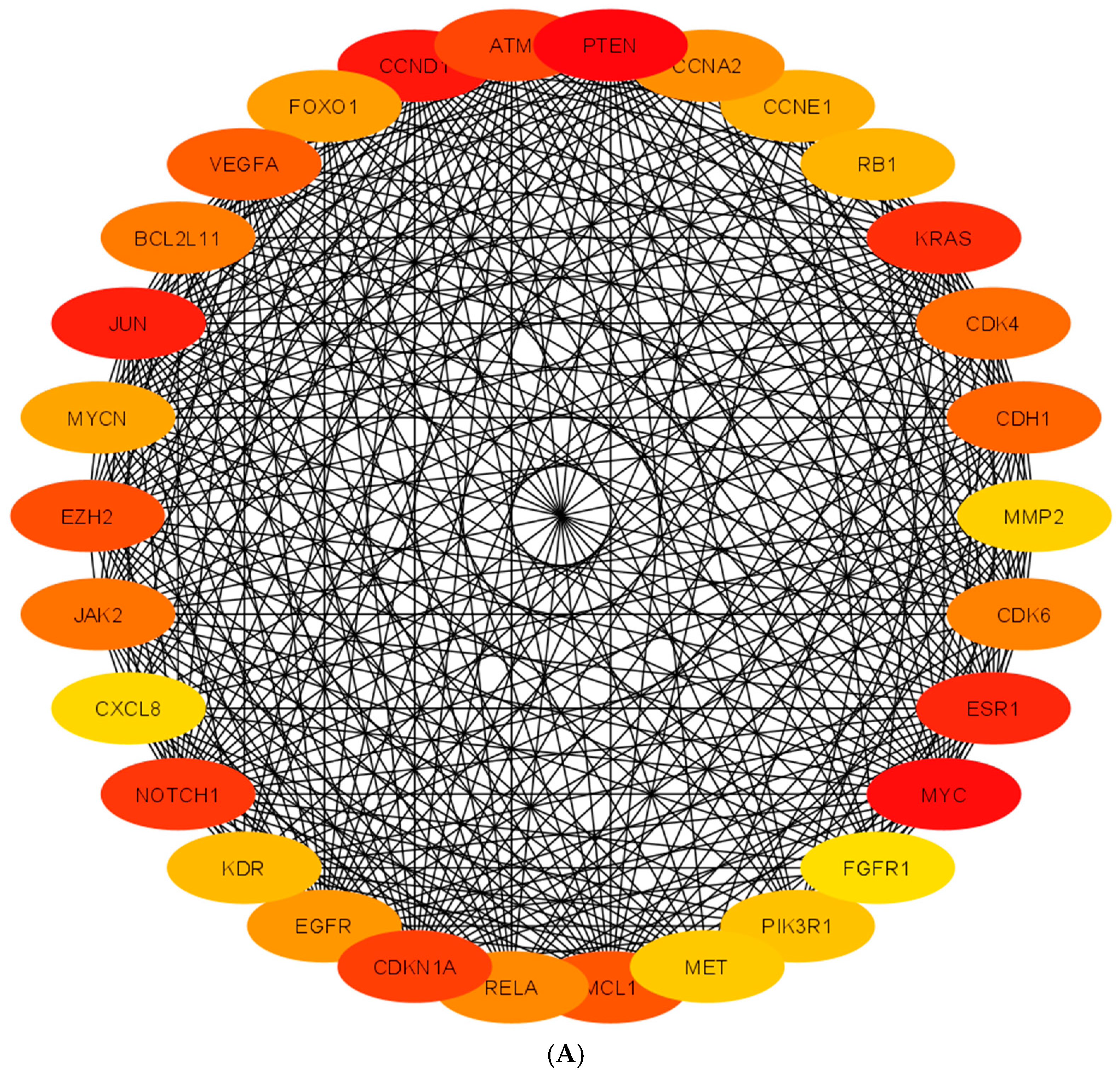



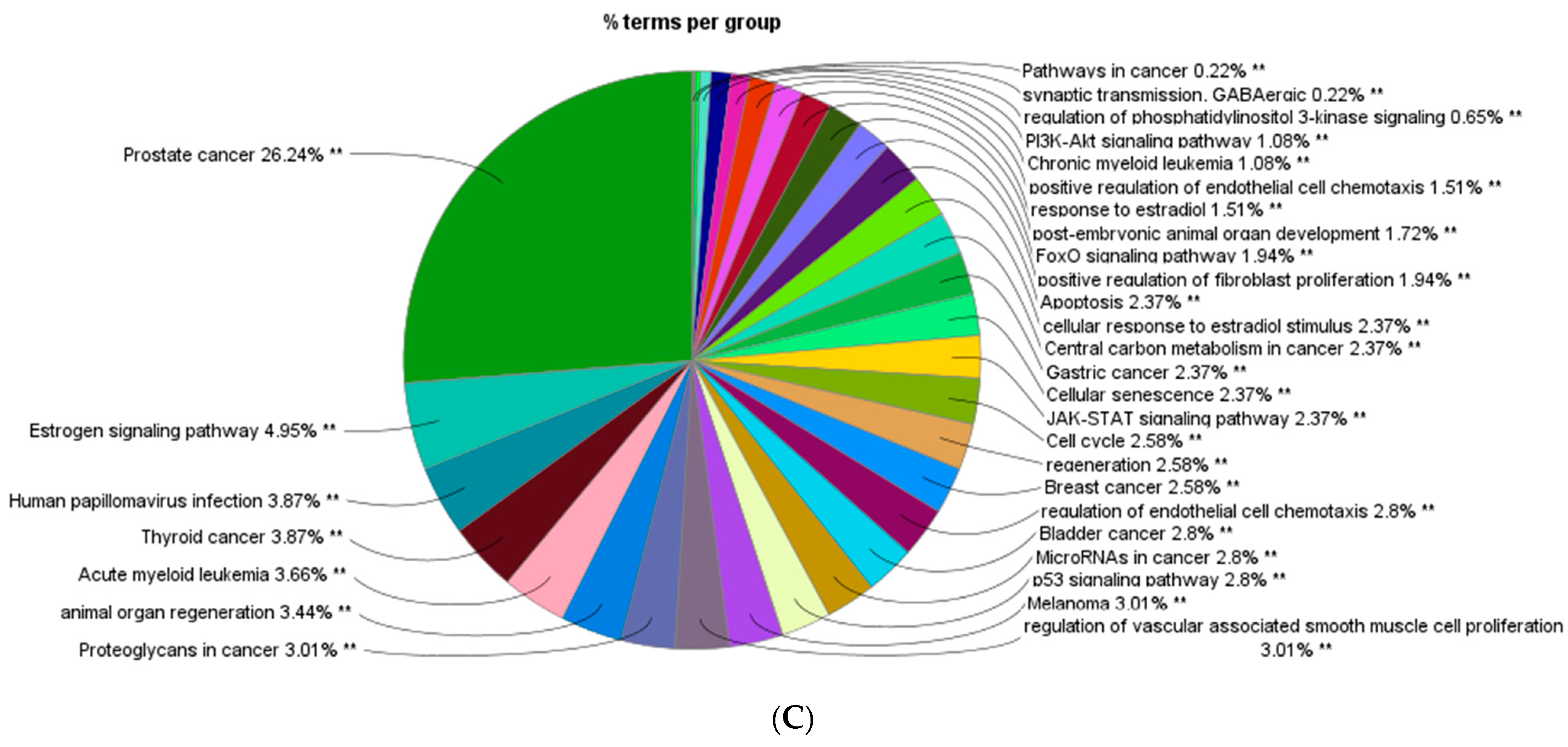
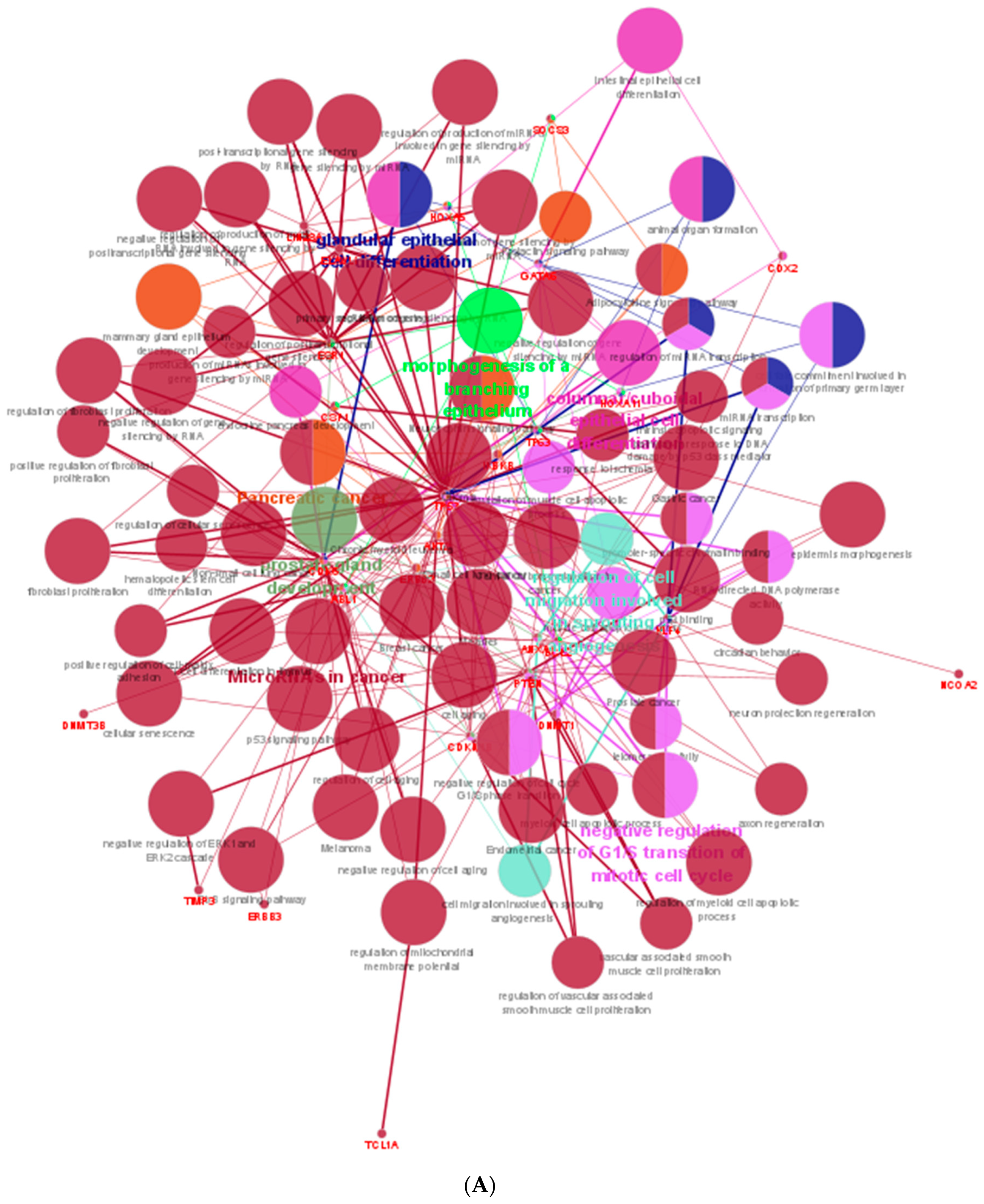

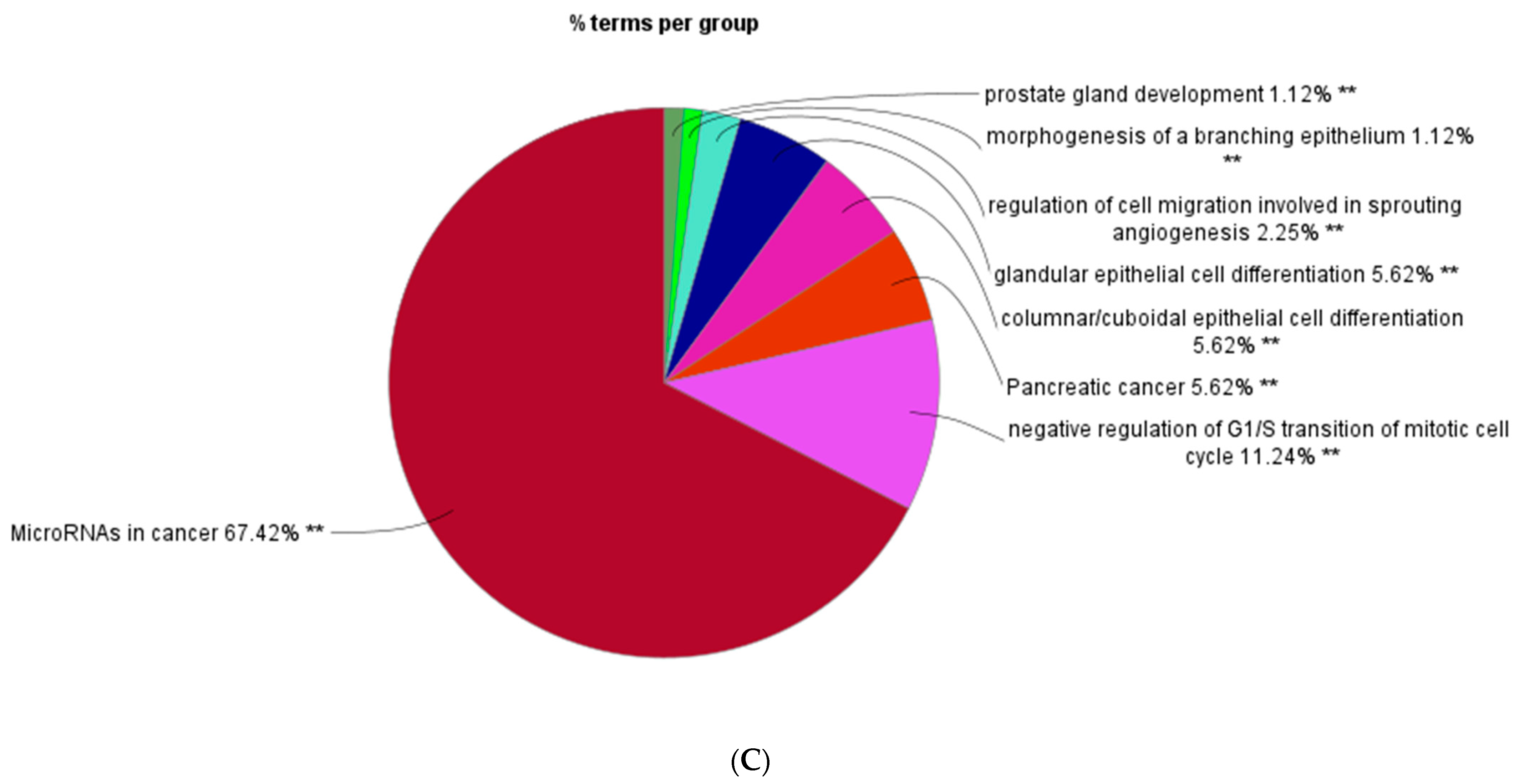

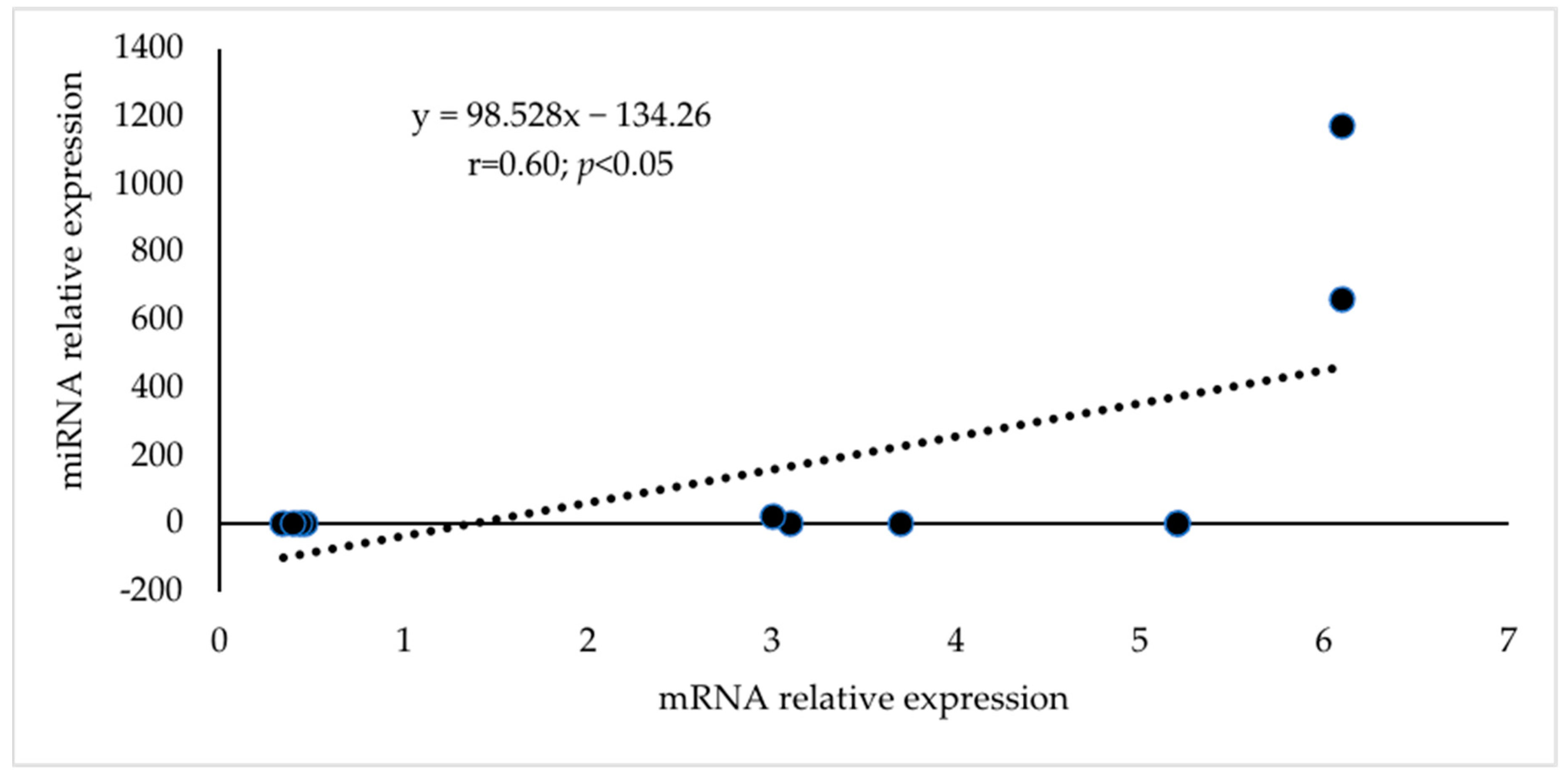
| Well | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | cfa-let-7a | cfa-let-7b | cfa-let-7c | cfa-let-7f | cfa-let-7g | cfa-miR-1 | cfa-miR-101 | cfa-miR-103 | cfa-miR-106a | cfa-miR-106b | cfa-miR-10b | cfa-miR-122 |
| B | cfa-miR-124 | cfa-miR-125a | cfa-miR-125b | cfa-miR-126 | cfa-miR-130a | cfa-miR-133a | cfa-miR-133b | cfa-miR-137 | cfa-miR-141 | cfa-miR-143 | cfa-miR-145 | cfa-miR-146a |
| C | cfa-miR-146b | cfa-miR-148a | cfa-miR-150 | cfa-miR-15a | cfa-miR-15b | cfa-miR-16 | cfa-miR-17 | cfa-miR-181a | cfa-miR-181b | cfa-miR-182 | cfa-miR-183 | cfa-miR-184 |
| D | cfa-miR-18a | Cfa-miR-191 | Cfa-miR-192 | Cfa-miR-195 | Cfa-miR-196a | Cfa-miR-19a | Cfa-miR-200a | Cfa-miR-200b | Cfa-miR-200c | Cfa-miR-203 | Cfa-miR-204 | Cfa-miR-205 |
| E | Cfa-miR-20a | cfa-miR-21 | cfa-miR-210 | cfa-miR-214 | cfa-miR-218 | cfa-miR-22 | cfa-miR-222 | cfa-miR-223 | cfa-miR-224 | cfa-miR-23a | cfa-miR-23b | cfa-miR-24 |
| F | cfa-miR-25 | cfa-miR-26a | cfa-miR-27a | cfa-miR-27b | cfa-miR-29b | cfa-miR-29c | cfa-miR-30b | cfa-miR-30c | cfa-miR-30d | cfa-miR-31 | cfa-miR-335 | cfa-miR-342 |
| G | cfa-miR-34a | cfa-miR-34b | cfa-miR-34c | cfa-miR-375 | cfa-miR-378 | cfa-miR-451 | cfa-miR-499 | cfa-miR-7 | cfa-miR-9 | cfa-miR-92a | cfa-miR-93 | cfa-miR-96 |
| H | cel-miR-39-3p | cel-miR-39-3p | SNORD61 | SNORD68 | SNORD72 | SNORD95 | SNORD96A | RNU6-2 | miRTC | miRTC | PPC | PPC |
| Gene | Primer Sequence (5′–3′) | Product Length | Accession Number |
|---|---|---|---|
| CDKN1A | F: CCTCGGAGGAGGTGCCAT | 187 | XM_038683340.1 |
| R: CGTCTCGGTGACGAAGTCAA | |||
| EGFR | F: TAGGATCAGGGCCCGCAG | 187 | XM_038423676.1 |
| R: GCAACTTCCTGGATGGTCTTT | |||
| JUN | F: CCTTCTACGACGATGCCCTC | 101 | XM_038666089.1 |
| R: GTTCAGGGTCATGCTCTGCT | |||
| NOTCH1 | F: CAGTGCAATGAGGGACCAGT | 274 | XM_038438708.1 |
| R: AGCATCCTCCACTCTCTGTCT | |||
| PIK3R1 | F: CACAACCTGCAAACATTGCC | 160 | XM_038659066.1 |
| R: AGGTCCCATCGGCTGTATC | |||
| DNMT1 | F: CTCTACGGTGTGTGCAGTGT | 209 | XM_038428673.1 |
| R: CAGGTGACCACGCTTACAGT | |||
| PTEN | F: CATCATCAAGGAGATCGTCAGCAG | 217 | NM_001003192.1 |
| R: ATGTCTTTCAGCACACAGATTGTA | |||
| ESR1 | F: CACGGAGCTACACGCACAT | 74 | NM_001286958.2 |
| R: GGCTTGTAGAAGTCAAGGGCT | |||
| TIMP3 | F: CCTCCAAGAACGAGTGCCTT | 161 | NM_001284439.1 |
| R: GGGGTCTGTGGCATTGATGA | |||
| GAPDH | F: AACATCATCCCTGCTTCCAC | 234 | NM_001003142.2 |
| R: GACCACCTGGTCCTCAGTGT |
| miRNA ID | Sequence |
|---|---|
| cfa-miR-34b | AGGCAGUGUAAUUAGCUGAUUG |
| hsa-miR-34b | UAGGCAGUGUCAUUAGCUGAUUG |
| cfa-miR-34c | AGGCAGUGUAGUUAGCUGAUUGC |
| hsa-miR-34c | AGGCAGUGUAGUUAGCUGAUUGC |
| cfa-miR-146b | UGAGAACUGAAUUCCAUAGGCU |
| hsa-miR-146b | UGAGAACUGAAUUCCAUAGGCUG |
| cfa-miR-29b | UAGCACCAUUUGAAAUCAGUGUU |
| hsa-miR-29b | UAGCACCAUUUGAAAUCAGUGUU |
| cfa-miR-122 | UGGAGUGUGACAAUGGUGUUUG |
| hsa-miR-122 | UGGAGUGUGACAAUGGUGUUUG |
| cfa-miR-29c | UAGCACCAUUUGAAAUCGGUUA |
| hsa-miR-29c | AUCUCUUACACAGGCUGACCGAUUUCUCCUGGUGUUCAGAGUCUGUUUUUGUCUAGCACCAUUUGAAAUCGGUUAUGAUGUAGGGGGA |
| cfa-miR-375 | GCCCCGCGACGAGCCCCUCGCACAAACCGGACCUGAGCGUUUUGUUCGUUCGGCUCGCGUGAGGCAGGGG |
| hsa-miR-375 | GCGACGAGCCCCUCGCACAAACC |
| cfa-miR-9 | UCUUUGGUUAUCUAGCUGUAUGA |
| hsa-miR-9 | UCUUUGGUUAUCUAGCUGUAUGA |
| cfa-miR-7 | UGGAAGACUAGUGAUUUUGUUGU |
| hsa-miR-7 | UGGAAGACUAGUGAUUUUGUUGUU |
| cfa-miR-18a | UAAGGUGCAUCUAGUGCAGAUA |
| hsa-miR-18a | UAAGGUGCAUCUAGUGCAGAUAG |
| cfa-miR-335 | UCAAGAGCAAUAACGAAAAAUGU |
| hsa-miR-335 | UCAAGAGCAAUAACGAAAAAUGU |
| cfa-miR-15a | UAGCAGCACAUAAUGGUUUGU |
| hsa-miR-15a | UAGCAGCACAUAAUGGUUUGUG |
| cfa-miR-200b | CAUCUUACUGGGCAGCAUUGGA |
| hsa-miR-200b | CAUCUUACUGGGCAGCAUUGGA |
| cfa-miR-22 | AAGCUGCCAGUUGAAGAACUGU |
| hsa-miR-22 | AAGCUGCCAGUUGAAGAACUGU |
| cfa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA |
| hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA |
| cfa-miR-15b | UAGCAGCACAUCAUGGUUUA |
| hsa-miR-15b | UAGCAGCACAUCAUGGUUUACA |
| cfa-miR-16 | UAGCAGCACGUAAAUAUUGGCG |
| hsa-miR-16 | UAGCAGCACGUAAAUAUUGGCG |
| cfa-miR-192 | CUGACCUAUGAAUUGACAGCC |
| hsa-miR-192 | CUGACCUAUGAAUUGACAGCC |
| cfa-miR-106a | AAAGUGCUUACAGUGCAGGUAG |
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAG |
| cfa-miR-96 | UUUGGCACUAGCACAUUUUUGCU |
| hsa-miR-96 | UUUGGCACUAGCACAUUUUUGCU |
| cfa-miR-210 | ACUGUGCGUGUGACAGCGGCUGA |
| hsa-miR-210 | CUGUGCGUGUGACAGCGGCUGA |
| cfa-miR-133b | UUUGGUCCCCUUCAACCAGCUA |
| hsa-miR-133b | UUUGGUCCCCUUCAACCAGCUA |
| cfa-miR-378 | ACUGGACUUGGAGUCAGAAGGC |
| hsa-miR-378 | ACUGGACUUGGAGUCAGAAGGC |
| cfa-miR-200a | CAUCUUACCGGACAGUGCUGGA |
| hsa-miR-200a | CAUCUUACCGGACAGUGCUGGA |
| cfa-miR-141 | AACACUGUCUGGUAAAGAUGG |
| hsa-miR-141 | UAACACUGUCUGGUAAAGAUGG |
| cfa-miR-124 | UAAGGCACGCGGUGAAUGCCA |
| hsa-miR-124 | UAAGGCACGCGGUGAAUGCCAA |
| cfa-miR-143 | UGAGAUGAAGCACUGUAGCUC |
| hsa-miR-143 | UGAGAUGAAGCACUGUAGCUC |
| cfa-miR-191 | CAACGGAAUCCCAAAAGCAGCU |
| hsa-miR-191 | CAACGGAAUCCCAAAAGCAGCUG |
| cfa-miR-1 | UGGAAUGUAAAGAAGUAUGUA |
| hsa-miR-1 | UGGAAUGUAAAGAAGUAUGUAU |
| cfa-miR-20a | UAAAGUGCUUAUAGUGCAGGUAG |
| hsa-miR-20a | UAAAGUGCUUAUAGUGCAGGUAG |
| cfa-miR-145 | GUCCAGUUUUCCCAGGAAUCCCU |
| hsa-miR-145 | GUCCAGUUUUCCCAGGAAUCCCU |
| cfa-miR-101 | UACAGUACUGUGAUAACUGA |
| hsa-miR-101 | UACAGUACUGUGAUAACUGAA |
| cfa-miR-137 | UUAUUGCUUAAGAAUACGCGU |
| hsa-miR-137 | UUAUUGCUUAAGAAUACGCGUAG |
| cfa-miR-203 | GUGAAAUGUUUAGGACCACUAG |
| hsa-miR-203 | GUGAAAUGUUUAGGACCACUAG |
| cfa-miR-184 | UGGACGGAGAACUGAUAAGGGU |
| hsa-miR-184 | UGGACGGAGAACUGAUAAGGGU |
| cfa-miR-214 | ACAGCAGGCACAGACAGGCAGU |
| hsa-miR-214 | ACAGCAGGCACAGACAGGCAGU |
| cfa-miR-130a | CAGUGCAAUGUUAAAAGGGCAU |
| hsa-miR-130a | CAGUGCAAUGUUAAAAGGGCAU |
| cfa-miR-181b | AACAUUCAUUGCUGUCGGUG |
| hsa-miR-181b | AACAUUCAUUGCUGUCGGUGGGU |
| cfa-miR-148a | UCAGUGCACUACAGAACUUUGU |
| hsa-miR-148a | UCAGUGCACUACAGAACUUUGU |
| cfa-miR-196a | UAGGUAGUUUCAUGUUGUUGGG |
| hsa-miR-196a | UAGGUAGUUUCAUGUUGUUGGG |
| cfa-miR-125a | UCCCUGAGACCCUUUAACCUGU |
| hsa-miR-125a | UCCCUGAGACCCUUUAACCUGUGA |
| cfa-miR-224 | CAAGUCACUAGUGGUUCCGUUU |
| hsa-miR-224 | UCAAGUCACUAGUGGUUCCGUUUAG |
| cfa-miR-342 | UCUCACACAGAAAUCGCACCCGU |
| hsa-miR-342 | UCUCACACAGAAAUCGCACCCGU |
| cfa-miR-181a | AACAUUCAACGCUGUCGGUGAG |
| hsa-miR-181a | AACAUUCAACGCUGUCGGUGAGU |
| Top Hub Genes | Roles | Tissue Expressions | PPIs |
|---|---|---|---|
| (A) | |||
| PTEN | Regulation of cell division and growth; Sertoli cell and spermatogonial stem cells | testis, prostate | CSNK2A1, NEDD4, PDGFRB, SLC9A3R1, SPOP, USP7 |
| CCNA2 | Regulation of cell cycle | testis, prostate | CDC6, CDK2, CDKN1A, CDKN1B, E2F1, SKP2, |
| CCNE1 | Cell cycle regulation and progression; cell proliferation | testis, prostate | CDK2, CDC25A, CDKN1A, CDKN1B, FBXW7, SKP2, |
| KRAS | relays signals from outside the cell to the cell’s nucleus | testis, prostate | ARAF, CALM1, PIK3CG. RAF1, RALGDS. RASSF5 |
| VEGFA | endothelial cell proliferation, promotion of cell migration, inhibition of apoptosis | testis, prostate | FLT1, KDR, DLL4, HIF1A, MYOD1, STAT3, RUNX2, MYC, |
| CDK4 | Regulation of cell cycle progression, G1 phase | testis, prostate | CCND1, CCND2, CCND3, CDKN1A, CDNK1B, RB1 |
| CDH1 | Regulation of cell–cell adhesions, mobility and proliferation; spermatogenic stem cells and type A spermatogonia | testis, prostate | CBLL1, CDC27, CTNNA1, CTNNB1, CTNND1, EGFR |
| MMP2 | Regulate space between cells, cell architecture. | testis, prostate | CCL7, COL1A1, COL5A1, TIMP2, TIMP3, TIMP4 |
| CDK6 | Regulation of cell cycle progression, G1 phase | testis, prostate | CCND1, CCND2, CDKN2A, CDKN2B, CDKN2C, RB1 |
| ESR1 | maintenance of gluconeogenesis and lipid metabolism; regulate cell proliferation; growth sexual development | testis, prostate | EP300, NCOA1, NCOA2, NR2F1, CREBBP, TRIM24, |
| MYC | Regulates cell cycle, and proliferation and apoptosis | testis, prostate | BRCA1, FBXW7, MAX, RAF1, RUVBL1, SMARCA4 |
| FGFR1 | Cell proliferation, differentiation, survival and migration | testis, prostate | FGF1, FRS2, GRB14, NCAM1, NEDD4, PLCG1, |
| PIK3R1 | Cell proliferation and survival | testis, prostate | ERBB3, GAB1, GRB2, IRS1, KHDRBS1, PIK3CA |
| MET | Cell growth and survival | testis, prostate | CBL, EGFR, GAB1, GRB2, HGF, PLXNB1, SRC |
| MCL1 | Regulates cell apoptosis | testis, prostate | BAD, BAX, BCL2L11, BIK, BMF, TPT1 |
| RELA | Regulates all types of cellular processes, including cellular metabolism, chemotaxis | testis, prostate | CREBBP, HDAC1, NFKB1, NFKB2, NFKBIA, NR3C1 |
| CDKN1A | Regulates cell cycle progression at G1-S phase | testis, prostate | CCND1, CDK2, CDK4, GADD45G, PCNA, TSG101 |
| EGFR | Directs the behavior of epithelial cells; regulates cell migration | testis, prostate | EGF, GRB2, PTPN1, SHC1, SOS1, SRC |
| KDR | Promotes proliferation, survival, migration and differentiation of endothelial cells | testis, prostate | CDH5, SHC1, SHC2, SRC, VEGFA, VEGFC |
| NOTCH1 | cellular fate determination, cell proliferation, cell differentiation and cellular apoptosis | testis, prostate | DTX1, FBXW7, JAG1, PSEN1, RBPJ, SMAD3 |
| CXCL8 | protein coding gene attracts neutrophils, basophils, and T-cells | testis, prostate | ACKR1, CCL5, CXCR2, RELA, SDC1, TNFAIP6 |
| JAK2 | protein coding gene regulates cell growth | testis, prostate | EPOR, PTPN1, IRS3, PTPN11, SH2B1, SOCS1, STAT5B |
| EZH2 | Regulates cell fate determination | testis, prostate | DNMT1, EED, HDAC1, RBBP4, SUZ12, VAV1 |
| MYCN | Regulates cell growth and division and apoptosis | testis, prostate | AURKA, EZH2, FBXW7, MAX, SP1, ZBTB17 |
| JUN | Protein coding gene regulates cell proliferation and apoptosis | testis, prostate | ATF3, FOS, FOSL1, FOSL2, JDP2, MAPK8 |
| BCL2L11 | Regulates anti- and pro-apoptic regulators | testis, prostate | BCL2, BCL2A1, BCL2L1, BCL2L2, DYNLL1, MCL1 |
| FOXO1 | Protects cell from oxidative stress; regulates cell proliferation | testis, prostate | AKT1, AR, CREBBP, ESR1, SIRT1, YWHAZ, |
| CCND1 | Regulates cell cycle progression at G1-S phase | testis, prostate | AR, CDK2, CDK4, CDK6, CDKN1A, CDKN1B |
| ATM | Regulates cell proliferation | testis, prostate | ABL1, AP1B1, BRCA1, FANCD2, TRFF1, TP53, |
| (B) | |||
| LIN28 | Posttranscriptional regulator of genes involved in developmental timing and self-renewal in stem cells | testis, prostate | DHX36, IGF2BP3, LARP1, L1TD1, ZCCHC11 |
| NLK | Negative regulator in cell proliferation | testis, prostate | FAM222A, LEF1, MYB, PKM, MAP3K7, SMAD4 |
| GATA6 | Regulation of cellular differentiation and organogenesis | testis, prostate | CDK9, EP300, EGLN3, KLF2, NKX2-1, EP300, |
| KRT5 | Regulation of cell structural framework | testis, prostate | ALOX12, EGFR, KRT14, LARP7, PKP2, SUMO2 |
| CDX2 | Regulation of cell growth and differentiation | testis, prostate | CREBBP, EP300, GSK3B, HNF1A, PAX6, RELA |
| DNMT1 | Regulates DNA methylation; maintain a transcriptionally repressive state of genes in undifferentiated stem cells | testis, prostate | DNMT3B, HDAC1, PCNA, RB1, UHRF1, USP7 |
| TIMP3 | regulation of cell growth, cell death, angiogenesis, and invasion | testis, prostate | ADAM17, AGTR2, ASGR2, IFI30, MMP2, MMP3 |
| CDKN1B | Oppose cell cycle progression; regulators of cell proliferation | testis, prostate | CCND1, CCND3, CCND2, CDK2, CDK4, STMN1 |
| NCOA2 | Regulates cell growth, development, and homeostasis | testis, prostate | AR, ESR1, NR3C1, PPARG, RXRA, VDR |
| ESR1 | maintenance of gluconeogenesis and lipid metabolism; regulate cell proliferation; growth sexual development | testis, prostate | EP300, NCOA1, NCOA2, NR2F1, CREBBP, TRIM24, |
| TP53 | regulates cell division by keeping cells from proliferating in an uncontrolled way | testis, prostate | BCL2L1, DAXX, HSPA9, HMGB1, MDM2, TOPORS |
| HOXA5 | Regulates morphogenesis and differentiation | testis, prostate | ELAVL1, FOXO1, FOXA2, MEIS1, PRMT6, TWIST1 |
| BCL2 | Regulation of apoptosis | testis, prostate | BAD, BAX, BBC3, BCL2L11, BID, BIK |
| AKT2 | Regulation of cell proliferation, growth and survival | testis, prostate | APPL1, CHUK, HSP90AA1, SH3RF1, TTC3, TCL1A |
| TCL1A | Co-activator of AKT kinases Enhances cell proliferation, stabilizes mitochondrial membrane potential and promotes cell survival | testis, prostate | AKT1, AKT2, EP300, FOS, JUN, JUNB |
| KLF4 | Prevents differentiation of stem cells | ovary, uterus, placenta | CREBBP, CTBP1, EP300, HDAC2, KLF6, SP1 |
| SOCS3 | maintenance of cell shape and integrity | ovary, uterus, placenta | EGFR, IL6ST, JAK2, LEPR, PTPN11 |
| IKBKB | Regulation of cell growth and apoptosis | testis, prostate | CDC37, CHUK, IKBKG, NFKB1, NFKB1A, TRAF2 |
| PTEN | Regulation of cell division and growth; sertoli cell and spermatogonial stem cells | testis, prostate | CSNK2A1, NEDD4, PDGFRB, SLC9A3R1, SPOP, USP7 |
| TP63 | Regulation of epithelial morphogenesis, and adult stem/progenitor cell | testis, prostate | DAXX, HNRNPAB, HIPK2, ITCH, TP53, TP73 |
| ERBB2 | Regulation of cell membrane; regulates cell proliferation and anti-apoptosis | testis, prostate | EGFR, ERBB3, ERBB4, ERBIN, GRB2, PIK3R1 |
| ABL1 | Regualtes cell growth, survival, cell adhesion, cell migration; cytoskeleton remodeling | testis, prostate | CRK, ABI1, DOK1, NCK1, RAD51, RIN1 |
| PIK3CB | cell adhesion; immune (PIK3) and inflammatory responses | testis, prostate | PIK3R1, PIK3R2, PIK3R3, HCK, IRS1 |
| CDCK6 | prevents cell proliferation and regulates negatively cell differentiation | testis, prostate | CDKN2A, CDKN2B, RB1, CDKN2D, CCND1, CCND3, |
| E2F6 | regulation of DNA replication, DNA repair, mitosis, and cell fate. | testis, prostate | KDM5C, PCGF6, RING1, RYBP, TFDP1, TFDP2 |
| ELAVL1 | Anti-proliferation of cell, negatively affects meiotic division | testis, prostate | AGO2, CHEK2, HNRNPA1, IGF2BP1, RBM3, TNPO2 |
| HOXA11 | Regulates cell proliferation and differentiation | testis, prostate | FOXO1, HDAC1, HDAC2, MEIS1, PGBD3, YY1 |
| DNMT3B | Regulation of DNA methylation | testis, prostate | DNMT1, DNMT3A, EED, EZH2, HDAC1, UBE2I |
| miRNA | Hub Gene |
|---|---|
| (A) | |
| cfa-miR-1 | MET |
| cfa-miR-7 | EGFR |
| cfa-miR-9 | CDH1, FOXO1 |
| cfa-miR-15a | CCND1, JUN, MCL1, VEGFA |
| cfa-miR-15b | CCNE1 |
| cfa-miR-16 | CCND1, CCNE1, FGFR1, JUN, MCL1, VEGFA |
| cfa-miR-18a | ESR1 |
| cfa-miR-19a | CCND1, BCL2L11, ESR1, PTEN |
| cfa-miR-20a | CCND1, VEGFA |
| cfa-miR-22 | ESR1, PTEN |
| cfa-miR-29b | MCL1, MMP2, PIK3R1 |
| cfa-miR-29c | PIK3R1 |
| cfa-miR-34b | MYC, NOTCH1 |
| cfa-miR-34c | NOTCH1 |
| cfa-miR-96 | FOXO1 |
| cfa-miR-101 | ATM, EZH2, MCL1, MYCN |
| cfa-miR-106a | CDKN1A, RB1, VEGFA |
| cfa-miR-124 | CDK4, CDK6, EZH2, RELA |
| cfa-miR-143 | KRAS |
| cfa-miR-145 | CCNA2, MYC |
| cfa-miR-192 | RB1 |
| cfa-miR-375 | JAK2 |
| cfa-miR-378 | VEGFA |
| (B) | |
| cfa-miR-125a-5p | ERBB2, ERBB3, LIN28, TP53 |
| cfa-miR-130a | CSF1, HOXA4 |
| cfa-miR-137 | CDK6, E2F6, KLF4, NCOA2 |
| cfa-miR-148a | DNMT1, DNMT3B |
| cfa-miR-181a | BCL2, CDKN1B, CDX2, ELAVL1, ESR1, GATA6, HOXA11, NLK |
| cfa-miR-181b | CDX2, ESR1, GATA6, NLK, TCL1A, TIMP3 |
| cfa-miR-184 | AKT4 |
| cfa-miR-196a | ANXA1, IKBKB, KRT5 |
| cfa-miR-203 | ABL1, SOCS3, TP63 |
| cfa-miR-214 | PTEN |
| cfa-miR-342 | DNMT1 |
| Upregulated Hub Genes Associated KEGG Pathway | Down Regulated Hub Genes Associated KEGG Pathway |
|---|---|
| ErbB signaling pathway | ErbB signaling pathway |
| Cell cycle | p53 signaling pathway |
| p53 signaling pathway | Neurotrophin signaling pathway |
| Mitophagy | Prolactin signaling pathway |
| PI3K-Akt signaling pathway | Adipocytokine signaling pathway |
| Apoptosis | |
| Longevity regulating pathway | |
| Cellular senescence | |
| VEGF signaling pathway | |
| Adherens junction | |
| JAK-STAT signaling pathway | |
| Th1 and Th2 cell differentiation | |
| T cell receptor signaling pathway | |
| B cell receptor signaling pathway | |
| GnRH signaling pathway | |
| Estrogen signaling pathway | |
| Prolactin signaling pathway | |
| Thyroid hormone signaling pathway | |
| Relaxin signaling pathway | |
| AGE-RAGE signaling pathway in diabetic complications | |
| Cancer |
| mRNA | miRNA | mRNA Relative Expression | miRNA Relative Expression |
|---|---|---|---|
| CDKN1A | cfa-miR-106a | 3.1 | 2.63 |
| EGFR | cfa-miR-7 | 3.7 | 4.35 |
| JUN | cfa-miR-15a | 5.2 | 3.43 |
| JUN | cfa-miR-16 | 5.2 | 2.75 |
| NOTCH1 | cfa-miR-34b | 6.1 | 1175.42 |
| NOTCH1 | cfa-miR-34c | 6.1 | 662.79 |
| PIK3R1 | cfa-miR-29b | 3 | 23.59 |
| DNMT1 | cfa-miR-148a | 0.46 | 0.32 |
| PTEN | cfa-miR-214 | 0.34 | 0.2 |
| ESR1 | cfa-miR-181b | 0.43 | 0.29 |
| TIMP3 | cfa-miR-181b | 0.4 | 0.29 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kasimanickam, V.R.; Kasimanickam, R.K. In Silico Analysis of miRNA-Mediated Genes in the Regulation of Dog Testes Development from Immature to Adult Form. Animals 2023, 13, 1520. https://doi.org/10.3390/ani13091520
Kasimanickam VR, Kasimanickam RK. In Silico Analysis of miRNA-Mediated Genes in the Regulation of Dog Testes Development from Immature to Adult Form. Animals. 2023; 13(9):1520. https://doi.org/10.3390/ani13091520
Chicago/Turabian StyleKasimanickam, Vanmathy R., and Ramanathan K. Kasimanickam. 2023. "In Silico Analysis of miRNA-Mediated Genes in the Regulation of Dog Testes Development from Immature to Adult Form" Animals 13, no. 9: 1520. https://doi.org/10.3390/ani13091520
APA StyleKasimanickam, V. R., & Kasimanickam, R. K. (2023). In Silico Analysis of miRNA-Mediated Genes in the Regulation of Dog Testes Development from Immature to Adult Form. Animals, 13(9), 1520. https://doi.org/10.3390/ani13091520






