Simple Summary
In pre-weaned ruminants, the colonization of rumen microbiota is crucial for rumen development and host metabolism. Hence, understanding the colonization process of the rumen microbiota in neonatal ruminants and the factors that affect it may have a positive impact on host health and growth. Rumen microbiota colonizes, and are rapidly established, after birth and constantly interacts with the host. The developing microbial community is more malleable and this may provide an opportunity for nutritional interventions to improve rumen fermentation and increase animal productivity. This paper reviews the latest advances in the colonization of rumen microbiota while providing insights into the most suitable time for manipulating rumen microbial colonization in early life. Furthermore, this manuscript also presents a number of potential factors that influence the establishment of rumen microbiota in early life to optimally enhance rumen development.
Abstract
In pre-weaned ruminants, the microbiota colonizes rapidly in the rumen after birth and constantly interacts with the host to sustain health and metabolism. The developing microbial community is more malleable, so its manipulation may improve ruminant health and productivity as well as may have long-term effects on ruminants. Hence, understanding the process of rumen microbiota establishment is helpful for nutritional interventions of rumen microbiota in pre-weaned ruminants. This paper reviews the latest advances in the colonization of rumen microbiota while providing insights into the most suitable time for manipulating rumen microbial colonization in early life. In addition, different factors that affect rumen microbiota establishment during the pre-weaned ruminants are discussed in the current manuscript. The purpose of this review is to aid in the development of guidelines for manipulating rumen microbiota to improve animal productivity and health.
1. Introduction
The rumen has the unique ability to utilize various feeds, which is mainly attributed to the complex, diverse, and non-pathogenic microorganisms inhabiting the rumen, including bacteria, fungi, protozoa, and archaea [1]. These microbes work closely together to break down plant organic matter and provide volatile fatty acids (VFAs) to the ruminants. Thus, ruminants have a special ability to convert roughage into nutrient-rich meat and milk through microbial fermentation.
The world’s population is expected to peak at 9.7 billion in 2050 [2] when demand for meat and milk will increase significantly [1]. As a result, meeting the demand for meat and milk will become a priority for global food security. Researchers have been working for decades on nutritional manipulations of the rumen microbiota to improve fiber utilization or decrease methane production [3]. Nevertheless, the utility of such nutritional manipulation is often inefficient, principally due to redundancy (overlap of physiological capabilities among various microbial taxa) and resilience (capacity to recover its structure following perturbation) within the rumen of adult animals [4]. In newborn ruminants, the developing microbial community is more malleable, so the effects of nutritional manipulation on the rumen microbial community structure may persist for some time after the manipulation is stopped [5]. Consequently, the newborn ruminants’ microbiota provides a suitable opportunity for interventions in such a complicated microbial environment of the rumen.
Previous studies have revealed that the rumen development process could be divided into three phases: the non-rumination phase (from birth to week 3); the transition phase (from week 3 to 8); and the rumination phase (from week 8 onwards) [6,7]. In the process of rumen development, once the rumen microorganisms begin to colonize, the host-associated commensal microbiota is very important for rumen development [8]. To date, there are many studies on the bacterial and archaea establishment process in early life [9,10,11]. Nevertheless, the establishment process of the other rumen microbiota has rarely been reported. This paper focuses on the colonization of rumen microbiota while providing insights into the factors that affect their establishment during the pre-weaned ruminants. In addition, this manuscript will also provide information about the most suitable time for manipulating rumen microbial colonization in early life. The purpose of this review is to aid in the development of guidelines for manipulating rumen microbiota to improve animal productivity and host health.
2. Establishment of Rumen Microbiota in Pre-Weaned Ruminants
2.1. Establishment of Rumen Bacteria
It is generally believed that the gastrointestinal tract (GIT) of most ruminants is free of microbes at birth [7]. Then, the microbiota from the maternal vaginal, milk, and the surrounding environment rapidly colonize the rumen after birth [6]. However, Bi et al. [12] revealed that a microbiome with low diversity and biomass could be detected in the prenatal gut of lambs, indicating that the prenatal gut harbors a microbiome and that microbial colonization of the fetal gut begins in utero. In contrast, some studies have suggested that microbes detected in utero may be due to DNA present in laboratory reagents and equipment contaminants since the microbiome does not show immune activity or virulence [13,14]. Thus, there is no consensus as to whether the gut microbial community begins colonization in the fetal gut before delivery.
Bacteria essential for mature rumen function can be detected in the rumen of 1-day-old ruminants. For example, Malmuthuge et al. [15] found that Veillonella, followed by Prevotella, Bacteroides, Eubacterium, Streptococcus, Acidaminococcus, Clostridium, Bifidobacterium, and Ruminococcus were predominant (account for 88.7%) in the calf rumen at birth. In addition, the typical rumen bacteria Eubacterium ruminantium, Ruminococcus ruminicola, Ruminococcus flavefaciens, and Ruminococcus albus, which can degrade plant polysaccharides, colonize the rumen during the first week of life when calves are fed milk only [15]. The presence of typical rumen bacteria during the first week of life suggests that cellulolytic bacteria colonization occurs in the absence of solid feed. In line with the results of Malmuthuge et al. [15]. Yin et al. [9] found that bacteria responsible for digesting solid feed colonized the rumen before lambs were fed solid feed.
Several studies have found that bacteria colonize and are rapidly established in the rumen in a defined and progressive sequence during the pre-weaning period [7,10,16,17]. Rey et al. [18] found that the bacterial community of calves was established in three successive stages, namely 2 days of age, 3–12 days of age, and 15–83 days of age. Specifically, at 2 days of age, Proteobacteria (70%) and Bacteroidetes (14%) were the dominant phyla. During postnatal 3 to 12 days, Proteobacteria was gradually replaced by Bacteroidetes as the main phyla. The most abundant genera were Bacteroides (21%), followed by Prevotella (11%), Fusobacterium (5%), and Streptococcus (4%). During postnatal 15 to 83 days, the Bacteroidetes phylum was mainly composed of Prevotella (42%). During this stage, the composition structure of rumen bacterial flora at the phylum level did not change significantly with time, but only the relative abundance at the genus level still changed accordingly [18]. Similar to the pattern of rumen bacterial colonization in calves, rumen bacterial colonization in lambs is also divided into three consecutive stages, namely 0–3, 10–20, and 20–60 days of age [9]. The Proteobacteria were replaced by Bacteroidetes as the dominant phylum at the initial colonization process of lambs, the genera Bacteroides was the main transitional taxa, and Prevotella were the main mature taxa [10,17]. However, there are still some differences in the colonization process of rumen microbes between calves and goat kids. Li et al. [10] and Zhang et al. [17] reported that the colonization process of the rumen bacteria in goat kids can be divided into three stages: the initial phase (0–14 days after birth), transition phase (14–28 days after birth), and relative stable phase (28–56 days after birth). During the initial phase, the genera Bacillus and Lactococcus were predominant in newborns. As the dominant maternal vaginal flora, the genera Bacillus and Lactococcus were delivered to the next generation and play a key role in host-immune regulation [19]. However, the colonization of Bacillus and Lactococcus was transient and it was rapidly replaced by the genera known to readily degrade milk nutrients (e.g., Bacteroides). During the transition phase, the genera Bacteroides, Bacteroidales BS11 gut group, Alloprevotella, and Ruminococcaceae NK4A214 group were the main transitional taxa. During the relatively stable phase, the genera capable of degrading plant fibers were increased, such as Prevotella, Treponema, Ruminococcus, and the unclassified Prevotellaceae were the main mature taxa.
Taken together, the above studies suggest that changes in age may be the main factor affecting the microbial colonization of the rumen. Keeping in view the above studies, the early rumen microbial establishment process can be divided into three stages (Figure 1): (1) In the initial stage, there is a switch from aerobic or facultative anaerobic to strictly anaerobic bacterial community [20]. At the phylum level, the Proteobacteria and Firmicutes were the dominant phyla [10,18]. The ruminal bacteria come from their dams (including the maternal vaginal, milk, and saliva) and external environments [21], such as the genera Bacillus and Lactococcus. (2) In the transition stage, with the intake of breast milk gradually increased, Proteobacteria were replaced by Bacteroidetes as the dominant phylum. At the genus level, Bacteroides, capable of using some components of milk, were the dominant genus. In addition, some rumen bacteria that are commonly found in the mature rumen were already established in the rumens, such as Prevotella and Ruminococcus [17,22]. (3) In the relatively stable stage, with the intake of solid feed increased, Bacteroidetes and Firmicutes were the dominant phylum. At the genus level, some rumen bacteria that can be capable of utilizing starch and fiber gradually increased, and maintained the relative stable abundance in the rumen, such as Prevotella, Ruminococcus, and Treponema [9,10,18].
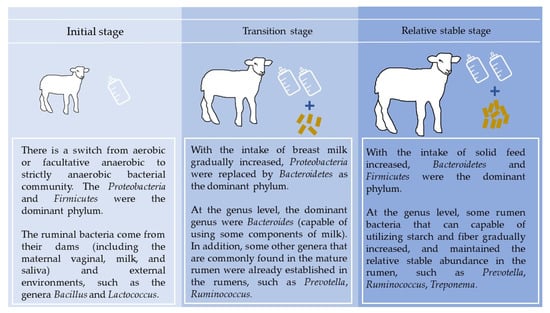
Figure 1.
Establishment of rumen bacteria in per-weaned ruminants.
2.2. Establishment of Rumen Methanogenic Archaea
Typically, archaeal communities contribute to only 3–4% of the rumen microbiota, while methanogenic archaea are a phylogenetically diverse group of archaea [23]. Early studies have found that methanogens begin to colonize the rumen as early as 2–4 days of age in lambs, and the abundance of methanogens reaches adult ruminant levels by 10–14 days of age [24]. Nevertheless, the viability and metabolic activity of methanogens in the first days of life remained unknown. Using real-time quantification PCR (qRT-PCR), the existence of metabolically active methanogens (Methanobrevibacter mobile, Methanobrevibacter votae, and Methanobrevibacter spp.) were found in the rumen of calves within 20 min after birth [25]. Friedman et al. [26] investigated the metabolic potential and taxonomic composition of the methanogenic archaeal communities across different rumen developmental stages. The results indicate that methanogens in newborn calves have metabolic functions and the activity of methylotrophic methanogens was higher in the first 2 months after birth. In contrast, the activity of hydrogenotrophic methanogens was higher in adult ruminants. However, Malmuthuge et al. [15] discovered that no archaea colonization was detected in calves at birth and the methyl coenzyme M reductase (mcrA) gene was observed in weeks 3 and 6 after birth, but not in weeks 1 of calves. Since the mcrA gene encodes the α-subunit of methyl coenzyme M reductase, which catalyzes the last step of methanogenesis [27], suggesting that the methanogens had no metabolic activity in weeks 1 of calves. Therefore, studies are still needed on methanogen’s initial colonization and its actual metabolic functionality in the newborn ruminant. It has been reported that the rumen bacterial composition of the goat changes remarkably with age. In contrast, archaeal communities respond less to age [23]. The dominant phyla in the rumen fluid of goats in different age groups were Euryarchaeota (82%) and Thaumarchaeota (15%) [23]. The abundance of Euryarchaeota increased gradually from 1 to 15 days of age and then tended to be stable, while the abundance of Thaumarchaeota decreased gradually from birth to 15 days of age and then tended to be stable [23]. Other studies have suggested that feeding system and age were the two main factors affecting methanogen diversity [11,28]. For example, Jiao [28] explored the variation in feeding system (supplemental feeding versus grazing) and the changes related to age in rumen microbial colonization, the results showed that microbial colonization in the rumen is achieved at 1 month and methanogens alpha diversity indices were less for supplemental feeding versus grazing; in addition, within the genus, Methanobrevibacter, Methanobrevibacter ruminantium clade, and Methanobrevibacter gottschalkii clade surged in abundance after the solid feed was offered, while Methanobrevibacter acididurans clade only dominated on 70 days after birth. Methanimicrococcus, Methanomicrobium, and Methanosphaera are minor genera while Rumen cluster C is dominant [28]. Wang et al. [11] used qRT-PCR and 16S rRNA sequencing to study the initial colonization and subsequent changes of metabolically active methanogens in the solid-attached, liquid-associated, protozoal-associated, and epithelial-associated rumen fractions during rumen development from 1 to 60 d after birth in goats, and the results showed that the Chao 1 index increased in an age-dependent manner only in the rumen epithelium-associated methanogens. Methanogens colonized in the rumen liquid-associated and epithelium-associated on the first day of life, and the methanogens densities in four fractions stabilized as the starter diets were introduced. Furthermore, the rumen solid attached accommodated the most methanogens, while the lowest density of methanogens was observed in rumen epithelium-associated. Data obtained from the 16S rRNA sequencing indicated that Methanobrevibacter, Candidatus Methanomethylophilus, and Methanosphaera were basically the top three genera in the four fractions and together represented from 89.8% to 98.3% of total methanogens.
2.3. Establishment of Rumen Fungi
It is well known that anaerobic fungi have a critical role in the degradation of plant fiber, primarily due to the enzymes produced by fungi used to degrade plant structural polymers. Moreover, their rhizoids can penetrate plant structural barriers [1]. Compared to bacterial communities, the colonization of rumen fungi seems to appear later [7,29]. For example, the colonization of anaerobic fungi began within 8–10 days after birth in ruminants [29]. However, recent studies have found that the rumen fungal community was rapidly established after the birth of the lambs, and the process of the fungi establishment in the rumen of lambs can be divided into three phases: the initial phase, the transition phase, and the relatively stable phase [17,30]. A study of ruminal fungal community colonization in goats suggested that during the initial colonization of anaerobic fungi before 14 days of age, the Ascomycota phylum was the main dominant flora, but after 14 days of age, the Neocallimastigomycota phylum gradually became the dominant flora. At the genus level, the abundance of Aspergillus reached 47% on d 0, but then it dropped from d 3 to 14, and the abundance of Neocallimastix_sp, Orpinomyces_sp, and Caecomyces are the predominant fungal genera after d14 [17]. Another study showed that the Ascomycota phylum dominated on d 0 and 7, while the Neocallimastigomycota phylum dominated on d 28, 42, and 70. Moreover, Arthrinium, Aspergillus, Boeremia, Candida, Neurospora, and Purpureocillium genera dominated on d 0, while the Candida genus dominated on d 7 [28].
2.4. Establishment of Rumen Protozoa
The rumen is dominated by bacteria, and protozoa account for about 20% or up to 50% of the total rumen microbial population [1]. The main protozoa are ciliates and flagellates [31]. Flagellates mainly exist in the rumen of newborn ruminants. With the increase of age, the number of flagellates gradually decreases and ciliates dominate the protozoa group in the rumen after adulthood. The colonization of protozoa began within 15 days after birth, with small Entodinia colonizing before large Endomorphs and Holotrich protozoa [32,33]. However, if neonatal ruminants are rapidly separated from other ruminants after birth, no protozoa are established in the rumen. This is due to rumen protozoa can only be transmitted from animal to animal through saliva [34].
2.5. Rumen Epithelial Microbiota
The rumen epithelial microbiota, defined as the epimural microbiota, is the tissue-attached microorganism of the rumen epithelium. Recent studies have reported that the epimural community is more diverse than the content-associated microbial community [35] and diet can change its composition [36]. In addition, the changes in the epimural bacterial population are associated with host gene expression [37] and ruminal acidosis [38]. The bacterial community is the main member of the epimural microbiota, and the epimural bacterial community has a greater variation in richness among individual ruminants compared to rumen content- and fluid-associated bacteria [39,40,41]. Through a meta-analysis, a study showed that the genera Campylobacter, Prevotella, Christensenellaceae R-7 group, Butyrivibrio, and the uncultured genus of the family Neisseriaceae being dominant, and the genera Methanobrevibacter and Methanomethylophilaceae were also identified as the core epimural archaeal taxa [41]. Although several studies have been reported on the diversity and relative abundance of epimural microbial communities [41,42], further studies are needed to determine the establishment process of epimural microbial communities. Moreover, although the epimural microbiota is less than 1% of the total rumen microbiota and has a low contribution to VFA production [43], they are the ones likely to interact directly with the host [44], suggesting that the epimural microbiota plays an important role in rumen epithelial function [45]. To date, the understanding of the epimural microbial community is very limited, although the functions of the epimural microbiota are speculated to be involved in oxygen scavenging, urea recycling, and tissue recycling [45,46]. Therefore, future studies are essential to identify the role of epimural microbiota in rumen function.
2.6. The Window of Time for Manipulating Rumen Microbial Colonization in Early Life
There are many studies on the manipulation of rumen microbial colonization in the early stage of life, however, further confirmation of whether nutritional interventions in early life have long-term effects on adult ruminants is still needed. In addition to the potential manipulation strategies described above, it is also important to grasp the most sensitive time window for intervention. The bacterial communities were unstable and more malleable before 20 days of age until weaning (60 days after birth). Therefore, Yin et al. [9] suggested that the first 20 days of life may be the best opportunity for interventions. Different from Yin et al. [9], Li et al. [10] suggested that probiotic intervention during the weaning transition (6–8 weeks) is the best opportunity to manipulate microbiota community composition. As mentioned earlier, the colonization of bacterial, archaeal, and fungal communities occurs before the introduction of a solid diet, and the main microbial communities commonly found in the mature rumen are established in the rumen of young ruminants at 1 week of age [15,22]. Therefore, we suggest that the most suitable time for microbiota manipulation could be right after birth, which agrees with Dias et al. [22] and Abecia et al. [47]. However, the most suitable time for microbiota manipulation and the lasting effects of early nutritional intervention on enhancing rumen fermentation needs to be studied further.
3. Factors That Influence Early Life Rumen Microbiota Colonization
Dietary changes, host genotype, age, host immunity, rumen microbial transplantation, and other factors all affect the colonization of rumen microorganisms in young ruminants (Table 1). The effect of the host age on the rumen microbiota has already been discussed.

Table 1.
Factors influencing the rumen microbiota.
3.1. Dietary Changes
As a substrate for rumen microbial fermentation, the nutrient composition and physical characteristics of the diet directly affect microbial colonization. Dias et al. [22] investigated the effect of a pre-weaned diet (milk versus milk plus starter feeds) on the rumen microbiota of dairy calves, and the results showed that the inclusion of starter feeds facilitated the bacteria, capable of utilizing carbohydrates in the rumen (e.g., Succinivribrio, Sharpea, and Megasphaera); the milk-fed group showed the bacteria (e.g., Parabacteroides, Bacteroides, and Lactobacillus) known to degrade milk nutrients were dominant in the rumen. Recent studies have found that feeding neonatal ruminants with only concentrate can easily cause symptoms such as decreased rumen pH, agglutination of rumen nipples, and incomplete keratinization while adding hay based on concentrate can increase rumen pH, increased rumen volume, and maintaining normal morphology of rumen nipples [54,55,56]. During this process of adding hay based on concentrate, the pre-weaning and post-weaning rumen microbiota were similar, and the fluctuations in bacterial abundance between pre- and post-weaning periods were reduced, promoting the adaptation of neonatal ruminants to the post-weaning diet and reducing weaning stress [48]. This provides a clue for strategies to improve rumen function by manipulating the rumen microbiota in early life. The addition of concentrate or concentrate plus alfalfa pellets to the feed of lambs from 20 to 60 days of age improves the abundance of cellulolytic bacteria and proteolytic bacteria in the rumen content, Ruminococcus unclassified drives the rumen epithelium transcriptome and signaling pathways in cell metabolism, and subsequently promote the development of the rumen epithelium, and may be an important genus in influencing lambs’ rumen development [57].
The rumen epithelium is a unique site of interaction between the rumen microbial metabolism and the host [58]. A growing body of research suggests that rumen epithelium development is influenced by lifelong metabolic communication between the rumen microbiota and the host, which develops and changes with diet [8,59]. Therefore, many studies have explored the potential mechanisms of microbiome-host interactions in stimulating ruminal epithelium development through different dietary niches. Wu et al. [60] investigated the diet-rumen microbiota-host interaction in pre-weaned yak calves, and the 16S rRNA sequencing results indicated that butyrate-producing genera were enhanced by the addition of the calf starter or alfalfa hay; transcriptomic results have revealed that alfalfa hay or the starter supplementation upregulated the PIK3-Akt signaling pathway that regulates the development of the ruminal epithelium; the correlation analysis showed that several altered genera (Christensenella, Sphingomonas, Vulcaniibacterium, shigella, Kandleria, Escherichia Aquabacterium, and Limnobacter) were positively correlated with butyrate production, and all of them were increased by the starter supplementation. These findings suggest that diet-rumen microbiota-host interactions stimulate the development of rumen epithelium in pre-weaned calves. Lin et al. [59] revealed the mechanism by which starter feeding promotes rumen epithelial development in lambs from the perspective of rumen microbiota-host interaction. The 16S rRNA and 18S rRNA sequencing results showed that the abundance of acetate-producing Mitsuokella spp., lactate-producing Sharpea spp., lactate-utilizing Megasphaera spp., and Entodinium spp. were enriched in the rumen microbial community in the starter-feed group; metagenomic results indicated that starter feeding significantly increased the GH13 encoding α-amylase; transcriptome results showed that gene expression of MAPK1, PIK3CB, TNFSF10, ITGA6, SNAI2, SAV1, and DLG, which are associated with growth-associated signal pathways, were significantly up-regulated, while gene expression of BAD, which is involved in the apoptotic process, was down-regulated; correlation analysis revealed a high correlation between the expressions of these genes and acetate and butyrate concentrations. The data suggest that the increased acetate and butyrate production by the microbiota mediates the expression of growth-related genes in the growth-related signaling pathway in the ruminal epithelium. Overall, these results suggest that the diets not only induce changes in the colonization process of rumen microorganisms but also affect the metabolic function and interaction patterns of rumen microorganisms through diet-rumen microbiota-host interactions.
3.2. Host Genotype
During rumen development, changes in rumen structure and physiological properties are associated with the rumen microbiota [61]. Microbial colonization can cause a range of physiological changes in the rumen of newborn ruminants. On the contrary, the host’s physiological changes also cause a series of changes in the microbial composition of the rumen [1]. However, there is no conclusive understanding of whether microbial community alterations lead to functional changes in the host or whether these changes are the result of changes in the host physiology [8]. Pan et al. [62] investigated the rumen transcriptomic and metagenomic data to tease the developmental reprogramming of rumen functions, microbiota colonization, and their functional interactions in seven-time points from postnatal goats. The results showed that both the temporal dynamics of the rumen transcription profile and the microbial metagenome profile exhibit two distinct phases during pre-weaning rumen development; transcriptional profiles of the rumen were divided into the immune-related response phase (d 1–14 after birth) and nutrient-related metabolism phase (d 21–56 after birth); microbial metagenomic profiles of the rumen were divided into bacteriocin biosynthesis phase (d 7–28 after birth) and glycolysis/gluconeogenesis activity phase (d 42–56 after birth); the development shift in the rumen transcriptome (on d 21) was earlier than the feed stimulus (on d 25) and the shift of the rumen microbiome (on d 42). In addition, two newly evolved genes (LYZ1 and DEFB1) showed antibacterial activity against gram-positive bacteria. Taken together, these results suggest that first-stage rumen development is more likely to be a programmed process rather than a result of diet and microbial stimulation and that the newly evolved genes may have specific functions in regulating the composition of the microbiota [62,63]. A study of high- and low-efficiency Holstein cows found that after a near total exchange of rumen contents between the cows, the host restored the rumen bacterial community to its pre-exchange state, suggesting that the rumen microbial community is to some extent controlled by the host genes [64]. In addition to the host genetics, differences in rumen microbiota composition may be due to other factors, including higher cesarean delivery rates in some breeds of cattle, such as Belgian Blue cattle [65]. Moreover, the physiology of the rumen also likely plays an influential role in the rumen microbiota. Henderson et al. [66] identified diet as a major driver of rumen microbiota composition, but they also found some differences between certain bacterial communities in different ruminants, such as the higher abundance of unclassified Veillonellaceae in sheep, deer, and camelids compared to cattle, which may be related to differences in the rumen and camelid foregut size, physiology, and feeding frequencies, thus explaining the host specificity of the rumen microbiota. In the early stages of rumen development, there is significant heterogeneity in the composition of the rumen microbial community, and rumen development significantly affects the diversity of rumen bacteria. This is mainly due to substantial changes in the anatomical structure of the rumen, followed by changes in physiological and metabolic functions [67]. For example, the VFA produced by the rumen microbiota affects the size and shape of the rumen papillae, and these rumen papillae structures alter microbial colonization as they provide a niche environment for a particular rumen microbiota [68]. There is increasing evidence that host genetics, age, early life events, and rumen physiology have important effects on the microbial community in the rumen. Nonetheless, our current understanding of the host effect on the establishment of rumen microbiota is still incomplete.
3.3. Host Immunity
Due to the nature of fermentation and the constant exposure of the rumen epithelium to microorganisms, physiological factors in the rumen may differ from those in other parts of the GIT. The rumen epithelium consists of up to 15 cell layers, which limits permeability to large molecules, and the rumen epithelium has no organized lymphoid tissue. As stated by Yáñez-Ruiz et al. [7], the microbial equilibrium in the rumen is achieved through a combination of the immunoglobulins, IgG and IgA in saliva, Toll-like receptors (TLRs), genetically encoded pattern recognition receptors, peptidoglycan recognition proteins, and antimicrobial peptides defensins. Newborn ruminants have no immunity at birth, and the immunoglobulins, IgG, IgA, and IgM in colostrum can induce immunity and modulate the intestinal bacterial community composition, maintaining a balance between the intestinal microbiota and the host [7]. Moreover, the colonization of microbiota plays a key role in the development of host innate immunity [69]. Pan et al. [62] revealed that the host immune regulation could be activated after the microbiota colonization on d 1, the bacteriocin produced by rumen microbiota is crucial for the rumen immune response and subsequent microbiota colonization, and the expressions of some immune-related genes (CEBPE, SOCS1, SOCS3, CCDC3, and S100A9) in the rumen have a lasting effect on the establishment of microbiota. Secreted immunoglobulin A, which favors symbiotic bacteria in the gut [70], has been demonstrated to coat rumen bacteria [71] and regulate the host’s recognition of certain microbial species.
Members of the TLRs gene family are important regulatory elements of the rumen epithelium, and under normal physiological conditions, they may respond to changes in the structure of the rumen microbial community and make a corresponding regulatory response to maintain the immune tolerance of the rumen epithelium to symbiotic microorganisms [72]. In monogastric animals, some members of the TLRs gene family can inhibit the activation of epithelial NF-kB (nuclear factor-kappa B) inflammatory pathway by binding to specific symbiotic secreted proteins, and thus play an important regulatory role in the maintenance of digestive tract epithelial immune tolerance [73]. Shen et al. [74] found that non-fiber carbohydrates (NFC) induced the expansion of commensal bacteria (Verrucomicrobia subdivision 5), and promoted the immune tolerance of rumen epithelium by upregulation of TLR10 expression. In addition, Chen et al. [37] showed that, compared with acidosis-sensitive cattle, acidosis-resistant cattle had different epimural bacteria diversity and higher expressions of TLR2 and TLR4 in the rumen epithelium, suggesting that the susceptibility to subacute rumen acidosis may be related to the immune activity of the epithelial tissue, which is induced by the epimural microbiota. Therefore, the epimural microbiota plays a critical role in ruminal immune function through direct interactions with the host [45]. However, there has been little research on the immune system’s response to different microbial colonization patterns. Therefore, further studies are needed to illustrate the impact of different microbial colonization patterns on the immune system and the long-term effects.
3.4. Rumen Microbial Transplantation, Probiotics, and Other Factors
Rumen microbial transplantation is an effective method for reconstructing the structure of the rumen microbiota. As carriers of mature rumen function, adult rumen microorganisms are a donor source for microbial transplantation in young ruminants. Compared to specific microbial preparations, rumen microbial transplantation may accelerate the colonization process of rumen microorganisms and may even affect the host’s immune function. Belanche et al. [75] showed that inoculation of neonatal goats with adult goat rumen fluid could facilitate protozoa colonization and promote forage intake, VFA production, and nutrient absorption during the pre-weaning period. The researchers used young lambs as models, rumen fluid transplantation from adult sheep was performed orally before and during weaning, and it has been found that inoculation with fresh rumen fluid promotes bacterial colonization of lambs, such as Succiniclasticum, Prevotella, and Proteobacteria S24-7; rumen fluid transplantation before weaning promoted more microbial colonization than rumen fluid transplantation during weaning [52]. Feeding lyophilized rumen fluid to lambs changed the composition of Selemonas ruminantium and Megasphaera elsdenii, and reduced the relative abundance of Ruminococcus flavescens [76]. The studies indicate the importance of early intervention in rumen microflora. In addition, probiotic supplementation before weaning ruminants can also alter the process of rumen microbial colonization. For instance, when fibrolytic bacteria isolated from the rumen of the moose were used as probiotics in neonatal lambs for 9 weeks, the acetate/propionate ratio decreased and the abundance of Prevotella, Butyrivibrio, and Ruminococcus increased [77]. Supplementation with Saccharomyces cerevisiae in the diet of lambs between 1 and 180 days of age promoted cellulolytic bacterial colonization in the rumen, decreased the rumen ammonia concentration, and increase the VFA concentration [51]. In addition to the above factors, different feeding patterns (nursing with mother versus artificial feeding milk replacer), culture conditions (grazing versus house feeding), ruminant species, and environment affect the colonization of rumen microorganisms [34].
4. Conclusions
In summary, the establishment of the rumen microbiota in early life is crucial for the improvement of rumen function and immune system during pre-weaned ruminants. Several studies have found that early nutritional interventions can manipulate the establishment of rumen microbiota. However, whether early nutritional interventions have a lasting effect on adult animal growth and phenotypes needs to be investigated further. Furthermore, there is still a lack of understanding of the mechanisms that regulate rumen microbiome-host interactions during rumen development, which limits the development of new and effective feed additives for ruminants.
Author Contributions
K.L.: writing—original draft preparation, drawing pictures and sorting out forms; R.N. and B.S.: writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (No. 31760690).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors acknowledge the funding support by National Natural Science Foundation of China.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Huws, S.A.; Creevey, C.J.; Oyama, L.B.; Mizrahi, I.; Denman, S.E.; Popova, M.; Muñoz-Tamayo, R.; Forano, E.; Waters, S.M.; Hess, E.; et al. Addressing global ruminant agricultural challenges through understanding the rumen microbiome: Past, present, and future. Front. Microbiol. 2018, 9, 2161–2193. [Google Scholar] [CrossRef] [PubMed]
- United Nations. Global Issues: Population. Available online: https://www.un.org/en/global-issues/population (accessed on 30 January 2023).
- Beauchemin, K.A.; Ungerfeld, E.M.; Eckard, R.J.; Wang, M. Review: Fifty years of research on rumen methanogenesis: Lessons learned and future challenges for mitigation. Animal 2020, 14, s2–s16. [Google Scholar] [CrossRef] [PubMed]
- Weimer, P.J. Redundancy, resilience, and host specificity of the ruminal microbiota: Implications for engineering improved ruminal fermentations. Front. Microbiol. 2015, 6, 296–311. [Google Scholar] [CrossRef] [PubMed]
- Lyons, T.; Boland, T.; Storey, S.; Doyle, E. Linseed oil supplementation of lambs’ diet in early life leads to persistent changes in rumen microbiome structure. Front. Microbiol. 2017, 8, 1656–1667. [Google Scholar] [CrossRef] [PubMed]
- Jiao, J.; Li, X.; Beauchemin, K.A.; Tan, Z.; Tang, S.; Zhou, C. Rumen development process in goats as affected by supplemental feeding v. grazing: Age-related anatomic development, functional achievement and microbial colonisation. Br. J. Nutr. 2015, 113, 888–900. [Google Scholar] [CrossRef]
- Yáñez-Ruiz, D.R.; Abecia, L.; Newbold, C.J. Manipulating rumen microbiome and fermentation through interventions during early life: A review. Front. Microbiol. 2015, 6, 1133–1144. [Google Scholar] [CrossRef]
- Malmuthuge, N.; Guan, L.L. Understanding host-microbial interactions in rumen: Searching the best opportunity for microbiota manipulation. J. Anim. Sci. Biotechnol. 2017, 8, 8–14. [Google Scholar] [CrossRef]
- Yin, X.; Ji, S.; Duan, C.; Tian, P.; Ju, S.; Yan, H.; Zhang, Y.; Liu, Y. Age-related changes in the ruminal microbiota and their relationship with rumen fermentation in lambs. Front. Microbiol. 2021, 12, 679135–679146. [Google Scholar] [CrossRef]
- Li, B.; Zhang, K.; Li, C.; Wang, X.; Chen, Y.; Yang, Y. Characterization and comparison of microbiota in the gastrointestinal tracts of the goat (Capra hircus) during preweaning development. Front. Microbiol. 2019, 10, 2125–2140. [Google Scholar] [CrossRef]
- Wang, Z.; Elekwachi, C.O.; Jiao, J.; Wang, M.; Tang, S.; Zhou, C.; Tan, Z.; Forster, R.J. Investigation and manipulation of metabolically active methanogen community composition during rumen development in black goats. Sci. Rep. 2017, 7, 422–435. [Google Scholar] [CrossRef]
- Bi, Y.; Zhang, N.; Wang, S.; Diao, Q. Multiomics analysis reveals the presence of a microbiome in the gut of fetal lambs. Gut 2021, 70, 853–864. [Google Scholar] [CrossRef]
- de Goffau, M.C.; Charnock-Jones, D.S.; Smith, G.C.S.; Parkhill, J. Batch effects account for the main findings of an in utero human intestinal bacterial colonization study. Microbiome 2021, 9, 6–12. [Google Scholar] [CrossRef]
- Walter, J.; Hornef, M.W. A philosophical perspective on the prenatal in utero microbiome debate. Microbiome 2021, 9, 5–13. [Google Scholar] [CrossRef]
- Malmuthuge, N.; Liang, G.; Guan, L.L. Regulation of rumen development in neonatal ruminants through microbial metagenomes and host transcriptomes. Genome Biol. 2019, 20, 172–187. [Google Scholar] [CrossRef]
- Jiao, J.; Huang, J.; Zhou, C.; Tan, Z. Taxonomic identification of ruminal epithelial bacterial diversity during rumen development in goats. Appl. Environ. Microb. 2015, 81, 3502–3509. [Google Scholar] [CrossRef]
- Zhang, K.; Li, B.; Guo, M.; Liu, G.; Yang, Y.; Wang, X.; Chen, Y.; Zhang, E. Maturation of the goat rumen microbiota involves three stages of microbial colonization. Animals 2019, 9, 1028–1043. [Google Scholar] [CrossRef]
- Rey, M.; Enjalbert, F.; Combes, S.; Cauquil, L.; Bouchez, O.; Monteils, V. Establishment of ruminal bacterial community in dairy calves from birth to weaning is sequential. J. Appl. Microbiol. 2014, 116, 245–257. [Google Scholar] [CrossRef]
- Shakhov, A.G.; Sashnina, L.Y.; Yerina, T.A. Optimization of the process of forming an intestinal microbiocenose in newborn calves to prevent gastrointestinal diseases. Russ. Agric. Sci. 2015, 41, 55–58. [Google Scholar] [CrossRef]
- Jami, E.; Israel, A.; Kotser, A.; Mizrahi, I. Exploring the bovine rumen bacterial community from birth to adulthood. ISME J. 2013, 7, 1069–1079. [Google Scholar] [CrossRef]
- Yeoman, C.J.; Ishaq, S.L.; Bichi, E.; Olivo, S.K.; Lowe, J.; Aldridge, B.M. Biogeographical differences in the influence of maternal Microbial sources on the early successional development of the bovine neonatal gastrointestinal tract. Sci. Rep. 2018, 8, 3197–3200. [Google Scholar] [CrossRef]
- Dias, J.; Marcondes, M.I.; Noronha, M.F.; Resende, R.T.; Machado, F.S.; Mantovani, H.C.; Dill-McFarland, K.A.; Suen, G. Effect of pre-weaning diet on the ruminal archaeal, bacterial, and fungal communities of dairy calves. Front. Microbiol. 2017, 8, 1553–1569. [Google Scholar] [CrossRef]
- Wang, L.; Xu, Q.; Kong, F.; Yang, Y.; Wu, D.; Mishra, S.; Li, Y. Exploring the goat rumen microbiome from seven days to two years. PLoS ONE 2016, 11, e0154354–e0154366. [Google Scholar] [CrossRef] [PubMed]
- Morvan, B.; Dore, J.; Rieu-Lesme, F.; Foucat, L.; Fonty, G.; Gouet, P. Establishment of hydrogen-utilizing bacteria in the rumen of the newborn lamb. FEMS. Microbiol. Lett. 1994, 117, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Guzman, C.E.; Bereza-Malcolm, L.T.; De Groef, B.; Franks, A.E. Presence of selected methanogens, fibrolytic bacteria, and proteobacteria in the gastrointestinal tract of neonatal dairy calves from birth to 72 hours. PLoS ONE 2015, 10, e0133048. [Google Scholar] [CrossRef] [PubMed]
- Friedman, N.; Jami, E.; Mizrahi, I. Compositional and functional dynamics of the bovine rumen methanogenic community across different developmental stages. Environ. Microbiol. 2017, 19, 3365–3373. [Google Scholar] [CrossRef]
- Dong, L.; Ma, J.; Tu, Y.; Diao, Q. Weaning methods affect ruminal methanogenic archaea composition and diversity in Holstein calves. J. Integr. Agric. 2019, 18, 1080–1092. [Google Scholar] [CrossRef]
- Jiao, J.Z. Postnatal Microbial Succession and Fermentation Capacity Establishment of the Rumen and Hindgut in Supplemental Feeding and Grazing Goats. Ph.D. Thesis, University of Chinese Academy of Sciences, Beijing, China, 2015. [Google Scholar]
- Fonty, G.; Gouet, P.; Jouany, J.P.; Senaud, J. Establishment of the microflora and anaerobic fungi in the rumen of lambs. J. Gen. Microbiol. 1987, 133, 1835–1843. [Google Scholar] [CrossRef]
- Yin, X.; Ji, S.; Duan, C.; Tian, P.; Ju, S.; Yan, H.; Zhang, Y.; Liu, Y. Dynamic change of fungal community in the gastrointestinal tract of growing lambs. J. Integr. Agric. 2022, 21, 3314–3328. [Google Scholar] [CrossRef]
- Cholewińska, P.; Czyż, K.; Nowakowski, P.; Wyrostek, A. The microbiome of the digestive system of ruminants—A review. Anim. Health. Res. Rev. 2020, 21, 3–14. [Google Scholar] [CrossRef]
- Naga, M.A.; Akkada, A.R.A.; El-Shazly, K. Establishment of rumen ciliate protozoa in cow and water buffalo (Bos bubalus L.) calves under late and early weaning systems. J. Dairy Sci. 1969, 52, 110–112. [Google Scholar] [CrossRef]
- Eadie, J.M. The development of rumen microbial populations in lambs and calves under various conditions of management. J. Gen. Microbiol. 1962, 29, 563–578. [Google Scholar] [CrossRef]
- Zhang, Y.; Choi, S.H.; Nogoy, K.M.; Liang, S. Review: The development of the gastrointestinal tract microbiota and intervention in neonatal ruminants. Animal 2021, 15, 100316–100324. [Google Scholar] [CrossRef]
- Li, M.; Zhou, M.; Adamowicz, E.; Basarab, J.A.; Guan, L.L. Characterization of bovine ruminal epithelial bacterial communities using 16S rRNAsequencing, PCR-DGGE, and qRT-PCR analysis. Vet. Microbiol. 2012, 155, 72–80. [Google Scholar] [CrossRef]
- Chen, Y.; Penner, G.B.; Li, M.; Oba, M.; Guan, L.L. Changes in bacterial diversity associated with epithelial tissue in the beef cow rumen during the transition to a high-grain diet. Appl. Environ. Microbiol. 2011, 77, 5770–5781. [Google Scholar] [CrossRef]
- Chen, Y.; Oba, M.; Guan, L.L. Variation of bacterial communities and expression of Toll-like receptor genes in the rumen of steers differing in susceptibility to subacute ruminal acidosis. Vet. Microbiol. 2012, 159, 451–459. [Google Scholar] [CrossRef]
- Petri, R.M.; Schwaiger, T.; Penner, G.B.; Beauchemin, K.A.; Forster, R.J.; Mckinnon, J.J.; McAllister, T.A. Characterization of the core rumen microbiome in cattle during transition from forage to concentrate as well as during and after an acidotic challenge. PLoS ONE 2013, 8, e83424. [Google Scholar] [CrossRef]
- Holman, D.B.; Gzyl, K.E. A meta-analysis of the bovine gastrointestinal tract microbiota. FEMS Microbiol. Ecol. 2019, 95, fiz072. [Google Scholar] [CrossRef]
- De Mulder, T.; Goossens, K.; Peiren, N.; Vandaele, L.; Haegeman, A.; De Tender, C.; Ruttink, T.; de Wiele, T.V.; De Campeneere, S. Exploring the methanogen and bacterial communities of rumen environments: Solid adherent, fluid and epimural. FEMS Microbiol. Ecol. 2017, 93, fiw251. [Google Scholar] [CrossRef]
- Anderson, C.J.; Koester, L.R.; Schmitz-Esser, S. Rumen epithelial communities share a core bacterial microbiota: A meta-analysis of 16s rRNA gene Illumina Miseq sequencing datasets. Front. Microbiol. 2021, 12, 539. [Google Scholar] [CrossRef]
- Ishaq, S.L.; AlZahal., O.; Walker, N.; McBride, B. An investigation into rumen fungal and protozoal diversity in three rumen fractions, during high-fiber or grain-induced sub-acute ruminal acidosis conditions, with or without active dry yeast supplementation. Front. Microbiol. 2017, 8, 1943–1958. [Google Scholar] [CrossRef]
- Sadet, S.; Martin, C.; Meunier, B.; Morgavi, D.P. PCR-DGGE analysis reveals a distinct diversity in the bacterial population attached to the rumen epithelium. Animal 2007, 1, 939–944. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Bian, G.; Sun, D.; Zhu, W.; Mao, S. Starter feeding altered ruminal epithelial bacterial communities and some key immune-related genes’ expression before weaning in lambs. J. Anim. Sci. 2017, 95, 910–921. [Google Scholar] [CrossRef] [PubMed]
- Na, S.W.; Guan, L.L. Understanding the role of rumen epithelial host-microbe interactions in cattle feed efficiency. Anim. Nutr. 2022, 10, 41–53. [Google Scholar] [CrossRef] [PubMed]
- Cheng, K.; McCowan, R.; Costerton, J. Adherent epithelial bacteria in ruminants and their roles in digestive tract function. Am. J. Clin. Nutr. 1979, 32, 139–148. [Google Scholar] [CrossRef]
- Abecia, L.; Martín-García, A.I.; Martínez, G.; Newbold, C.J.; Yáñez-Ruiz, D.R. Nutritional intervention in early life to manipulate rumen microbial colonization and methane output by kid goats postweaning. J. Anim. Sci. 2013, 91, 4832–4840. [Google Scholar] [CrossRef]
- Yang, B.; Le, J.; Wu, P.; Liu, J.; Guan, L.L.; Wang, J. Alfalfa intervention alters rumen microbial community development in Hu lambs during early life. Front. Microbiol. 2018, 9, 574. [Google Scholar] [CrossRef]
- Ribeiro, G.O.; Oss, D.B.; He, Z.; Gruninger, R.J.; Elekwachi, C.; Forster, R.J.; Yang, W.Z.; Beauchemin, K.A.; McAllister, T.A. Repeated inoculation of cattle rumen with bison rumen contents alters the rumen microbiome and improves nitrogen digestibility in cattle. Sci. Rep. 2017, 7, 1276. [Google Scholar] [CrossRef]
- Li, F.; Li, C.; Chen, Y.; Liu, J.; Zhang, C.; Irving, B.; Fitzsimmons, C.; Plastow, G.; Guan, L.L. Host genetics influence the rumen microbiota and heritable rumen microbial features associate with feed efficiency in cattle. Microbiome 2019, 7, 92–108. [Google Scholar] [CrossRef]
- Chaucheyras-Durand, F.; Fonty, G. Establishment of cellulolytic bacteria and development of fermentative activities in the rumen of gnotobiotically-reared lambs receiving the microbial additive Saccharomyces cerevisiae CNCM I-1077. Reprod. Nutr. Dev. 2001, 41, 57–68. [Google Scholar] [CrossRef]
- Yu, S.; Zhang, G.; Liu, Z.; Wu, P.; Yu, Z.; Wang, J. Repeated inoculation with fresh rumen fluid before or during weaning modulates the microbiota composition and cooccurrence of the rumen and colon of lambs. BMC. Microbiol. 2020, 20, 29–44. [Google Scholar] [CrossRef]
- Ma, T.; Wu, W.; Tu, Y.; Zhang, N.; Diao, Q. Resveratrol affects in vitro rumen fermentation, methane production and prokaryotic community composition in a time- and diet-specific manner. Microb. Biotechnol. 2020, 13, 1751–1764. [Google Scholar] [CrossRef]
- Castells, L.; Bach, A.A.; Terré, M. Effects of forage provision to young calves on rumen fermentation and development of the gastrointestinal tract. J. Dairy Sci. 2013, 96, 5226–5236. [Google Scholar] [CrossRef]
- Khan, M.A.; Weary, D.M.; von Keyserlingk, M.A.G. Hay intake improves performance and rumen development of calves fed higher quantities of milk. J. Dairy Sci. 2011, 94, 3547–3553. [Google Scholar] [CrossRef]
- Connor, E.E.; Baldwin, R.L., VI; Li, C.; Li, R.W.; Chung, H. Gene expression in bovine rumen epithelium during weaning identifies molecular regulators of rumen development and growth. Funct. Integr. Genom. 2013, 13, 133–142. [Google Scholar] [CrossRef]
- Lv, X.K. Effects of Different Diets on Rumen Development of 20~60 Day-Old Goat Kids. Master’s Thesis, Chinese Academy of Agricultural Sciences, Beijing, China, 2019. [Google Scholar]
- Liu, K.; Zhang, Y.; Yu, Z.; Xu, Q.; Zheng, N.; Zhao, S.; Huang, G.; Wang, J. Ruminal microbiota-host interaction and its effect on nutrient metabolism. Anim. Nutr. 2021, 7, 49–55. [Google Scholar] [CrossRef]
- Lin, L.; Xie, F.; Sun, D.; Liu, J.; Zhu, W.; Mao, S. Ruminal microbiome-host crosstalk stimulates the development of the ruminal epithelium in a lamb model. Microbiome 2019, 7, 83–98. [Google Scholar] [CrossRef]
- Wu, S.; Cui, Z.; Chen, X.; Zheng, L.; Ren, H.; Wang, D.; Yao, J. Diet-ruminal microbiome-host crosstalk contributes to differential effects of calf starter and alfalfa hay on rumen epithelial development and pancreatic α-amylase activity in yak calves. J. Dairy Sci. 2021, 104, 4326–4340. [Google Scholar] [CrossRef]
- Diao, Q.; Zhang, R.; Fu, T. Review of strategies to promote rumen development in calves. Animals 2019, 9, 490–505. [Google Scholar] [CrossRef]
- Pan, X.; Li, Z.; Li, B.; Zhao, C.; Wang, Y.; Chen, Y.; Jiang, Y. Dynamics of rumen gene expression, microbiome colonization, and their interplay in goats. BMC Genom. 2021, 22, 288–303. [Google Scholar] [CrossRef]
- Pan, X.; Cai, Y.; Li, Z.; Chen, X.; Heller, R.; Wang, N.; Wang, Y.; Zhao, C.; Wang, Y.; Xu, H.; et al. Modes of genetic adaptations underlying functional innovations in the rumen. Sci. China Life Sci. 2020, 63, 1–21. [Google Scholar] [CrossRef]
- Weimer, P.J.; Cox, M.S.; De Paula, T.V.; Lin, M.; Hall, M.B.; Suen, G. Transient changes in milk production efficiency and bacterial community composition resulting from near-total exchange of ruminal contents between high- and low-efficiency Holstein cows. J. Dairy Sci. 2017, 100, 7165–7182. [Google Scholar] [CrossRef] [PubMed]
- De Mulder, T.; Peiren, N.; Vandaele, L.; Ruttink, T.; De Campeneere, S.; Van de Wiele, T.; Goossens, K. Impact of breed on the rumen microbial community composition and methane emission of Holstein Friesian and Belgian Blue heifers. Livestock Sci. 2018, 207, 38–44. [Google Scholar] [CrossRef]
- Henderson, G.; Cox, F.; Ganesh, S.; Jonker, A.; Young, W. Global Rumen Census Collaborators and Janssen P H. Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci. Rep. 2015, 5, 14567–14581. [Google Scholar] [CrossRef] [PubMed]
- Li, R.W.; Connor, E.E.; Li, C.; Baldwin, R.L., VI; Sparks, M.E. Characterization of the rumen microbiota of pre-ruminant calves using metagenomic tools. Environ. Microbiol. 2012, 14, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Rieu, F.; Fonty, G.; Gaillard, B.; Gouet, P. Electron microscopy study of the bacteria adherent to the rumen wall in young conventional lambs. Can. J. Microbiol. 1990, 36, 140–144. [Google Scholar] [CrossRef]
- Arshad, M.A.; Hassan, F.; Rehman, M.S.; Huws, S.A.; Cheng, Y.; Din, A.U. Gut microbiome colonization and development in neonatal ruminants: Strategies, prospects, and opportunities. Anim. Nutr. 2021, 7, 883–895. [Google Scholar] [CrossRef]
- Gutzeit, C.; Magri, G.; Cerutti, A. Intestinal IgA production and its role in host-microbe interaction. Immun. Rev. 2014, 260, 76–85. [Google Scholar] [CrossRef]
- Fouhse, J.M.; Smiegielski, L.; Tuplin, M.; Guan, L.L.; Willing, B.P. Host immune selection of rumen bacteria through salivary secretory IgA. Front. Microbiol. 2017, 8, 848–856. [Google Scholar] [CrossRef]
- Zhang, R.; Zhu, W.; Mao, S. High-concentrate feeding upregulates the expression of inflammation-related genes in the ruminal epithelium of dairy cattle. J. Anim. Sci. Biotechnol. 2016, 7, 42. [Google Scholar] [CrossRef]
- Xiang, R.; Oddy, V.H.; Archibald, A.L.; Vercoe, P.E.; Dalrymple, B.P. Epithelial, metabolic and innate immunity transcriptomic signatures differentiating the rumen from other sheep and mammalian gastrointestinal tract tissues. Peer J. 2016, 4, 1762–1792. [Google Scholar] [CrossRef]
- Shen, H.; Lu, Z.; Chen, Z.; Wu, Y.; Shen, Z. Rapid fermentable substance modulates interactions between ruminal commensals and Toll-like receptors in promotion of immune tolerance of goat rumen. Front. Microbiol. 2016, 7, 1812–1821. [Google Scholar] [CrossRef]
- Belanche, A.; Palma-Hidalgo, J.M.; Nejjam, I.; Jiménez, E.; Yáñez-Ruiz, D.R. Inoculation with rumen fluid in early life as a strategy to optimize the weaning process in intensive dairy goat systems. J. Dairy Sci. 2020, 103, 5047–5060. [Google Scholar] [CrossRef]
- Yu, S.; Shi, W.; Yang, B.; Gao, G.; Chen, H.; Cao, L.; Yu, Z.; Wang, J. Effects of repeated oral inoculation of artificially fed lambs with lyophilized rumen fluid on growth performance, rumen fermentation, microbial population and organ development. Anim. Feed Sci. Tech. 2020, 264, 114465. [Google Scholar] [CrossRef]
- Ishaq, S.L.; Kim, C.J.; Reis, D.; Wright, A.-D.G. Fibrolytic bacteria isolated from the rumen of North American Moose (Alces alces) and their use as a probiotic in neonatal lambs. PLoS ONE 2015, 10, e0144804–e0144828. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).