Hepatopancreas Proteomic Analysis Reveals Key Proteins and Pathways in Regulatory of Ovary Maturation of Macrobrachium nipponense
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Samples Preparation
2.2. Total Protein Extraction, Digestion and TMT Labeling
2.3. Fraction Separation and LC-MS/MS Analysis
2.4. Data Processing and Bioinformatics Analysis
2.5. Real-Time PCR Validation and Statistical Analysis
3. Results
3.1. Proteins Identification and Analysis
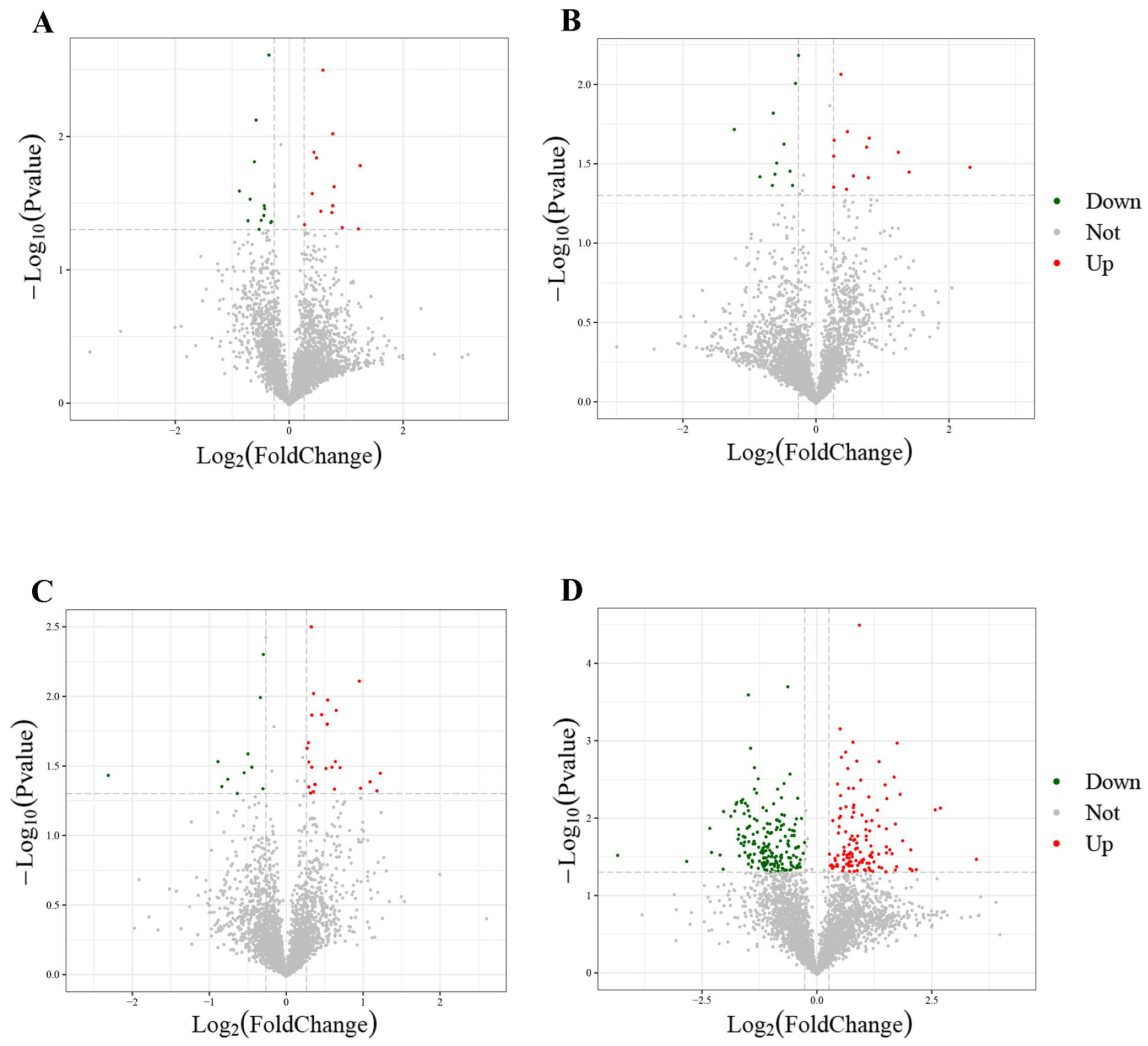
3.2. Comparison of Proteome Profiles and DEP Discovered
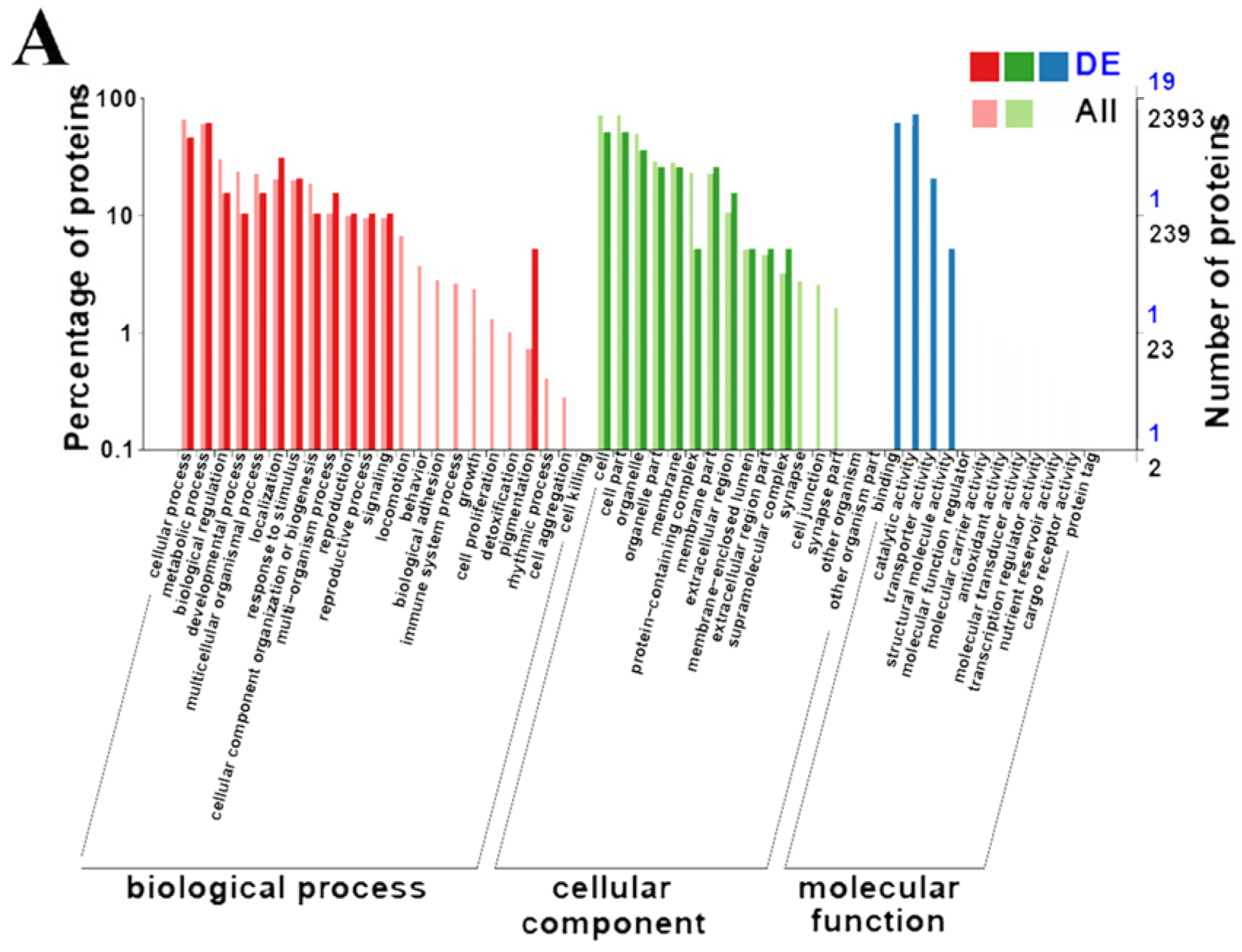
3.3. GO Enrichment
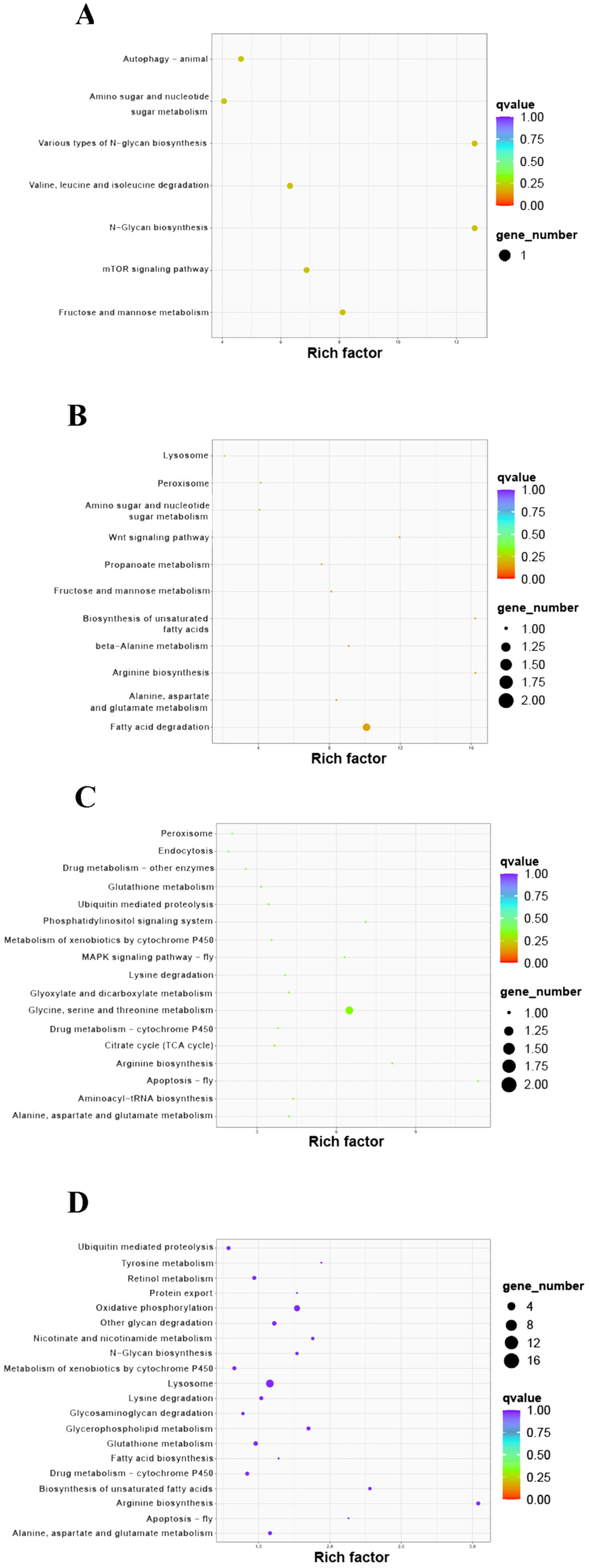
3.4. KEGG Enrichment
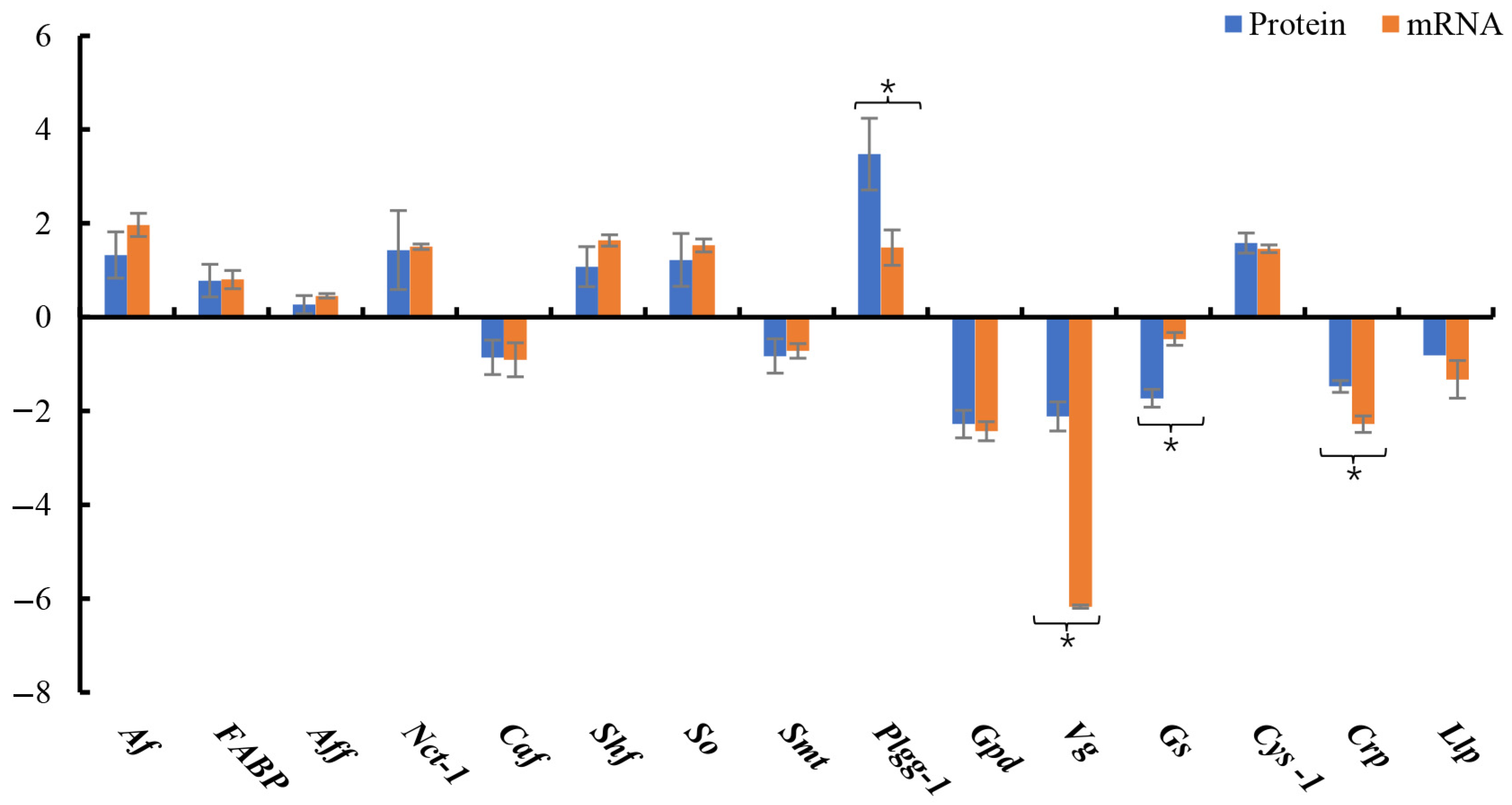
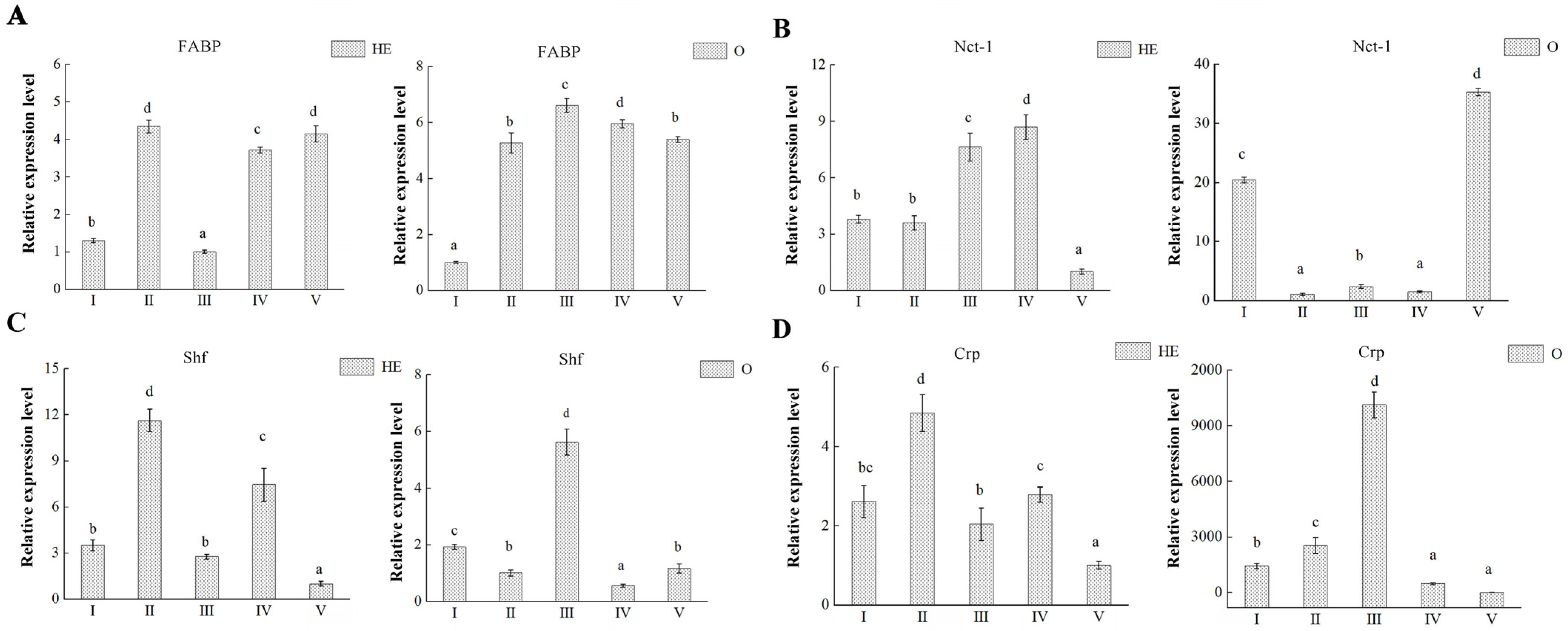
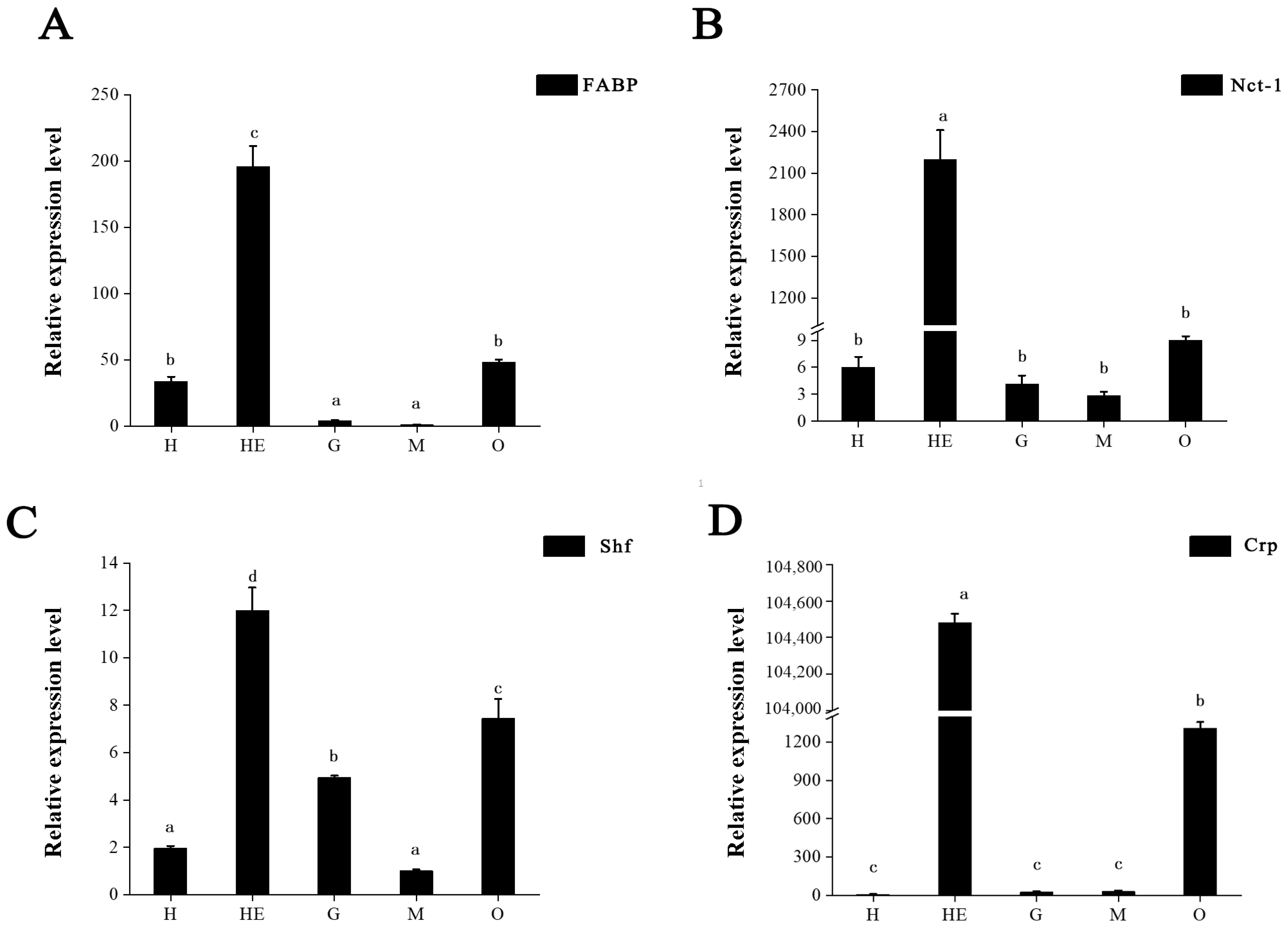
3.5. DEP Validation
3.6. Key Proteins Involved in Ovary Maturation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rodríguez-González, H.; Hernández-Llamas, A.; Villarreal, H.; Saucedo, P.E.; García-Ulloa, M.; Rodríguez-Jaramillo, C. Gonadal development and biochemical composition of female crayfish Cherax quadricarinatus (Decapoda: Parastacidae) in relation to the Gonadosomatic Index at first maturation. Aquaculture 2006, 254, 637–645. [Google Scholar] [CrossRef]
- Haefner, P.A., Jr.; Spaargaren, D.H. Interactions of ovary and hepatopancreas during the reproductive cycle of Crangon crangon (L.): I. weight and volume relationships. J. Crustacean Biol. 1993, 13, 523–531. [Google Scholar] [CrossRef]
- Spaargaren, D.H.; Haefner, P.A., Jr. Interactions of ovary and hepatopancreas during the reproductive cycle of Crangon crangon (L.). II. Biochemical relationships. J. Crustacean Biol. 1994, 14, 6–19. [Google Scholar] [CrossRef]
- Zara, F.J.; Gaeta, H.H.; Costa, T.M.; Toyama, M.H.; Caetano, F.H. The ovarian cycle histochemistry and its relationship with hepatopancreas weight in the blue crab Callinectes danae (Crustacea: Portunidae). Acta Zool. 2013, 94, 134–146. [Google Scholar] [CrossRef]
- Magalhães, T.; Mossolin, E.C.; Mantelatto, F.L. Gonadosomatic and hepatosomatic indexes of the freshwater shrimp Macrobrachium olfersii (Decapoda, Palaemonidae) from São Sebastião Island, Southeastern Brazil. Pan-Am. J. Aquat. Sci. 2012, 7, 1–9. [Google Scholar]
- Tseng, D.Y.; Chen, Y.N.; Kou, G.H.; Lo, C.F.; Kuo, C.M. Hepatopancreas is the extraovarian site of vitellogenin synthesis in black tiger shrimp, Penaeus monodon. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2001, 129, 909–917. [Google Scholar] [CrossRef]
- Tiu, S.H.K.; Benzie, J.; Chan, S.M. From hepatopancreas to ovary: Molecular characterization of a shrimp vitellogenin receptor involved in the processing of vitellogenin. Biol. Reprod. 2008, 79, 66–74. [Google Scholar] [CrossRef]
- Wang, X.; Jin, M.; Cheng, X.; Hu, X.; Zhao, M.; Yuan, Y.; Zhou, Q. Hepatopancreas transcriptomic and lipidomic analyses reveal the molecular responses of mud crab (Scylla paramamosain) to dietary ratio of docosahexaenoic acid to eicosapentaenoic acid. Aquaculture 2022, 551, 737903. [Google Scholar] [CrossRef]
- Yang, F.; Xu, H.T.; Dai, Z.M.; Yang, W.J. Molecular characterization and expression analysis of vitellogenin in the marine crab Portunus trituberculatus. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2005, 142, 456–464. [Google Scholar] [CrossRef]
- Zmora, N.; Trant, J.; Chan, S.M.; Chung, J.S. Vitellogenin and its messenger RNA during ovarian development in the female blue crab, Callinectes sapidus: Gene expression, synthesis, transport, and cleavage. Biol. Reprod. 2007, 77, 138–146. [Google Scholar] [CrossRef]
- Rankin, S.M.; Bradfield, J.Y.; Keeley, L.L. Ovarian protein synthesis in the South American white shrimp, Penaeus vannamei, during the reproductive cycle. Invertebr. Reprod. Dev. 1989, 15, 27–33. [Google Scholar] [CrossRef]
- Tom, M.; Fingerman, M.; Hayes, T.K.; Johnson, V.; Kerner, B.; Lubzens, E. A comparative study of the ovarian proteins from two penaeid shrimps, Penaeus semisulcatus de Haan and Penaeus vannamei (Boone). Comp. Biochem. Physiol. Part B Comp. Biochem. 1992, 102, 483–490. [Google Scholar] [CrossRef]
- Boucard, C.G.V.; Levy, P.; Ceccaldi, H.J.; Brogren, C.H. Developmental changes in concentrations of vitellin, vitellogenin, and lipids in hemolymph, hepatopancreas, and ovaries from different ovarian stages of Indian white prawn Fenneropenaeus indicus. J. Exp. Mar. Biol. Ecol. 2002, 281, 63–75. [Google Scholar] [CrossRef]
- Vazquez, F.J.; Tropea, C.; Lopez Greco, L.S. Development of the female reproductive system in the freshwater crayfish Cherax quadricarinatus (Decapoda, Parastacidae). Invertebr. Biol. 2008, 127, 433–443. [Google Scholar] [CrossRef]
- Alava, V.R.; Quinitio, E.T.; De Pedro, J.B.; Priolo, F.M.P.; Orozco, Z.G.A.; Wille, M. Lipids and fatty acids in wild and pond-reared mud crab Scylla serrata (Forsskål) during ovarian maturation and spawning. Aquac. Res. 2007, 38, 1468–1477. [Google Scholar] [CrossRef]
- Kong, Y.; Ding, Z.; Zhang, Y.; Zhou, P.; Wu, C.; Zhu, M.; Ye, J. Types of carbohydrate in feed affect the growth performance, antioxidant capacity, immunity, and activity of digestive and carbohydrate metabolism enzymes in juvenile Macrobrachium nipponense. Aquaculture 2019, 512, 734282. [Google Scholar] [CrossRef]
- Ministry of Agriculture Fisheries Bureau. China Fishery Statistical Yearbook; Chinese Agricultural Press: Beijing, China, 2021. [Google Scholar]
- Jiang, S.; Xiong, Y.; Zhang, W.; Zhu, J.; Cheng, D.; Gong, Y.; Fu, H. Molecular Characterization of a Novel Cathepsin L in Macrobrachium nipponense and its Function in Ovary Maturation. Front. Endocrinol. 2021, 12, 816813. [Google Scholar] [CrossRef]
- Qiao, H.; Fu, H.; Xiong, Y.; Jiang, S.; Sun, S.; Jin, S.; Wu, Y. Molecular insights into reproduction regulation of female Oriental River prawns Macrobrachium nipponense through comparative transcriptomic analysis. Sci. Rep. 2017, 7, 12161. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Fu, H.; Qiao, H.; Jin, S.; Zhang, W.; Jiang, S.; Xiong, Y. Expression and functional analysis of cathepsin L1 in ovarian development of the oriental river prawn, Macrobrachium nipponense. Aquac. Rep. 2021, 20, 100724. [Google Scholar] [CrossRef]
- Zhang, Y.; Jiang, S.; Qiao, H.; Xiong, Y.; Fu, H.; Zhang, W.; Gong, Y.; Jin, S.; Wu, Y. Transcriptome analysis of five ovarian stages reveals gonad maturation in female Macrobrachium nipponense. BMC Genom. 2021, 22, 510. [Google Scholar] [CrossRef]
- Jiang, S.; Zhang, W.; Xiong, Y.; Cheng, D.; Wang, J.; Jin, S.; Fu, H. Hepatopancreas transcriptome analyses provide new insights into the molecular regulatory mechanism of fast ovary maturation in Macrobrachium nipponense. BMC Genom. 2022, 23, 625. [Google Scholar] [CrossRef] [PubMed]
- Qiao, H.; Xiong, Y.; Zhang, W.; Fu, H.; Jiang, S.; Sun, S.; Bai, H.; Jin, S.; Gong, Y. Characterization, expression, and function analysis of gonad-inhibiting hormone in Oriental River prawn, Macrobrachium nipponense and its induced expression by temperature. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2015, 185, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Bian, C.; Jiang, S.; Han, K.; Xiong, Y.; Zhang, W.; Fu, H. A chromosome-level genome assembly of the oriental river prawn, Macrobrachium nipponense. GigaScience 2021, 10, giaa160. [Google Scholar] [CrossRef]
- Hu, Y.; Fu, H.; Qiao, H.; Sun, S.; Zhang, W.; Jin, S.; Wu, Y. Validation and evaluation of reference genes for quantitative real-time PCR in Macrobrachium Nipponense. Int. J. Mol. Sci. 2018, 19, 2258. [Google Scholar] [CrossRef]
- Yuan, H.; Zhang, W.; Fu, Y.; Jiang, S.; Xiong, Y.; Zhai, S.; Wu, Y. MnFtz-f1 Is Required for Molting and Ovulation of the Oriental River Prawn Macrobrachium nipponense. Front. Endocrinol. 2021, 12, 798577. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Chai, Y.M.; Yu, S.S.; Zhao, X.F.; Zhu, Q.; Wang, J.X. Comparative proteomic profiles of the hepatopancreas in Fenneropenaeus chinensis response to white spot syndrome virus. Fish Shellfish. Immunol. 2010, 29, 480–486. [Google Scholar] [CrossRef] [PubMed]
- Nagaraju, G.P.C. Is methyl farnesoate a crustacean hormone. Aquaculture 2007, 272, 39–54. [Google Scholar] [CrossRef]
- Liu, M.; Pan, J.; Liu, Z.; Cheng, Y.; Gong, J.; Wu, X. Effect of estradiol on vitellogenesis and oocyte development of female swimming crab, Portunus trituberculatus. Aquaculture 2018, 486, 240–245. [Google Scholar] [CrossRef]
- Montes-Dominguez, A.L.; Avena-Soto, J.A.; Lizarraga-Rodriguez, J.L.; de Jesus Perez-Gala, R.; Jimenez-Gutierrez, S.; Sotelo-Falomir, J.A.; Jimenez-Gutierrez, L.R. Comparison between cultured and wild Pacific white shrimp (Penaeus vannamei) vitellogenesis: Next-generation sequencing and relative expression of genes directly and indirectly related to reproduction. PeerJ 2021, 9, e10694. [Google Scholar] [CrossRef]
- Vogt, G. Life-cycle and functional cytology of the hepatopancreatic cells of Astacus astacus (Crustacea, Decapoda). Zoomorphology 1994, 114, 83–101. [Google Scholar] [CrossRef]
- Wang, L.; Yan, B.; Liu, N.; Li, Y.; Wang, Q. Effects of cadmium on glutathione synthesis in hepatopancreas of freshwater crab, Sinopotamon yangtsekiense. Chemosphere 2008, 74, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wu, X.; Liu, Z.; Zheng, H.; Cheng, Y. Insights into hepatopancreatic functions for nutrition metabolism and ovarian development in the crab Portunus trituberculatus: Gene discovery in the comparative transcriptome of different hepatopancreas stages. PloS ONE 2014, 9, e84921. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Lai, W. The lipid accumulations during the stages of the ovarian fast maturation and their effect on the spawning of Eriocheir sinensis. J. Fish. China 2000, 24, 113–118. [Google Scholar]
- Zhang, Y.; Fu, Y.; Jiang, S.; Qiao, H.; Xiong, Y.; Fu, H.; Wu, Y. Comparative metabolomics analysis of ovarian developmental stages in Macrobrachium nipponense. Comp. Biochem. Physiol. Part D Genom. Proteomics 2020, 34, 100648. [Google Scholar] [CrossRef] [PubMed]
- Domino, S.E.; Zhang, L.; Gillespie, P.J.; Saunders, T.L.; Lowe, J.B. Deficiency of reproductive tract α (1, 2) fucosylated glycans and normal fertility in mice with targeted deletions of the FUT1 or FUT2 α (1, 2) fucosyltransferase locus. Mol. Cell. Biol. 2001, 21, 8336–8345. [Google Scholar] [CrossRef] [PubMed]
- Litvinova, E.A.; Bets, V.D.; Feofanova, N.A.; Gvozdeva, O.V.; Achasova, K.M.; Alperina, E.L.; Kozhevnikova, E.N. Dietary fucose affects macrophage polarization and reproductive performance in mice. Nutrients 2021, 13, 855. [Google Scholar] [CrossRef]
- Tantibhedhyangkul, J.; Weerachatyanukul, W.; Carmona, E.; Xu, H.; Anupriwan, A.; Michaud, D.; Tanphaichitr, N. Role of sperm surface arylsulfatase A in mouse sperm-zona pellucida binding. Biol. Reprod. 2002, 67, 212–219. [Google Scholar] [CrossRef]
- Sayasith, K.; Sirois, J.; Lussier, J.G. Expression, regulation, and promoter activation of Vanin-2 (VNN2) in bovine follicles prior to ovulation. Biol. Reprod. 2013, 89, 1–11. [Google Scholar] [CrossRef]
- Del Collado, M.; da Silveira, J.C.; Sangalli, J.R.; Andrade, G.M.; Sousa, L.R.D.S.; Silva, L.A.; Perecin, F. Fatty acid binding protein 3 and transzonal projections are involved in lipid accumulation during in vitro maturation of bovine oocytes. Sci. Rep. 2017, 7, 2645. [Google Scholar] [CrossRef]
- Ding, L.Y.; Jin, M.; Sun, P.; Lu, Y.; Ma, H.N.; Yuan, Y.; Zhou, Q.C. Cloning, tissue expression of the fatty acid-binding protein (Pt-FABP1) gene, and effects of dietary phospholipid levels on fabp and vitellogenin gene expression in the female swimming crab Portunus trituberculatus. Aquaculture 2017, 474, 57–65. [Google Scholar] [CrossRef]
- Feng, Q.M.; Liu, M.M.; Cheng, Y.X.; Wu, X.G. Comparative proteomics elucidates the dynamics of ovarian development in the Chinese mitten crab Eriocheir sinensis. Comp. Biochem. Physiol. Part D Genom. Proteom. 2021, 40, 100878. [Google Scholar] [CrossRef] [PubMed]
- Dunning, K.R.; Russell, D.L.; Robker, R.L. Lipids and oocyte developmental competence: The role of fatty acids and b-oxidation. Reproduction 2014, 148, R15–R27. [Google Scholar] [CrossRef] [PubMed]
- Aoyama, T.; Souri, M.; Kamijo, T.; Ushikubo, S.; Hashimoto, T. Peroxisomal acyl-coenzyme A oxidase is a rate-limiting enzyme in a very long-chain fatty acid β-oxidation system. Biochem. Biophys. Res. Commun. 1994, 201, 1541–1547. [Google Scholar] [CrossRef] [PubMed]
- Madureira, T.V.; Castro, L.F.C.; Rocha, E. Acyl-coenzyme A oxidases 1 and 3 in brown trout (Salmo trutta f. fario): Can peroxisomal fatty acid β-oxidation be regulated by estrogen signaling. Fish Physiol. Biochem. 2016, 42, 389–401. [Google Scholar] [CrossRef]
- Gévry, N.Y.; Lopes, F.L.; Ledoux, S.; Murphy, B.D. Aberrant intracellular cholesterol transport disrupts pituitary and ovarian function. Mol. Endocrinol. 2004, 18, 1778–1786. [Google Scholar] [CrossRef]
- Pfeffer, S.R. NPC intracellular cholesterol transporter 1 (NPC1)-mediated cholesterol export from lysosomes. J. Biol. Chem. 2019, 294, 1706–1709. [Google Scholar] [CrossRef]
- Roger, P.; Pujol, P.; Lucas, A.; Baldet, P.; Rochefort, H. Increased immunostaining of fibulin-1, an estrogen-regulated protein in the stroma of human ovarian epithelial tumors. Am. J. Pathol. 1998, 153, 1579–1588. [Google Scholar] [CrossRef]
- Haendler, B.; Yamanouchi, H.; Lessey, B.A.; Chwalisz, K.; Hess-Stumpp, H. Cycle-dependent endometrial expression and hormonal regulation of the fibulin-1 gene. Mol. Reprod. Dev. Inc. Gamete Res. 2004, 68, 279–287. [Google Scholar] [CrossRef]
- Kritzer, M.F.; Kohama, S.G. Ovarian hormones differentially influence immunoreactivity for dopamine β-hydroxylase, choline acetyltransferase, and serotonin in the dorsolateral prefrontal cortex of adult rhesus monkeys. J. Comp. Neurol. 1999, 409, 438–451. [Google Scholar] [CrossRef]
- Bohacek, J.; Bearl, A.M.; Daniel, J.M. Long-term ovarian hormone deprivation alters the ability of subsequent oestradiol replacement to regulate choline acetyltransferase protein levels in the hippocampus and prefrontal cortex of middle-aged rats. J. Neuroendocrinol. 2008, 20, 1023–1027. [Google Scholar] [CrossRef] [PubMed]
- Rieger, D.; Loskutoff, N.M. Changes in the metabolism of glucose, pyruvate, glutamine and glycine during maturation of cattle oocytes in vitro. Reproduction 1994, 100, 257–262. [Google Scholar] [CrossRef] [PubMed]
- Lonergan, P.; Gutiérrez-Adán, A.; Rizos, D.; Pintado, B.; De La Fuente, J.; Boland, M.P. Relative messenger RNA abundance in bovine oocytes collected in vitro or in vivo before and 20 hr after the preovulatory luteinizing hormone surge. Mol. Reprod. Dev. Inc. Gamete Research 2003, 66, 297–305. [Google Scholar] [CrossRef]
- Gupta, R.; Yang, Q.; Dogra, S.K.; Wajapeyee, N. Serine hydroxymethyl transferase 1 stimulates pro-oncogenic cytokine expression through sialic acid to promote ovarian cancer tumor growth and progression. Oncogene 2017, 36, 4014–4024. [Google Scholar] [CrossRef]
- Bedford, M.T.; Richard, S. Arginine methylation: An emerging regulator of protein function. Mol. Cell 2005, 18, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Bazer, F.W.; Davis, T.A.; Kim, S.W.; Li, P.; Marc Rhoads, J.; Yin, Y. Arginine metabolism and nutrition in growth, health and disease. Amino Acids 2009, 37, 153–168. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Shi, H.; Liang, S.; Ning, G.; Xu, N.; Lu, J.; Lin, F. MoARG1, MoARG5, 6 and MoARG7 involved in arginine biosynthesis are essential for growth, conidiogenesis, sexual reproduction, and pathogenicity in Magnaporthe oryzae. Microbiol. Res. 2015, 180, 11–22. [Google Scholar] [CrossRef]
- Srivastava, A.S.; Oohara, I.; Suzuki, T.; Singh, S.N. Activity and expression of aspartate aminotransferase during the reproductive cycle of a fresh water fish, Clarias batrachus. Fish Physiol. Biochem. 1999, 20, 243–250. [Google Scholar] [CrossRef]
- Hattori, Y.; Shimoda, S.; Gross, S.S. Effect of lipopolysaccharide treatment in vivo on tissue expression of argininosuccinate synthetase and argininosuccinate lyase mRNAs: Relationship to nitric oxide synthase. Biochem. Biophys. Res. Commun. 1995, 215, 148–153. [Google Scholar] [CrossRef]
- Dixit, V.D.; Parvizi, N. Nitric oxide and the control of reproduction. Anim. Reprod. Sci. 2001, 65, 1–16. [Google Scholar] [CrossRef]
- Li, Y.; Wang, Z.; Andersen, C.L.; Ye, X. Functions of lysosomes in mammalian female reproductive system. Reprod. Dev. Med. 2020, 4, 109–122. [Google Scholar] [CrossRef]
- Jiang, S.; Xiong, Y.; Zhang, W.; Zhu, J.; Cheng, D.; Gong, Y.; Fu, H. A novel legumain-like proteases in Macrobrachium nipponense: Identification, characterization and function analysis in ovary maturation. Front. Endocrinol. 2022, 13, 858726. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Khawar, M.B.; Li, W. Autophagy in reproduction. In Autophagy: Biology and Diseases; Springer: Berlin/Heidelberg, Germany, 2019; pp. 453–468. [Google Scholar]

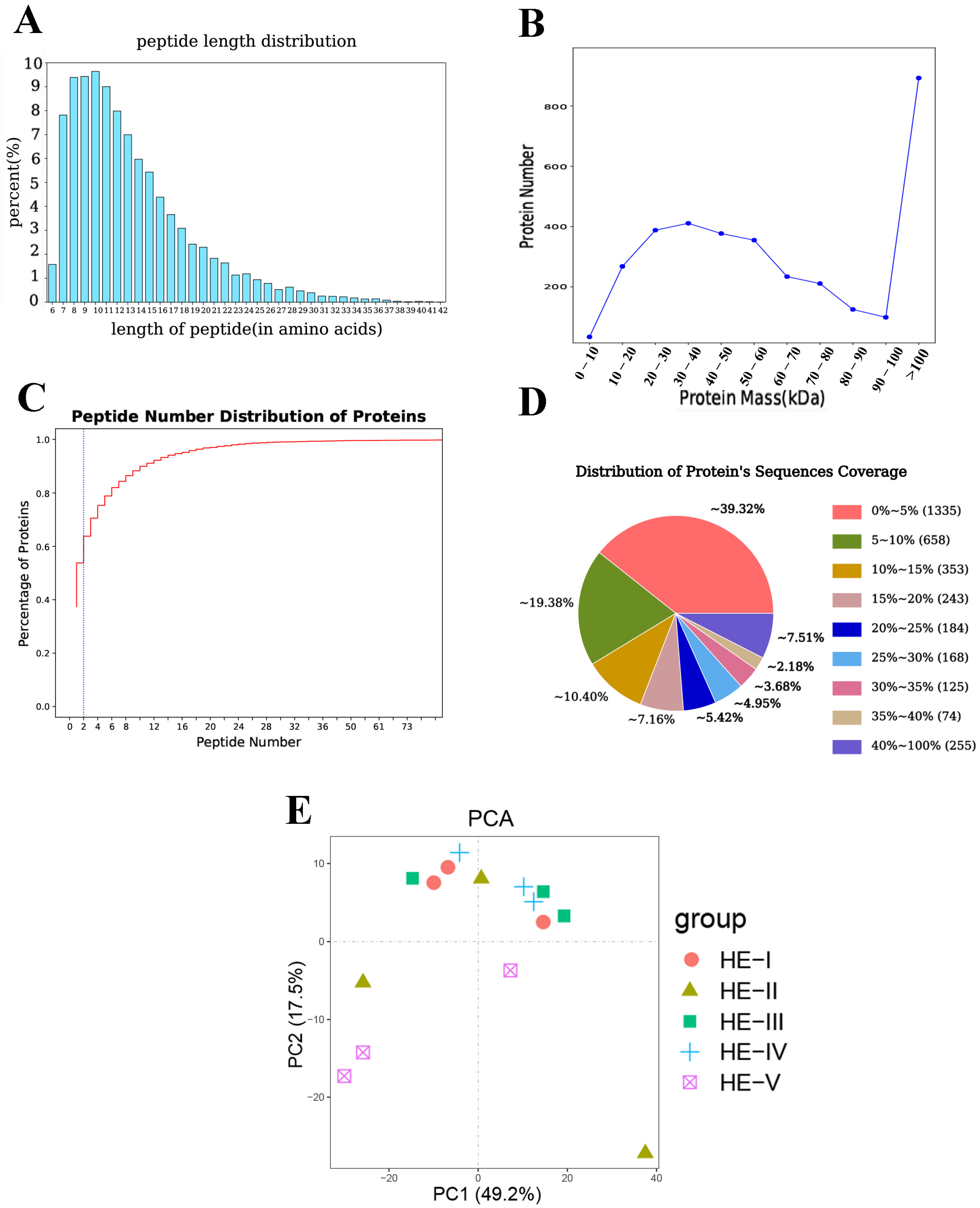


| Ovary Stages | The Description of Characteristics |
|---|---|
| OI | transparent, undeveloped stage |
| OII | yellow, developing stage |
| OIII | light green, nearly-ripe stage |
| OIV | dark green, ripe stage |
| OV | gray, spent stage |
| Comparison Group | Description | Regulation | Fold Change |
|---|---|---|---|
| HE-I vs. HE-II | Arylsulfatase | up | 2.37 |
| Vanin-like protein 2 | up | 2.32 | |
| Fatty acid-binding protein | up | 1.70 | |
| sodium-dependent dicarboxylate transporter | down | 0.54 | |
| Protein Hook homolog | down | 0.62 | |
| Probable 3-hydroxyisobutyrate dehydrogenase | down | 0.65 | |
| HE-II vs. HE-III | NPC intracellular cholesterol transporter 1 | up | 2.64 |
| Pteridine reductase 1 | up | 2.36 | |
| Fibulin-1 | up | 1.74 | |
| Choline O-acetyltransferase | down | 0.55 | |
| GDP-mannose 4,6 dehydratase | down | 0.63 | |
| Arylsulfatase | down | 0.64 | |
| HE-III vs. HE-IV | Sarcosine oxidase | up | 2.33 |
| Serine hydroxymethyltransferase | up | 2.13 | |
| Origin recognition complex subunit 1 | up | 1.95 | |
| sodium-dependent multivitamin transporter | down | 0.56 | |
| Myosin-VIIa | down | 0.59 | |
| ABC transporter C family member 10 | down | 0.70 | |
| HE-IV vs. HE-V | Protein lgg-1 | up | 11.10 |
| Epsin | up | 6.45 | |
| Aromatic amino acid aminotransferase | up | 5.59 | |
| NPC intracellular cholesterol transporter 1 | down | 0.04 | |
| Glyceraldehyde-3-phosphate dehydrogenase | down | 0.14 | |
| Vitellogenin | down | 0.20 |
| Comparison Group | Pathways | Pathway ID | DEP Number | Description | Regulation | Fold Change |
|---|---|---|---|---|---|---|
| HE-I vs. HE-II | N-Glycan biosynthesis | map00510 | 1 | Alpha-(1,6)-fucosyltransferase | up | 1.21 |
| Various types of N-glycan biosynthesis | map00513 | 1 | Alpha-(1,6)-fucosyltransferase | up | 1.21 | |
| HE-II vs. HE-III | Fatty acid degradation | map00071 | 2 | peroxisomal acyl-coenzyme A oxidase 1 | up | 1.20 |
| choline O-acetyltransferase | down | 0.55 | ||||
| HE-III vs. HE-IV | Glycine, serine and threonine metabolism | map00260 | 2 | sarcosine oxidase | up | 2.33 |
| serine hydroxymethyltransferase | up | 2.13 | ||||
| HE-IV vs. HE-V | Arginine biosynthesis | map00220 | 4 | aspartate aminotransferase | up | 1.49 |
| Glutamine synthetase | down | 0.31 | ||||
| Argininosuccinate synthase | down | 0.54 | ||||
| argininosuccinate lyase | down | 0.74 | ||||
| Lysosome | map04142 | 14 | V-type proton ATPase subunit d 1 | up | 2.84 | |
| Sphingomyelin phosphodiesterase | down | 0.41 | ||||
| ceroid-lipofuscinosis neuronal protein 7 | up | 1.31 | ||||
| Cystatin-1 | up | 3.01 | ||||
| NPC intracellular cholesterol transporter 1 | down | 0.04 | ||||
| Crustapin | down | 0.32 | ||||
| AP-1 complex subunit mu-1-I | down | 0.61 | ||||
| Arylsulfatase | down | 0.35 | ||||
| AP-3 complex subunit delta | down | 0.77 | ||||
| Mite group 2 allergen Lep d 2 | down | 0.33 | ||||
| Beta-galactosidase 1 | down | 0.45 | ||||
| legumain-like protein | down | 0.44 | ||||
| Cathepsin L | down | 0.52 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, S.; Qiao, H.; Fu, H.; Gu, Z. Hepatopancreas Proteomic Analysis Reveals Key Proteins and Pathways in Regulatory of Ovary Maturation of Macrobrachium nipponense. Animals 2023, 13, 977. https://doi.org/10.3390/ani13060977
Jiang S, Qiao H, Fu H, Gu Z. Hepatopancreas Proteomic Analysis Reveals Key Proteins and Pathways in Regulatory of Ovary Maturation of Macrobrachium nipponense. Animals. 2023; 13(6):977. https://doi.org/10.3390/ani13060977
Chicago/Turabian StyleJiang, Sufei, Hui Qiao, Hongtuo Fu, and Zemao Gu. 2023. "Hepatopancreas Proteomic Analysis Reveals Key Proteins and Pathways in Regulatory of Ovary Maturation of Macrobrachium nipponense" Animals 13, no. 6: 977. https://doi.org/10.3390/ani13060977
APA StyleJiang, S., Qiao, H., Fu, H., & Gu, Z. (2023). Hepatopancreas Proteomic Analysis Reveals Key Proteins and Pathways in Regulatory of Ovary Maturation of Macrobrachium nipponense. Animals, 13(6), 977. https://doi.org/10.3390/ani13060977







