Evolutionary and Genomic Diversity of True Polyploidy in Tetrapods
Abstract
Simple Summary
Abstract
1. Introduction
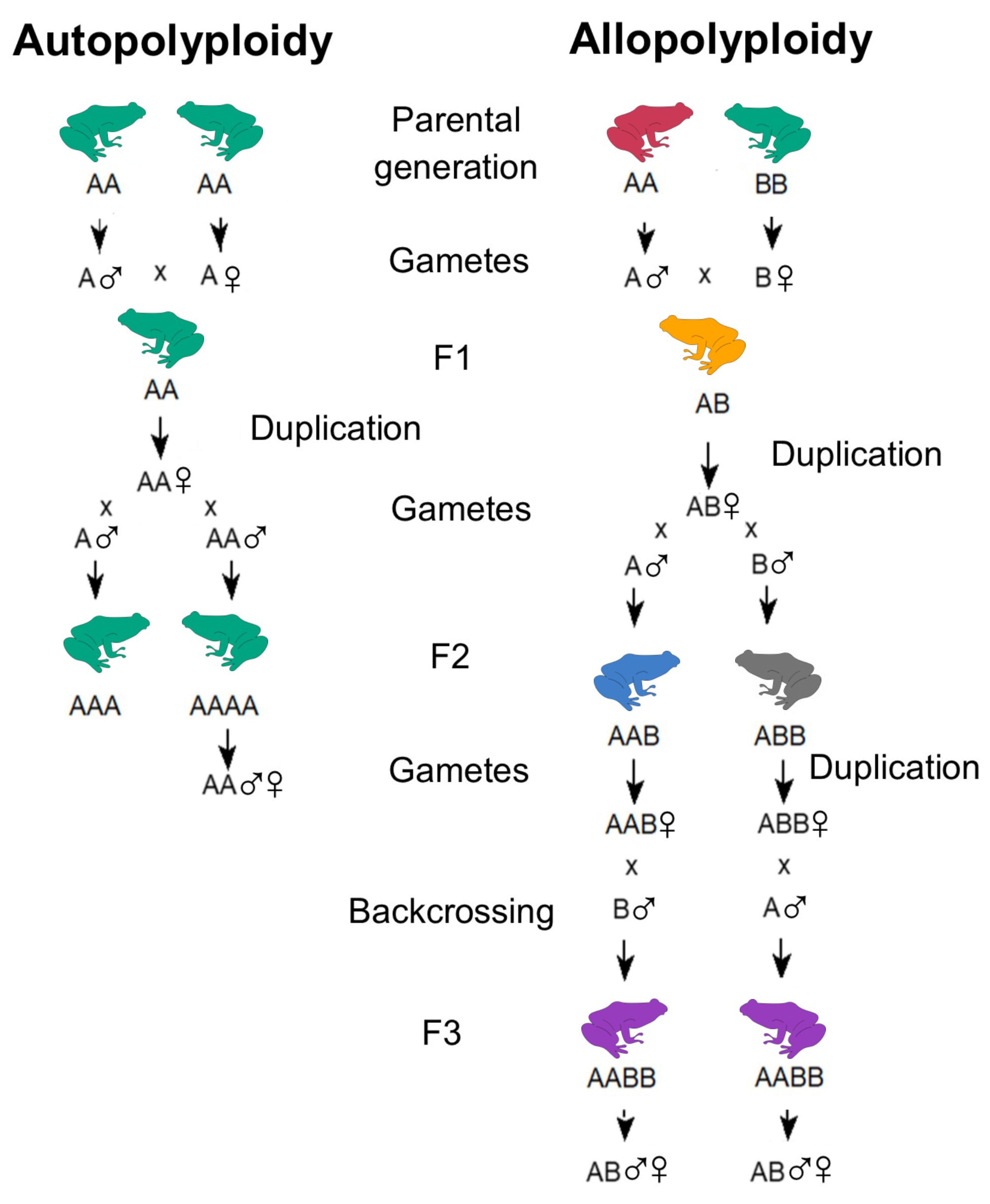
2. Classification and Mechanisms of Polyploidy
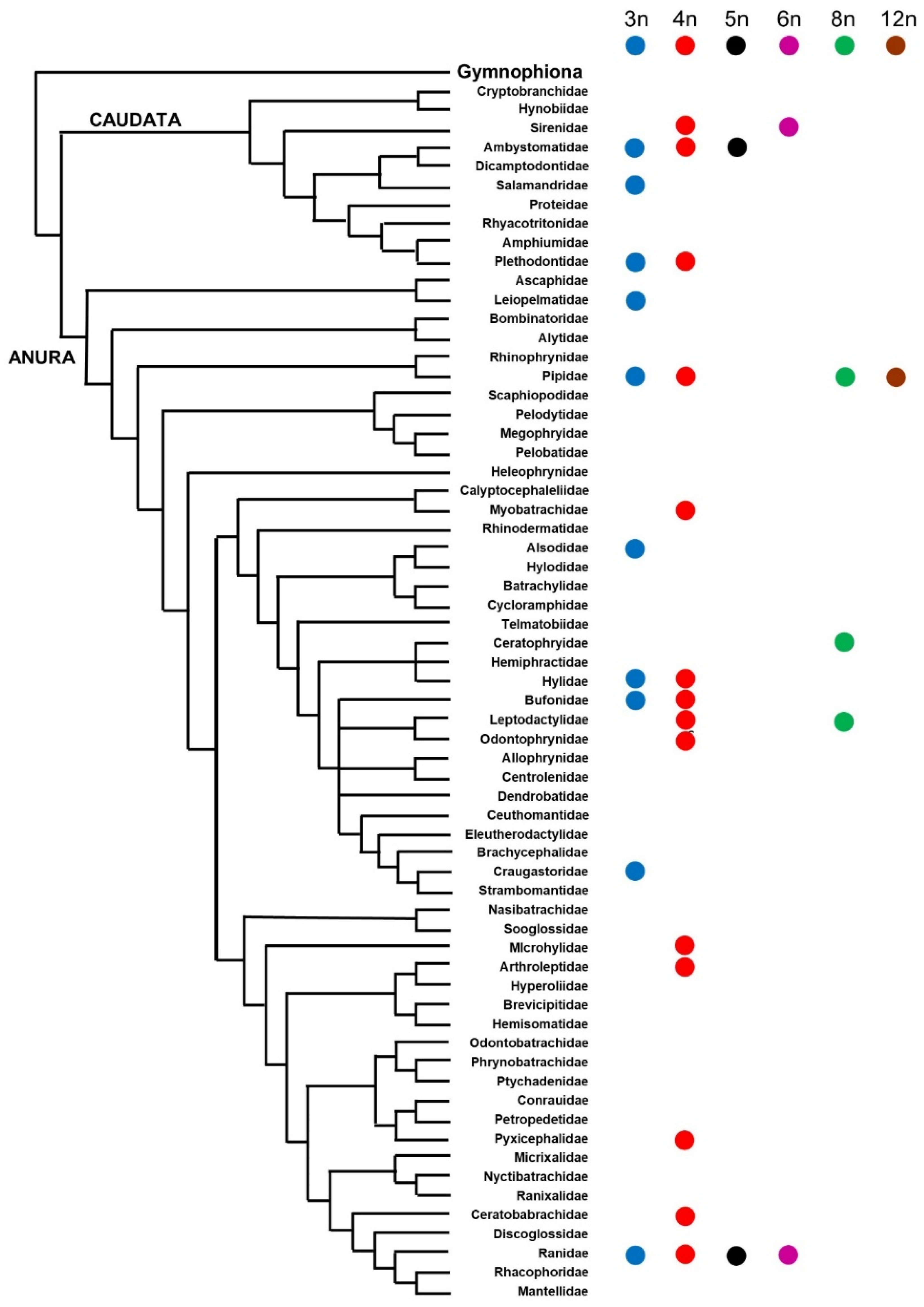
3. Amphibians
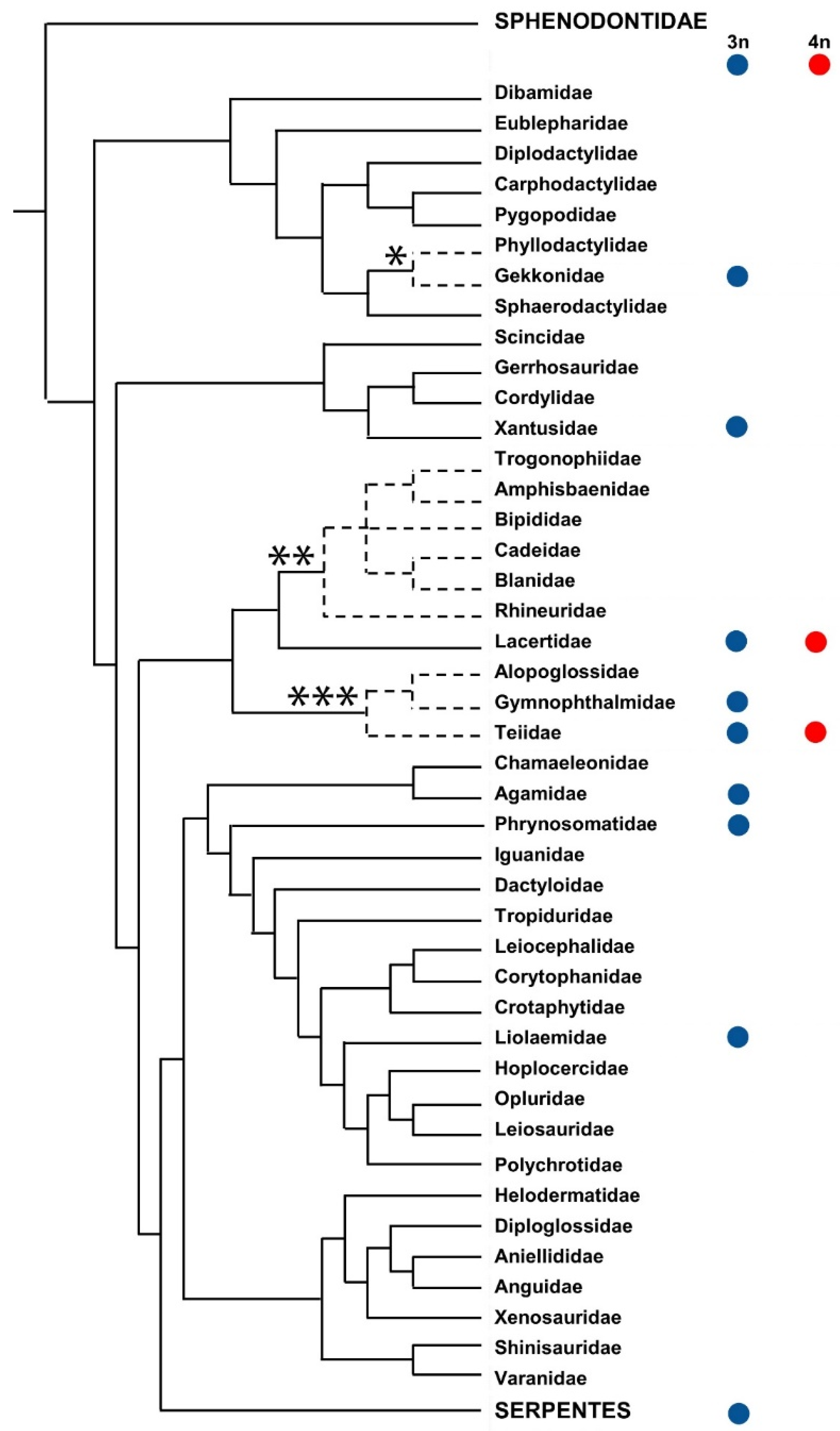
4. Reptiles
5. Birds
6. Mammals
7. Advantages and Disadvantages of Polyploidy
8. Conclusions and Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ramsey, J.; Schemske, D.W. Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu. Rev. Ecol. Syst. 1998, 29, 467–501. [Google Scholar] [CrossRef]
- Rieseberg, L.H.; Willis, J.H. Plant speciation. Science 2007, 317, 910–914. [Google Scholar] [CrossRef] [PubMed]
- Soltis, D.E.; Visger, C.J.; Soltis, P.S. The polyploidy revolution then…and now: Stebbins revisited. Am. J. Bot. 2014, 101, 1057–1078. [Google Scholar] [CrossRef]
- Van de Peer, Y.; Mizrachi, E.; Marchal, K. The evolutionary significance of polyploidy. Nat. Rev. Genet. 2017, 18, 411–424. [Google Scholar] [CrossRef] [PubMed]
- Mable, B.K. Why polyploidy is rarer in animals than in plants: Myths and mechanisms. Biol. J. Linn. Soc. 2004, 82, 453–466. [Google Scholar] [CrossRef]
- Otto, S.P.; Whitton, J. Polyploid incidence and evolution. Annu. Rev. Genet. 2000, 34, 401–437. [Google Scholar] [CrossRef]
- Mable, B.K.; Alexandrou, M.A.; Taylor, M.I. Genome duplication in amphibians and fish: An extended synthesis: Polyploidy in amphibians and fish. J. Zool. 2011, 284, 151–182. [Google Scholar] [CrossRef]
- Ohno, S. Evolution by Gene Duplication; Springer: Berlin, Germany, 1970. [Google Scholar]
- Kasahara, M. The 2R hypothesis: An update. Curr. Opin. Immunol. 2007, 19, 547–552. [Google Scholar] [CrossRef]
- Le Comber, S.C.; Smith, C. Polyploidy in fishes: Patterns and processes. Biol. J. Linn. Soc. 2004, 82, 431–442. [Google Scholar] [CrossRef]
- Schmid, M.; Evans, B.J.; Bogart, J.P. Polyploidy in Amphibia. Cytogenet. Genome Res. 2015, 145, 315–330. [Google Scholar] [CrossRef]
- Moreira, M.O.; Fonseca, C.; Rojas, D. Parthenogenesis is self-destructive for scaled reptiles. Biol. Lett. 2021, 17, 20210006. [Google Scholar] [CrossRef]
- Tiersch, T.R.; Beck, M.L.; Douglas, M. ZZW autotriploidy in a blue-and-yellow macaw. Genetica 1991, 84, 209–212. [Google Scholar] [CrossRef]
- Gallardo, M.H.; Bickham, J.W.; Honeycutt, R.L.; Ojeda, R.A.; Köhler, N. Discovery of tetraploidy in a mammal. Nature 1999, 401, 341. [Google Scholar] [CrossRef]
- Wertheim, B.; Beukeboom, L.W.; van de Zande, L. Polyploidy in animals: Effects of gene expression on sex determination, evolution and ecology. Cytogenet. Genome Res. 2013, 140, 256–269. [Google Scholar] [CrossRef]
- Kihara, H.; Ono, T. Chromosomenzahlen und systematische Gruppierung der Rumex-Arten. Z. Zellforsch. Microsk. Anat. 1926, 4, 475–481. [Google Scholar] [CrossRef]
- Stebbins, G.L. Types of polyploids: Their classification and significance. Adv. Genet. 1947, 1, 403–429. [Google Scholar] [PubMed]
- Comai, L. The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 2005, 6, 836–846. [Google Scholar] [CrossRef] [PubMed]
- Abel, S.; Becker, H.C. The effect of autopolyploidy on biomass production in homozygous lines of Brassica rapa and Brassica oleracea. Plant Breed. 2007, 126, 642–643. [Google Scholar] [CrossRef]
- Soltis, D.E.; Segovia-Salcedo, M.C.; Jordon-Thaden, I.; Majure, L.; Miles, N.M.; Mavrodiev, E.V.; Mei, W.; Cortez, M.B.; Soltis, P.S.; Gitzendanner, M.A. Are polyploids really evolutionary dead-ends (again)? A critical reappraisal of Mayrose et al. (2011). New Phytol. 2014, 202, 1105–1117. [Google Scholar] [CrossRef]
- Sybenga, J. Chromosome pairing affinity and quadrivalent formation in polyploids: Do segmental allopolyploids exist? Genome 1996, 39, 1176–1184. [Google Scholar] [CrossRef]
- Zhang, Z.; Gou, X.; Xun, H.; Bian, Y.; Ma, X.; Li, J.; Li, N.; Gong, L.; Feldman, M.; Liu, B.; et al. Homoeologous exchanges occur through intragenic recombination generating novel transcripts and proteins in wheat and other polyploids. Proc. Natl. Acad. Sci. USA 2020, 117, 14561–14571. [Google Scholar] [CrossRef]
- Wu, Y.; Lin, F.; Zhou, Y.; Wang, J.; Sun, S.; Wang, B.; Zhang, Z.; Li, G.; Lin, X.; Wang, X.; et al. Genomic mosaicism due to homoeologous exchange generates extensive phenotypic diversity in nascent allopolyploids. Natl. Sci. Rev. 2020, 8, nwaa277. [Google Scholar] [CrossRef] [PubMed]
- Edger, P.P.; McKain, M.R.; Bird, K.A.; VanBuren, R. Subgenome assignment in allopolyploids: Challenges and future directions. Curr. Opin. Plant Biol. 2018, 42, 76–80. [Google Scholar] [CrossRef] [PubMed]
- McClintock, B. The significance of responses of the genome to challenge. Science 1984, 226, 792–801. [Google Scholar] [CrossRef] [PubMed]
- Anatskaya, O.V.; Vinogradov, A.E. Polyploidy as a Fundamental Phenomenon in Evolution, Development, Adaptation and Diseases. Int. J. Mol. Sci. 2022, 23, 3542. [Google Scholar] [CrossRef]
- Madlung, A. Polyploidy and its effect on evolutionary success: Old questions revisited with new tools. Heredity 2013, 110, 99–104. [Google Scholar] [CrossRef]
- Storchova, Z.; Pellman, D. From polyploidy to aneuploidy, genome instability and cancer. Nat. Rev. Mol. Cell. Biol. 2004, 5, 45–54. [Google Scholar] [CrossRef]
- Dodsworth, S.; Chase, M.W.; Leitch, A.R. Is post-polyploidization diploidization the key to the evolutionary success of angiosperms? Bot. J. Linn. 2016, 180, 1–5. [Google Scholar] [CrossRef]
- Evans, B.J.; Pyron, R.A.; Wiens, J.J. Polyploidization and sex chromosome evolution in amphibians. In Polyploidy and Genome Evolution; Soltis, P.S., Soltis, D.E., Eds.; Springer: Berlin, Germany, 2012; pp. 385–410. [Google Scholar]
- Miura, I. Sex Determination and Sex Chromosomes in Amphibia. Sex. Dev. 2017, 11, 298–306. [Google Scholar] [CrossRef]
- Frost, D.R. Amphibian Species of the World, an Online Reference; Version 6.0; American Museum of Natural History: New York, NY, USA, 2014; Available online: http://research.amnh.org/herpetology/amphibia/index.html (accessed on 4 February 2014).
- Bogart, J.P. Gynogenetic diploids, tetraploids, or octoploids, and a path to polyploidy in anuran amphibians. Genome 2021, 64, 1053–1065. [Google Scholar] [CrossRef]
- AmphibiaWeb. 2023. Available online: https://amphibiaweb.org (accessed on 7 February 2023).
- Blackburn, D.C.; Wake, D.B. Class Amphibia Gray, 1825. In Animal Biodiversity: An Outline of Higher-Level Classification and Survey of Taxonomic Richness; Zhang, Z., Ed.; Magnolia Press: Auckland, New Zealand, 2011; Volume 3148, pp. 39–55. [Google Scholar]
- Feng, Y.J.; Blackburn, D.C.; Liang, D.; Hillis, D.M.; Wake, D.B.; Cannatella, D.C.; Zhang, P. Phylogenomics reveals rapid, simultaneous diversification of three major clades of Gondwanan frogs at the Cretaceous-Paleogene boundary. Proc. Natl. Acad. Sci. USA 2017, 114, E5864–E5870. [Google Scholar] [CrossRef] [PubMed]
- Jetz, W.; Pyron, R.A. The interplay of past diversification and evolutionary isolation with present imperilment across the amphibian tree of life. Nat. Ecol. Evol. 2018, 2, 850–858. [Google Scholar] [CrossRef]
- Streicher, J.W.; Miller, E.; Guerrero, P.; Correa, C.; Ortiz, J.C.; Crawford, A.J.; Pie, M.R.; Wiens, J.J. Evaluating methods for phylogenomic analyses, and a new phylogeny for a major frog clade (Hyloidea) based on 2214 loci. Mol. Phylogenet. Evol. 2018, 119, 128–143. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.-Y.; Zhang, B.-L.; Raxworthy, C.J.; Weisrock, D.W.; Hime, P.M.; Jin, J.-Q.; Lemmon, E.M.; Lemmon, A.R.; Holland, S.D.; Kortyna, M.L.; et al. Natatanuran frogs used the Indian Plate to step-stone disperse and radiate across the Indian Ocean. Natl. Sci. Rev. 2019, 6, 10–14. [Google Scholar] [CrossRef]
- Knytl, M.; Smolík, O.; Kubíčková, S.; Tlapáková, T.; Evans, B.J.; Krylov, V. Chromosome divergence during evolution of the tetraploid clawed frogs, Xenopus mellotropicalis and Xenopus epitropicalis as revealed by Zoo-FISH. PLoS ONE 2017, 12, e0177087. [Google Scholar] [CrossRef]
- Bogart, J.P.; Bartoszek, J.; Noble, D.W.A.; Bi, K. Sex in unisexual salamanders: Discovery of a new sperm donor with ancient affinities. Heredity 2009, 103, 483–493. [Google Scholar] [CrossRef]
- Neaves, W.B.; Baumann, P. Unisexual reproduction among vertebrates. Trends Genet. 2011, 27, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Bogart, J.P. A family study to examine clonal diversity in unisexual salamanders (genus Ambystoma). Genome 2019, 62, 549–561. [Google Scholar] [CrossRef]
- Myers, E.M.; Zamudio, K.R. Multiple paternity in an aggregate breeding amphibian: The effect of reproductive skew on estimates of male reproductive success. Mol. Ecol. 2004, 13, 1951–1963. [Google Scholar] [CrossRef] [PubMed]
- Saccucci, M.J.; Denton, R.D.; Holding, M.L.; Gibbs, H.L. Polyploid unisexual salamanders have higher tissue regeneration rates than diploid sexual relatives. J. Zool. 2016, 300, 77–81. [Google Scholar] [CrossRef]
- Fankhauser, G. Polyploidy in the salamander, Eurycea bislineata. J. Hered. 1939, 30, 379–388. [Google Scholar] [CrossRef]
- Fankhauser, G.; Crotta, R.; Perrot, M. Spontaneous and cold-induced triploidy in the Japanese newt, Triturus pyrrhogaster. J. Exp. Zool. 1942, 89, 167–181. [Google Scholar] [CrossRef]
- Fankhauser, G.; Watson, R.C. Heat-induced triploidy in the Newt, Triturus viridescens. Proc. Natl. Acad. Sci. USA 1942, 28, 436–440. [Google Scholar] [CrossRef] [PubMed]
- Litvinchuk, S.N.; Rosanov, J.M.; Borkin, L.J. A case of natural triploidy in a smooth newt Triturus vulgaris (Linneaus, 1958), from Russia (Caudata: Salamandridae). Herpetozoa 1998, 11, 93–95. [Google Scholar]
- Stöck, M.; Moritz, C.; Hickerson, M.; Frynta, D.; Dujsebayeva, T.; Eremchenko, V.; Macey, J.R.; Papenfuss, T.J.; Wake, D.B. Evolution of mitochondrial relationships and biogeography of Palearctic green toads (Bufo viridis subgroup) with insights in their genomic plasticity. Mol. Phylogenet. Evol. 2006, 41, 663–689. [Google Scholar] [CrossRef]
- Stöck, M.; Ustinova, J.; Lamatsch, D.K.; Schartl, M.; Perrin, N.; Moritz, C. A vertebrate reproductive system involving three ploidy levels: Hybrid origin of triploids in a contact zone of diploid and tetraploid palearctic green toads (Bufo viridis subgroup). Evolution 2010, 64, 944–959. [Google Scholar] [CrossRef]
- Stöck, M.; Ustinova, J.; Betto-Colliard, C.; Schartl, M.; Moritz, C.; Perrin, N. Simultaneous Mendelian and clonal genome transmission in a sexually reproducing, all-triploid vertebrate. Proc. Biol. Sci. 2012, 279, 1293–1299. [Google Scholar] [CrossRef]
- Christiansen, D.G. Gamete types, sex determination and stable equilibria of all-hybrid populations of di- and triploid water frogs (Pelophylax esculentus). BMC Evol. Biol. 2009, 9, 135. [Google Scholar] [CrossRef]
- Mahony, M.J.; Roberts, J.D. Two new species of desert burrowing frogs of the genus Neobatrachus (Anura: Myobatrachidae). Rec. West. Aust. Mus. 1986, 13, 155–170. [Google Scholar]
- Krylov, V.; Tlapakova, T. Xenopus Cytogenetics and Chromosomal Evolution. Cytogenet. Genome Res. 2015, 145, 192–200. [Google Scholar] [CrossRef]
- Mezzasalma, M.; Andreone, F.; Glaw, F.; Guarino, F.M.; Odierna, G.; Petraccioli, A.; Picariello, O. Changes in heterochromatin content and ancient chromosome fusion in the endemic Malagasy boid snakes Sanzinia and Acrantophis (Squamata: Serpentes). Salamandra 2019, 55, 140–144. [Google Scholar]
- Mezzasalma, M.; Visone, V.; Petraccioli, A.; Odierna, G.; Capriglione, T.; Guarino, F.M. Non-random accumulation of LINE1-like sequences on differentiated snake W chromosomes. J. Zool. 2016, 300, 67–75. [Google Scholar] [CrossRef]
- Mezzasalma, M.; Di Febbraro, M.; Guarino, F.M.; Odierna, G.; Russo, D. Cold-blooded in the Ice Age: “refugia within refugia”, inter-and intraspecific biogeographic diversification of European whipsnakes (Squamata, Colubridae, Hierophis). Zoology 2018, 127, 84–94. [Google Scholar] [CrossRef] [PubMed]
- Mezzasalma, M.; Brunelli, E.; Odierna, G.; Guarino, F.M. First insights on the karyotype diversification of the endemic Malagasy leaf-toed geckos (Squamata: Gekkonidae: Uroplatus). Animals 2022, 12, 2054. [Google Scholar] [CrossRef]
- Petraccioli, A.; Guarino, F.M.; Kupriyanova, L.; Mezzasalma, M.; Odierna, G.; Picariello, O.; Capriglione, T. Isolation and Characterization of Interspersed Repeated Sequences in the Common Lizard, Zootoca vivipara, and Their Conservation in Squamata. Cytogenet. Genome Res. 2019, 157, 65–76. [Google Scholar] [CrossRef]
- Sidhom, M.; Said, K.; Chatti, N.; Guarino, F.M.; Odierna, G.; Petraccioli, A.; Picariello, O.; Mezzasalma, M. Karyological characterization of the common chameleon (Chamaeleo chamaeleon) provides insights on the evolution and diversification of sex chromosomes in Chamaeleonidae. Zoology 2020, 141, 125738. [Google Scholar] [CrossRef]
- Avise, J.C. Evolutionary perspectives on clonal reproduction in vertebrate animals. Proc. Natl. Acad. Sci. USA 2015, 112, 8867–8873. [Google Scholar] [CrossRef] [PubMed]
- Fujita, M.K.; Singhal, S.; Brunes, T.O.; Maldonado, J.A. Evolutionary dynamics and consequences of parthenogenesis in vertebrates. Annu. Rev. Ecol. Evol. Syst. 2020, 51, 191–214. [Google Scholar] [CrossRef]
- Röll, B.; von Düring, M.U.G. Sexual characteristics and spermatogenesis in males of the parthenogenetic gecko Lepidodactylus lugubris (Reptilia, Gekkonidae). Zoology 2008, 111, 385–400. [Google Scholar] [CrossRef] [PubMed]
- Watts, P.C.; Buley, K.R.; Sanderson, S.; Boardman, W.; Ciofi, C.; Gibson, R. Parthenogenesis in Komodo dragons. Nature 2006, 444, 1021–1022. [Google Scholar] [CrossRef]
- Van der Kooi, C.J.; Schwander, T. Parthenogenesis: Birth of a new lineage or reproductive accident? Curr. Biol. 2015, 25, R659–R661. [Google Scholar] [CrossRef] [PubMed]
- Burbrink, F.T.; Grazziotin, F.G.; Pyron, R.A.; Cundall, D.; Donnellan, S.; Irish, F. Interrogating genomic-scale data for squamata (lizards, snakes, and amphisbaenians) shows no support for key traditional morphological relationships. Syst. Biol. 2020, 69, 502–520. [Google Scholar] [CrossRef] [PubMed]
- Gamble, T.; Bauer, A.M.; Greenbaum, E.; Jackman, T.R. Evidence for Gondwanan vicariance in an ancient clade of gecko lizards. J. Biogeogr. 2008, 35, 88–104. [Google Scholar] [CrossRef]
- Vidal, N.; Hedges, S.B. The molecular evolutionary tree of lizards, snakes, and amphisbaenians. C. R. Biol. 2009, 332, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Morales, C.; Sturaro, M.J.; Nunes, P.M.S.; Lotzkat, S.; Peloso, P.L.V. A species-level total evidence phylogeny of the microteiid lizard family Alopoglossidae (Squamata: Gymnophthalmoidea). Cladistics 2020, 36, 259–300. [Google Scholar] [CrossRef]
- Mezzasalma, M.; Guarino, F.M.; Odierna, G. Lizards as Model Organisms of Sex Chromosome Evolution: What We Really Know from a Systematic Distribution of Available Data? Genes 2021, 12, 1341. [Google Scholar] [CrossRef]
- Araya-Donoso, R.; Torres-Pérez, F.; Véliz, D.; Lamborot, M. Hybridization and polyploidy in the weeping lizard Liolaemus chiliensis (Squamata: Liolaemidae). Biol. J. Linn. Soc. 2019, 128, 963–974. [Google Scholar] [CrossRef]
- Moritz, C.; Uzzell, T.; Spolsky, C.; Hotz, H.; Darevsky, I.; Kupriyanova, L.; Danielyan, F. The material ancestry and approximate age of parthenogenetic species of Caucasian rock lizards (Lacerta: Lacertidae). Genetica 1992, 87, 53–62. [Google Scholar] [CrossRef]
- Spangenberg, V.; Arakelyan, M.; Galoyan, E.; Matveevsky, S.; Petrosyan, R.; Bogdanov, Y.; Danielyan, F.; Kolomiets, O. Reticulate Evolution of the Rock Lizards: Meiotic Chromosome Dynamics and Spermatogenesis in Diploid and Triploid Males of the Genus Darevskia. Genes 2017, 8, 149. [Google Scholar] [CrossRef]
- Cole, C.J.; Taylor, H.L.; Baumann, D.P.; Baumann, P. Neaves’ Whiptail Lizard: The First Known Tetraploid Parthenogenetic Tetrapod (Reptilia: Squamata: Teiidae). Breviora 2014, 539, 1–20. [Google Scholar] [CrossRef]
- Ota, H.; Hikida, T.; Matsui, M.; Mori, A.; Wynn, A.H. Morphological variation karyotype and reproduction of the parthenogenetic blind snake Ramphotyphlops braminus, from the insular region of East Asia and Saipan. Amphibia-Reptilia 1991, 12, 181–193. [Google Scholar] [CrossRef]
- Mezzasalma, M.; Andreone, F.; Glaw, F.; Petraccioli, A.; Odierna, G.; Guarino, F.M. A karyological study of three typhlopid species with some inferences on chromosome evolution in blindsnakes (Scolecophidia). Zool. Anz. 2016, 264, 34–40. [Google Scholar] [CrossRef]
- Patawang, I.; Tanomtong, A.; Kaewmad, P.; Chuaynkern, Y.; Duengkae, P. New record on karyological analysis and first study of NOR localization of parthenogenetic brahminy blind snake, Ramphotyphlops braminus (Squamata, Typhlopidae) in Thailand. Nucleus 2016, 59, 61–66. [Google Scholar] [CrossRef]
- Rovatsos, M.; Augstenová, B.; Altmanová, M.; Sloboda, M.; Kodym, P.; Kratochvíl, L. Triploid colubrid snake provides insight into the mechanism of sex determination in advanced snakes. Sex. Dev. 2018, 12, 251–255. [Google Scholar] [CrossRef] [PubMed]
- Alam, S.M.; Sarre, S.D.; Gleeson, D.; Georges, A.; Ezaz, T. Did lizards follow unique pathways in sex chromosome evolution? Genes 2018, 9, 239. [Google Scholar] [CrossRef] [PubMed]
- Bickham, J.W.; Tucker, P.K.; Legler, J.M. Diploid-triploid mosaicism: An unusual phenomenon in side-necked turtles (Platemys platycephala). Science 1985, 227, 1591–1593. [Google Scholar] [CrossRef] [PubMed]
- Bickham, J.W.; Hanks, B.G.; Hale, D.W.; Martin, J.E. Ploidy diversity and the production of balanced gametes in male twist-necked turtles (Platemys platycephala). Copeia 1993, 1993, 723–727. [Google Scholar] [CrossRef]
- Barros, R.M.; Sampaio, M.M.; Assis, M.F.; Ayres, M.; Cunha, O.R. General considerations on the karyotypic evolution of Chelonia from the Amazon region of Brazil. Cytologia 1976, 41, 559–565. [Google Scholar] [CrossRef]
- Bull, J.J. Evolution of Sex Determining Mechanisms; Benjamin-Cummings: London, UK, 1983. [Google Scholar]
- Bickam, J.; Hanks, B.G. Diploid-Triploid Mosaicism and Tissue Ploidy Diversity within Platemys platycephala from Suriname. Cytogen. Gen. Res. 2009, 127, 280–286. [Google Scholar] [CrossRef]
- Abdel-Hameed, F.; Shoffner, R.N. Intersexes and sex determination in chickens. Science 1971, 172, 962–964. [Google Scholar] [CrossRef]
- Bloom, S.E. Chromosome abnormalities in chicken (Gallus domesticus) embryos: Types, frequencies and phenotypic effects. Chromosoma 1972, 37, 309–326. [Google Scholar] [CrossRef]
- Fechheimer, N.S.; Jaap, R.G. The parental source of heteroploidy in chick embryos determined with chromosomally marked gametes. J. Reprod. Fertil. 1978, 52, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Orr, H.A. “Why polyploidy is rarer in animals than in plants” revisited. Am. Nat. 1990, 136, 759–770. [Google Scholar] [CrossRef]
- Yamazaki, W.; Takahashi, M.; Kawahara, M. Restricted development of mouse triploid fetuses with disorganized expression of imprinted genes. Zygote 2015, 23, 874–884. [Google Scholar] [CrossRef] [PubMed]
- Eglitis, M.A.; Wiley, L.M. Tetraploidy and early development: Effects on developmental timing and embryonic metabolism. J. Embryol. Exp. Morphol. 1981, 66, 91–108. [Google Scholar] [CrossRef] [PubMed]
- Muller, H.J. Why polyploidy is rarer in animals than in plants. Am. Nat. 1925, 59, 346–353. [Google Scholar] [CrossRef]
- Svartman, M.; Stone, G.; Stanyon, R. Molecular cytogenetics discards polyploidy in mammals. Genomics 2005, 85, 425–430. [Google Scholar] [CrossRef] [PubMed]
- Gallardo, M.H.; González, C.A.; Cebrián, I. Molecular cytogenetics and allotetraploidy in the red vizcacha rat, Tympanoctomys barrerae (Rodentia, Octodontidae). Genomics 2006, 88, 214–221. [Google Scholar] [CrossRef]
- Kopp, E.; Mayr, B.; Schleger, W. Species-specific non-expression in ribosomal RNA genes in a mammalian hybrid, the mule. Chromosoma 1986, 94, 346–352. [Google Scholar] [CrossRef]
- Pontes, O.; Lawrence, R.J.; Neves, N.; Silva, M.; Lee, J.H.; Chen, Z.J.; Viegas, W.; Pikaard, C.S. Natural variation in nucleolar dominance reveals the relationship between nucleolus organizer chromatin topology and rRNA gene transcription in Arabidopsis. Proc. Natl. Acad. Sci. USA 2003, 100, 11418–11423. [Google Scholar] [CrossRef]
- Gallardo, M.H.; Kausel, G.; Jiménez, A.; Bacquet, C.; González, C.; Figueroa, J.; Köhler, N.; Ojeda, R. Whole-genome duplications in South American desert rodents (Octodontidae). Biol. J. Linn. Soc. 2004, 82, 443–451. [Google Scholar] [CrossRef]
- Suárez-Villota, E.; Vargas, R.; Marchant, C.; Torres, J.; Köhler, N.; Núñez, J.; De La Fuente, R.; Page, J.; Gallardo, M. Distribution of repetitive DNAs and the hybrid origin of the red vizcacha rat (Octodontidae). Genome 2012, 55, 105–117. [Google Scholar] [CrossRef]
- Panopoulou, G.; Poustka, A.J. Timing and mechanism of ancient vertebrate genome duplications—The adventure of a hypothesis. Trends Genet. 2005, 21, 559–567. [Google Scholar] [CrossRef]
- Ren, R.; Wang, H.; Guo, C.; Zhang, N.; Zeng, L.; Chen, Y.; Ma, H.; Qi, J. Widespread whole genome duplications contribute to genome complexity and species diversity in angiosperms. Mol. Plant 2018, 11, 414–428. [Google Scholar] [CrossRef]
- Hedrick, P.W. Adaptive introgression in animals: Examples and comparison to new mutation and standing variation as sources of adaptive variation. Mol. Ecol. 2013, 22, 4606–4618. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Steinmetz, L.M.; Gu, X.; Scharfe, C.; Davis, R.W.; Li, W.-H. Role of duplicate genes in genetic robustness against null mutations. Nature 2003, 421, 63–66. [Google Scholar] [CrossRef] [PubMed]
- Herbst, R.H.; Bar-Zvi, D.; Reikhav, S.; Soifer, I.; Breker, M.; Jona, G.; Shimoni, E.; Schuldiner, M.; Levy, A.A.; Barkai, N. Heterosis as a consequence of regulatory incompatibility. BMC Biol. 2017, 15, 38. [Google Scholar] [CrossRef] [PubMed]
- Pandit, M.K.; Pocock, M.J.O.; Kunin, W.E. Ploidy influences rarity and invasiveness in plants. J. Ecol. 2011, 9, 1108–1115. [Google Scholar] [CrossRef]
- Baduel, P.; Bray, S.; Vallejo-Marin, M.; Kolář, F.; Yant, L. The “Polyploid Hop”: Shifting Challenges and Opportunities Over the Evolutionary Lifespan of Genome Duplications. Front. Ecol. Evol. 2018, 6, 117. [Google Scholar] [CrossRef]
- Van de Peer, Y.; Ashman, T.-L.; Soltis, P.S.; Soltis, D.E. Polyploidy: An evolutionary and ecological force in stressful times. Plant Cell 2021, 33, 11–26. [Google Scholar] [CrossRef] [PubMed]
- David, K.T. Global gradients in the distribution of animal polyploids. Proc. Natl. Acad. Sci. USA 2022, 119, e2214070119. [Google Scholar] [CrossRef]
- Stebbins, G.L. Variation and Evolution in Plants; Oxford University Press: Oxford, UK, 1950. [Google Scholar]
- Stebbins, G.L. Chromosomal Evolution in Higher Plants; Edward Arnold: London, UK, 1971. [Google Scholar]
- Wagner, W.H. Biosystematics and evolutionary noise. Taxon 1970, 19, 146–151. [Google Scholar] [CrossRef]
- Mayrose, I.; Zhan, S.H.; Rothfels, C.J.; Magnuson-Ford, K.; Barker, M.S.; Rieseberg, L.H.; Otto, S.P. Recently formed polyploid plants diversify at lower rates. Science 2011, 333, 1257. [Google Scholar] [CrossRef]
- Mayrose, I.; Zhan, S.H.; Rothfels, C.J.; Arrigo, N.; Barker, M.S.; Rieseberg, L.H.; Otto, S.P. Methods for studying polyploid diversification and the dead end hypothesis: A reply to Soltis et al. (2014). New Phytol. 2015, 206, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Soltis, D.E.; Buggs, R.J.A.; Doyle, J.J.; Soltis, P.S. What we still don’t know about polyploidy. Taxon 2010, 59, 1387–1403. [Google Scholar] [CrossRef]
- Otto, S.P. The evolutionary consequences of polyploidy. Cell 2007, 131, 452–462. [Google Scholar] [CrossRef] [PubMed]
- Borel, F.; Lohez, O.D.; Lacroix, F.B.; Margolis, R.L. Multiple centrosomes arise from tetraploidy checkpoint failure and mitotic centrosome clusters in p53 and RB pocket protein-compromised cells. Proc. Natl. Acad. Sci. USA 2002, 99, 9819–9924. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.J. Plant Cytogenetics; CRC Press: Boca Raton, FL, USA, 2003. [Google Scholar]
- Mason, A.S.; Wendel, J.F. Homoeologous Exchanges, Segmental Allopolyploidy, and Polyploid Genome Evolution. Front. Genet. 2020, 11, 1014. [Google Scholar] [CrossRef]
- Matzke, M.A.; Mittelsten Scheid, O.; Matzke, A.J. Rapid structural and epigenetic changes in polyploid and aneuploid genomes. Bioessays 1999, 21, 761–767. [Google Scholar] [CrossRef]
- Salmon, A.; Ainouche, M.L. Polyploidy and DNA methylation: New tools available. Mol. Ecol. 2010, 19, 213–215. [Google Scholar] [CrossRef]
- Rajkov, J.; Shao, Z.; Berrebi, P. Evolution of Polyploidy and Functional Diploidization in Sturgeons: Microsatellite Analysis in 10 Sturgeon Species. J. Hered. 2014, 105, 521–531. [Google Scholar] [CrossRef] [PubMed]
- Visser, V.; Molofsky, J. Ecological niche differentiation of polyploidization is not supported by environmental differences among species in a cosmopolitan grass genus. Am. J. Bot. 2015, 102, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Gaeta, R.T.; Chris Pires, J. Homoeologous recombination in allopolyploids: The polyploid ratchet. New Phytol. 2010, 186, 18–28. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mezzasalma, M.; Brunelli, E.; Odierna, G.; Guarino, F.M. Evolutionary and Genomic Diversity of True Polyploidy in Tetrapods. Animals 2023, 13, 1033. https://doi.org/10.3390/ani13061033
Mezzasalma M, Brunelli E, Odierna G, Guarino FM. Evolutionary and Genomic Diversity of True Polyploidy in Tetrapods. Animals. 2023; 13(6):1033. https://doi.org/10.3390/ani13061033
Chicago/Turabian StyleMezzasalma, Marcello, Elvira Brunelli, Gaetano Odierna, and Fabio Maria Guarino. 2023. "Evolutionary and Genomic Diversity of True Polyploidy in Tetrapods" Animals 13, no. 6: 1033. https://doi.org/10.3390/ani13061033
APA StyleMezzasalma, M., Brunelli, E., Odierna, G., & Guarino, F. M. (2023). Evolutionary and Genomic Diversity of True Polyploidy in Tetrapods. Animals, 13(6), 1033. https://doi.org/10.3390/ani13061033