Comparative Analyses of the Fecal Microbiome of Five Wild Black-Billed Capercaillie (Tetrao parvirostris) Flocks
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. DNA Extraction and High-Throughput Sequencing
2.3. Sequence Processing and Statistical Analyses
3. Results
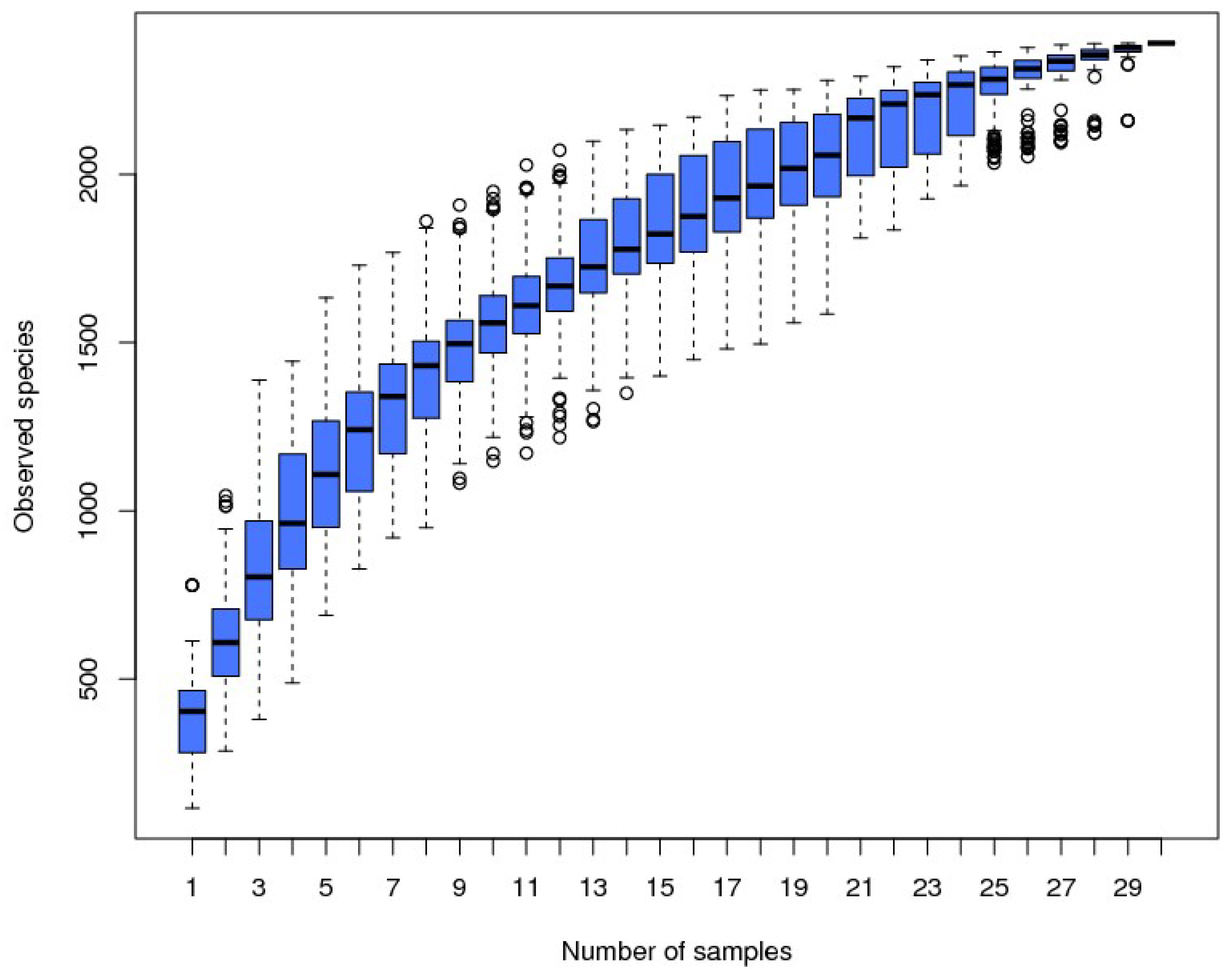
3.1. 16S rRNA Gene Data
3.2. Results for Fecal Microbiome
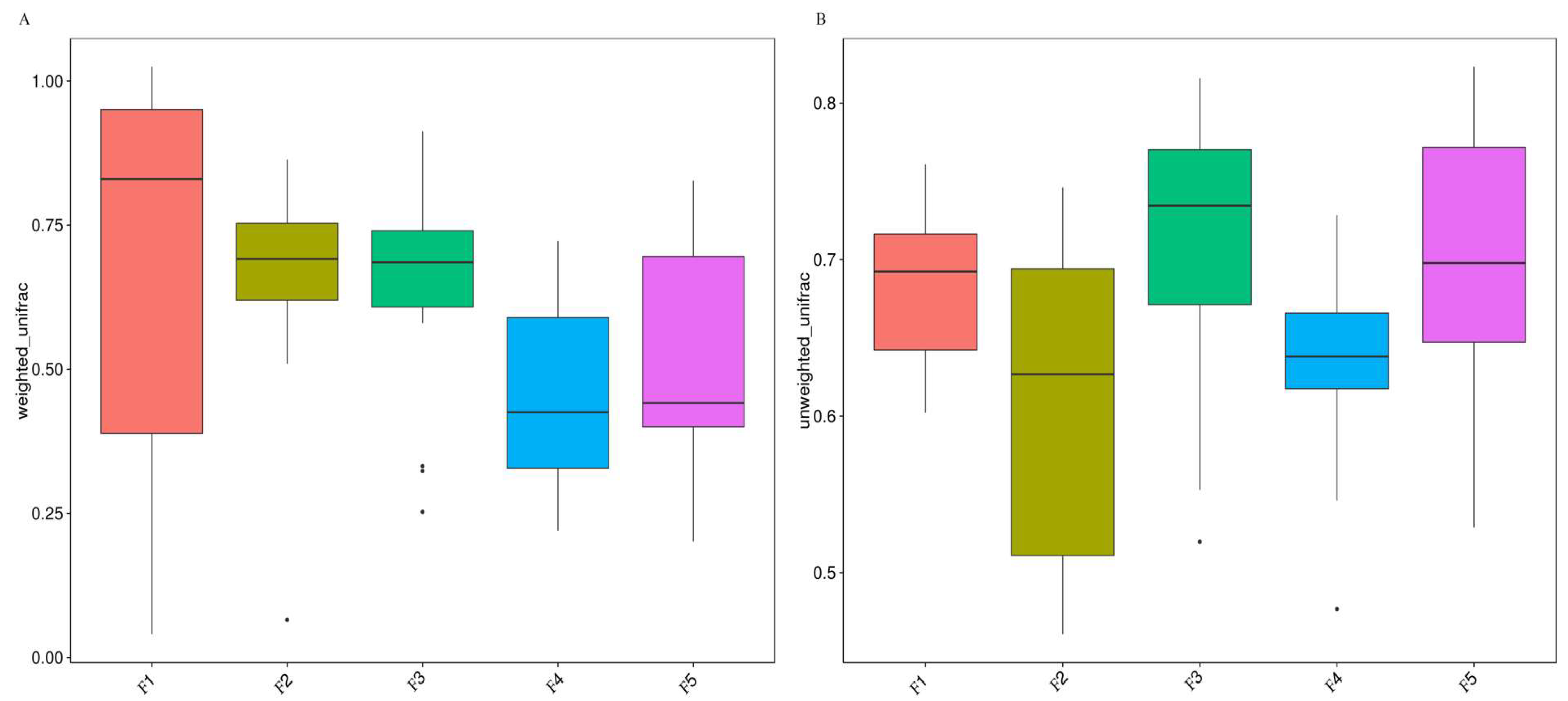
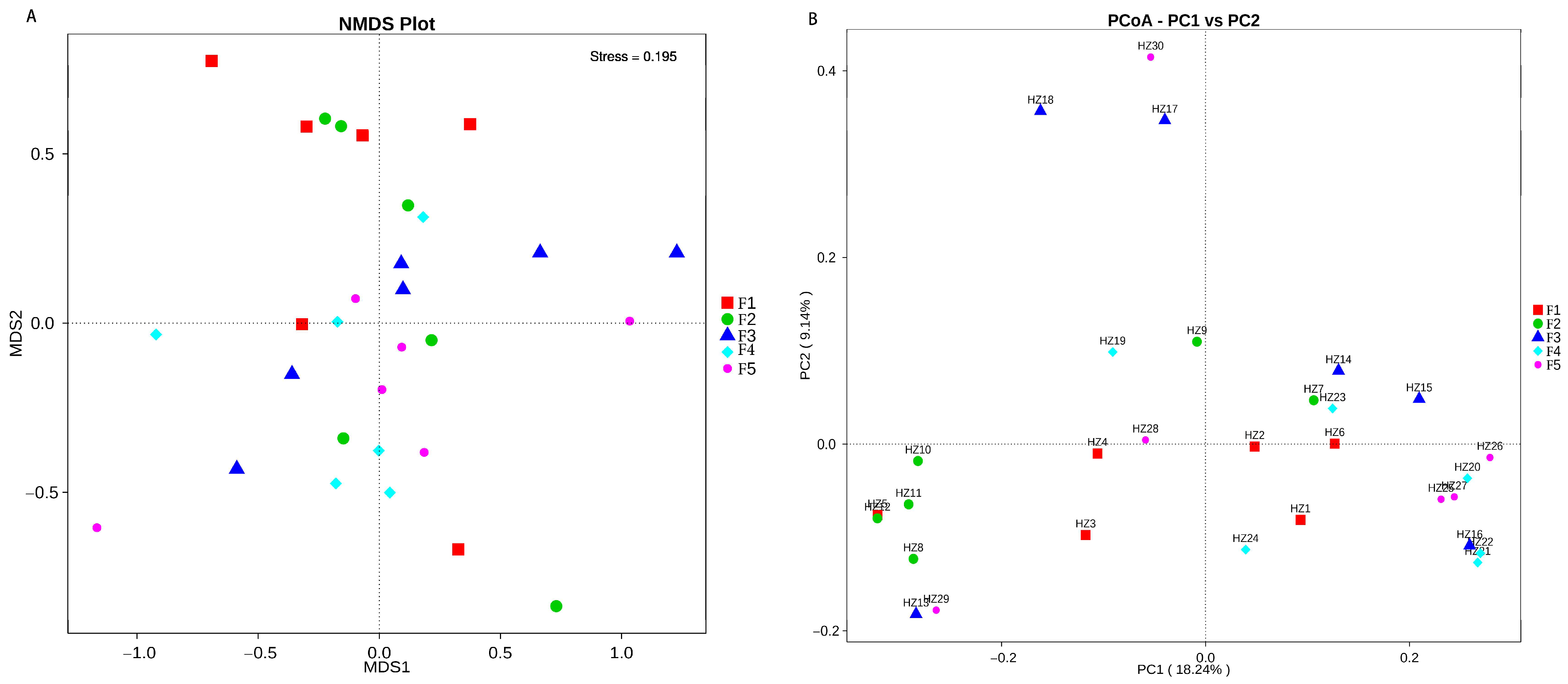
3.3. Analysis of Differences between the Flocks
3.4. Prediction of Gut Microbiome Function
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yang, L.; Zhang, C.; Chen, M.; Li, J.; Yang, L.; Huo, Z.; Ahmad, S.; Luan, X. Long-term ecological data for conservation: Range change in the black-billed capercaillie (Tetrao urogalloides) in northeast China (1970s–2070s). Ecol. Evol. 2018, 8, 3862–3870. [Google Scholar] [CrossRef] [PubMed]
- Jingli, Z.; Xueying, S.; Qi, Z.; Duoying, C.; Zhuo, X.; Paiyizulamu, S.; Qingming, W.; Hongfei, Z.; Ke, R. Population density and nocturnal habitat utilization of Black-billed Capercaillie in the northern area of Greater Khingan Range. Acta Ecol. Sin. 2019, 39, 6974–6980. [Google Scholar] [CrossRef]
- Andreev, A.V. Energetics and survival of birds in extreme environments. Ostrich 1999, 70, 13–22. [Google Scholar] [CrossRef]
- Liu, H.; Huang, H. Domesticated Feed of Tetrao parvirostris. Anim. Husb. Feed Sci. 2020, 12, 41–42. [Google Scholar] [CrossRef]
- Wang, X.; Shang, Y.; Wei, Q.; Wu, X.; Dou, H.; Zhang, H.; Zhou, S.; Sha, W.; Sun, G.; Ma, S.; et al. Comparative Analyses of the Gut Microbiome of Two Fox Species, the Red Fox (Vulpes vulpes) and Corsac Fox (Vulpes corsac), that Occupy Different Ecological Niches. Microb. Ecol. 2022, 83, 753–765. [Google Scholar] [CrossRef]
- Gil, J.C.; Hird, S.M. Multiomics Characterization of the Canada Goose Fecal Microbiome Reveals Selective Efficacy of Simulated Metagenomes. Microbiol. Spectr. 2022, 10, e0238422. [Google Scholar] [CrossRef]
- Sun, F.; Chen, J.; Liu, K.; Tang, M.; Yang, Y. The avian gut microbiota: Diversity, influencing factors, and future directions. Front. Microbiol. 2022, 13, 934272. [Google Scholar] [CrossRef]
- Ganz, H.H.; Doroud, L.; Firl, A.J.; Hird, S.M.; Eisen, J.A.; Boyce, W.M. Community-Level Differences in the Microbiome of Healthy Wild Mallards and Those Infected by Influenza A Viruses. mSystems 2017, 2, e00188-16. [Google Scholar] [CrossRef]
- Zhou, H.; Guo, W.; Zhang, T.; Xu, B.; Zhang, D.; Teng, Z.; Tao, D.; Lou, Y.; Gao, Y. Response of goose intestinal microflora to the source and level of dietary fiber. Poult. Sci. 2018, 97, 2086–2094. [Google Scholar] [CrossRef]
- Liu, G.; Luo, X.; Zhao, X.; Zhang, A.; Jiang, N.; Yang, L.; Huang, M.; Xu, L.; Ding, L.; Li, M.; et al. Gut microbiota correlates with fiber and apparent nutrients digestion in goose. Poult. Sci. 2018, 97, 3899–3909. [Google Scholar] [CrossRef]
- Berlow, M.; Wada, H.; Derryberry, E.P. Experimental Exposure to Noise Alters Gut Microbiota in a Captive Songbird. Microb. Ecol. 2021, 84, 1264–1277. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Wu, F.; Zhou, Q.; Zhao, D. Comparative Analysis of Gut Microbiota in Captive and Wild Oriental White Storks: Implications for Conservation Biology. Front. Microbiol. 2021, 12, 649466. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wu, X.; Shang, Y.; Mei, X.; Zhou, S.; Wei, Q.; Sun, G.; Dong, Y.; Zhang, H. Convergent evolution of the gut microbiome in marine carnivores. Ecol. Evol. 2022, 12, e9373. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chen, Y.; Shang, Y.; Wu, X.; Wei, Q.; Chen, J.; Yan, J.; Zhang, H. Comparison of the gut microbiome in red deer (Cervus elaphus) and fallow deer (Dama dama) by high-throughput sequencing of the V3-V4 region of the 16S rRNA gene. ScienceAsia 2019, 45, 515–524. [Google Scholar] [CrossRef]
- Xiao, K.; Fan, Y.; Zhang, Z.; Shen, X.; Li, X.; Liang, X.; Bi, R.; Wu, Y.; Zhai, J.; Dai, J.; et al. Covariation of the Fecal Microbiome with Diet in Nonpasserine Birds. mSphere 2021, 6, e00308-21. [Google Scholar] [CrossRef] [PubMed]
- Laviad-Shitrit, S.; Izhaki, I.; Lalzar, M.; Halpern, M. Comparative Analysis of Intestine Microbiota of Four Wild Waterbird Species. Front. Microbiol. 2019, 10, 1911. [Google Scholar] [CrossRef]
- Craft, J.; Eddington, H.; Christman, N.D.; Pryor, W.; Chaston, J.M.; Erickson, D.L.; Wilson, E. Increased Microbial Diversity and Decreased Prevalence of Common Pathogens in the Gut Microbiomes of Wild Turkeys Compared to Domestic Turkeys. Appl. Environ. Microbiol. 2022, 88, e0142321. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Mazcorro, J.F.; Castillo-Carranza, S.A.; Guard, B.; Gomez-Vazquez, J.P.; Dowd, S.E.; Brigthsmith, D.J. Comprehensive Molecular Characterization of Bacterial Communities in Feces of Pet Birds Using 16S Marker Sequencing. Microb. Ecol. 2017, 73, 224–235. [Google Scholar] [CrossRef]
- Hetland, H.; Svihus, B.; Olaisen, V. Effect of feeding whole cereals on performance, starch digestibility and duodenal particle size distribution in broiler chickens. Br. Poult. Sci. 2002, 43, 416–423. [Google Scholar] [CrossRef]
- Cockerham, S.; Lee, B.; Orben, R.A.; Suryan, R.M.; Torres, L.G.; Warzybok, P.; Bradley, R.; Jahncke, J.; Young, H.S.; Ouverney, C.; et al. Microbial Ecology of the Western Gull (Larus occidentalis). Microb. Ecol. 2019, 78, 665–676. [Google Scholar] [CrossRef]
- Berlow, M.; Phillips, J.N.; Derryberry, E.P. Effects of Urbanization and Landscape on Gut Microbiomes in White-Crowned Sparrows. Microb. Ecol. 2021, 81, 253–266. [Google Scholar] [CrossRef]
- Song, S.J.; Sanders, J.G.; Delsuc, F.; Metcalf, J.; Amato, K.; Taylor, M.W.; Mazel, F.; Lutz, H.L.; Winker, K.; Graves, G.R.; et al. Comparative Analyses of Vertebrate Gut Microbiomes Reveal Convergence between Birds and Bats. mBio 2020, 11, e02901-19. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wu, X.; Shang, Y.; Gao, Y.; Li, Y.; Wei, Q.; Dong, Y.; Mei, X.; Zhou, S.; Sun, G.; et al. High-Altitude Drives the Convergent Evolution of Alpha Diversity and Indicator Microbiota in the Gut Microbiomes of Ungulates. Front. Microbiol. 2022, 13, 953234. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Zhang, H.; Gao, X.; Shang, S.; Wu, X.; Chen, J.; Zhang, W.; Zhang, W.; Jiang, M.; Zhang, B.; et al. Comparison of the bacterial communities in feces from wild versus housed sables (Martes zibellina) by high-throughput sequence analysis of the bacterial 16S rRNA gene. AMB Express 2016, 6, 98. [Google Scholar] [CrossRef]
- Porebski, S.; Bailey, L.G.; Baum, B.R. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Rep. 1997, 15, 8–15. [Google Scholar] [CrossRef]
- Wu, X.; Zhang, H.; Chen, J.; Shang, S.; Wei, Q.; Yan, J.; Tu, X. Comparison of the fecal microbiota of dholes high-throughput Illumina sequencing of the V3-V4 region of the 16S rRNA gene. Appl. Microbiol. Biotechnol. 2016, 100, 3577–3586. [Google Scholar] [CrossRef] [PubMed]
- Yuzhu, C.; Jian, L.; Yufeng, Z.; Mingqian, Z.; Zheng, S.; Gongchao, J.; Shi, H.; Xiaoquan, S. Parallel-Meta Suite Interactive and rapid microbiome data analysis on multiple platforms. iMeta 2022, 1, e1. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef]
- Zhu, Y.; Li, Y.; Yang, H.; He, K.; Tang, K. Establishment of Gut Microbiome During Early Life and Its Relationship With Growth in Endangered Crested Ibis (Nipponia nippon). Front. Microbiol. 2021, 12, 723682. [Google Scholar] [CrossRef]
- Grond, K.; Sandercock, B.K.; Jumpponen, A.; Zeglin, L.H. The avian gut microbiota: Community, physiology and function in wild birds. J. Avian Biol. 2018, 49, e01788. [Google Scholar] [CrossRef]
- Kielak, A.M.; Barreto, C.C.; Kowalchuk, G.A.; van Veen, J.A.; Kuramae, E.E. The Ecology of Acidobacteria: Moving beyond Genes and Genomes. Front. Microbiol. 2016, 7, 744. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.; Yao, X.; Zuo, C.; Qi, Y.; Chen, D.; Ma, W. The gut bacterial diversity of sheep associated with different breeds in Qinghai province. BMC Vet. Res. 2020, 16, 254. [Google Scholar] [CrossRef] [PubMed]
- Hook, S.E.; Steele, M.A.; Northwood, K.S.; Dijkstra, J.; France, J.; Wright, A.D.; McBride, B.W. Impact of subacute ruminal acidosis (SARA) adaptation and recovery on the density and diversity of bacteria in the rumen of dairy cows. FEMS Microbiol. Ecol. 2011, 78, 275–284. [Google Scholar] [CrossRef]
- Rousk, K.; Jones, D.L.; Deluca, T.H. Moss-cyanobacteria associations as biogenic sources of nitrogen in boreal forest ecosystems. Front. Microbiol. 2013, 4, 150. [Google Scholar] [CrossRef]
- Murray, M.H.; Lankau, E.W.; Kidd, A.D.; Welch, C.N.; Ellison, T.; Adams, H.C.; Lipp, E.K.; Hernandez, S.M. Gut microbiome shifts with urbanization and potentially facilitates a zoonotic pathogen in a wading bird. PLoS ONE 2020, 15, e0220926. [Google Scholar] [CrossRef]
- De Maesschalck, C.; Van Immerseel, F.; Eeckhaut, V.; De Baere, S.; Cnockaert, M.; Croubels, S.; Haesebrouck, F.; Ducatelle, R.; Vandamme, P. Faecalicoccus acidiformans gen. nov., sp. nov., isolated from the chicken caecum, and reclassification of Streptococcus pleomorphus (Barnes et al. 1977), Eubacterium biforme (Eggerth 1935) and Eubacterium cylindroides (Cato et al. 1974) as Faecalicoccus pleomorphus comb. nov., Holdemanella biformis gen. nov., comb. nov. and Faecalitalea cylindroides gen. nov., comb. nov., respectively, within the family Erysipelotrichaceae. Int. J. Syst. Evol. Microbiol. 2014, 64, 3877–3884. [Google Scholar] [CrossRef]
- Ma, J.; Piao, X.; Mahfuz, S.; Long, S.; Wang, J. The interaction among gut microbes, the intestinal barrier and short chain fatty acids. Anim. Nutr. 2022, 9, 159–174. [Google Scholar] [CrossRef]
- LeBlanc, J.G.; Chain, F.; Martín, R.; Bermúdez-Humarán, L.G.; Courau, S.; Langella, P. Beneficial effects on host energy metabolism of short-chain fatty acids and vitamins produced by commensal and probiotic bacteria. Microb. Cell Fact. 2017, 16, 79. [Google Scholar] [CrossRef]
- Parche, S.; Amon, J.; Jankovic, I.; Rezzonico, E.; Beleut, M.; Barutçu, H.; Schendel, I.; Eddy, M.P.; Burkovski, A.; Arigoni, F.; et al. Sugar transport systems of Bifidobacterium longum NCC2705. Microb. Physiol. 2007, 12, 9–19. [Google Scholar] [CrossRef]
- Sharma, M.; Wasan, A.; Sharma, R.K. Recent developments in probiotics: An emphasis on Bifidobacterium. Food Biosci. 2021, 41, 100993. [Google Scholar] [CrossRef]
- Zhou, Y.; Li, R.; Gao, X.Y.; Lapidus, A.; Han, J.; Haynes, M.; Lobos, E.; Huntemann, M.; Pati, A.; Ivanova, N.N.; et al. High quality draft genome sequence of the slightly halophilic bacterium Halomonas zhanjiangensis type strain JSM 078169(T) (DSM 21076(T)) from a sea urchin in southern China. Stand. Genom. Sci. 2014, 9, 1020–1030. [Google Scholar] [CrossRef] [PubMed]
- Liao, X.; Shao, Y.; Sun, G.; Yang, Y.; Zhang, L.; Guo, Y.; Luo, X.; Lu, L. The relationship among gut microbiota, short-chain fatty acids, and intestinal morphology of growing and healthy broilers. Poult. Sci. 2020, 99, 5883–5895. [Google Scholar] [CrossRef] [PubMed]
- Lewis, W.B.; Moore, F.R.; Wang, S. Characterization of the gut microbiota of migratory passerines during stopover along the northern coast of the Gulf of Mexico. J. Avian Biol. 2016, 47, 659–668. [Google Scholar] [CrossRef]
- Thie, N.; Corl, A.; Turjeman, S.; Efrat, R.; Kamath, P.L.; Getz, W.M.; Bowie, R.C.K.; Nathan, R. Linking migration and microbiota at a major stopover site in a long-distance avian migrant. Mov. Ecol. 2022, 10, 46. [Google Scholar] [CrossRef]
- Chakraborty, P.; Gibbons, W.; Muthukumarappan, K. Conversion of volatile fatty acids into polyhydroxyalkanoate by Ralstonia eutropha. J. Appl. Microbiol. 2009, 106, 1996–2005. [Google Scholar] [CrossRef]
- Salanoubat, M.; Genin, S.; Artiguenave, F.; Gouzy, J.; Mangenot, S.; Arlat, M.; Billault, A.; Brottier, P.; Camus, J.C.; Cattolico, L.; et al. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 2002, 415, 497–502. [Google Scholar] [CrossRef]



| OTU | Abundance (%) | Phylum | Class | Order | Family | Genera |
|---|---|---|---|---|---|---|
| OTU_25228_508 | 13.80 | Cyanobacteria | Oxyphotobacteria | Unidentified | Unidentified | Unidentified |
| OTU_11392_1 | 12.56 | Bacillota | Erysipelotrichia | Erysipelotrichales | Erysipelotrichaceae | Faecalitalea |
| OTU_3504_102 | 10.58 | Camplyobacterota | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | Escherichia-Shigella |
| OTU_25228_2257 | 9.94 | Cyanobacteria | Oxyphotobacteria | unidentified | unidentified | unidentified |
| OTU_436_66 | 5.61 | Actinomycetota | Actinobacteria | Bifidobacteriales | Bifidobacteriaceae | Bifidobacterium |
| OTU_3327_4 | 3.37 | Camplyobacterota | Gammaproteobacteria | Aeromonadales | Succinivibrionaceae | Anaerobiospirillum |
| OTU_3616_27 | 2.72 | Camplyobacterota | Gammaproteobacteria | Oceanospirillales | Halomonadaceae | Halomonas |
| OTU_26443_5 | 2.69 | Camplyobacterota | Gammaproteobacteria | Betaproteobacteriales | Burkholderiaceae | Ralstonia |
| OTU_13901 | 2.62 | Actinomycetota | Coriobacteriia | Coriobacteriales | Coriobacteriaceae | Enorma |
| OTU_3400_101 | 2.24 | Camplyobacterota | Gammaproteobacteria | Alteromonadales | Shewanellaceae | Shewanella |
| OTU_24538_7 | 2.13 | Actinomycetota | Coriobacteriia | Coriobacteriales | Atopobiaceae | Olsenell |
| OTU_2185_14 | 1.92 | Bacillota | Negativicutes | Selenomonadales | Veillonellaceae | Megasphaera |
| OTU_2184_5 | 1.27 | Bacillota | Negativicutes | Selenomonadales | Veillonellaceae | Megasphaera |
| OTU_3616_340 | 1.04 | Camplyobacterota | Gammaproteobacteria | Oceanospirillales | Halomonadaceae | Halomonas |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, X.; Wang, X.; Wu, X.; Shang, Y.; Mei, X.; Zhou, S.; Wei, Q.; Sun, G.; Dong, Y.; Cui, W.; et al. Comparative Analyses of the Fecal Microbiome of Five Wild Black-Billed Capercaillie (Tetrao parvirostris) Flocks. Animals 2023, 13, 923. https://doi.org/10.3390/ani13050923
Gao X, Wang X, Wu X, Shang Y, Mei X, Zhou S, Wei Q, Sun G, Dong Y, Cui W, et al. Comparative Analyses of the Fecal Microbiome of Five Wild Black-Billed Capercaillie (Tetrao parvirostris) Flocks. Animals. 2023; 13(5):923. https://doi.org/10.3390/ani13050923
Chicago/Turabian StyleGao, Xiaodong, Xibao Wang, Xiaoyang Wu, Yongquan Shang, Xuesong Mei, Shengyang Zhou, Qinguo Wei, Guolei Sun, Yuehuan Dong, Weijia Cui, and et al. 2023. "Comparative Analyses of the Fecal Microbiome of Five Wild Black-Billed Capercaillie (Tetrao parvirostris) Flocks" Animals 13, no. 5: 923. https://doi.org/10.3390/ani13050923
APA StyleGao, X., Wang, X., Wu, X., Shang, Y., Mei, X., Zhou, S., Wei, Q., Sun, G., Dong, Y., Cui, W., & Zhang, H. (2023). Comparative Analyses of the Fecal Microbiome of Five Wild Black-Billed Capercaillie (Tetrao parvirostris) Flocks. Animals, 13(5), 923. https://doi.org/10.3390/ani13050923





