Simple Summary
Research on GP5 protein is of great significance in the diagnosis, prevention, and control of porcine reproductive and respiratory syndrome virus (PRRSV). We summarize its genetic variation, immunity, replication, apoptosis, virulence, interaction with viral protein and host proteins, which provides a theoretical foundation for exploring the PRRSV replication mechanisms and developing new vaccines.
Abstract
Porcine reproductive and respiratory syndrome (PRRS) is an acute, febrile, and highly contagious disease caused by the porcine reproductive and respiratory syndrome virus (PRRSV). Glycoprotein 5 (GP5) is a glycosylated envelope protein encoded by the PRRSV ORF5, which has good immunogenicity and can induce the body to produce neutralizing antibodies. Therefore, study of GP5 protein is of great significance in the diagnosis, prevention, and control of PRRSV and the development of new vaccines. We reviewed GP5 protein genetic variation, immune function, interaction with viral protein and host proteins, induction of cell apoptosis, and stimulation of neutralizing antibodies. GP5 protein’s influence on virus replication and virulence, as well as its use as a target for viral detection and immunization are reviewed.
1. Introduction
Porcine reproductive and respiratory syndrome (PRRS) is a severe infectious disease caused by the porcine reproductive and respiratory syndrome virus (PRRSV). The disease is characterized by adverse reproductive outcomes such as miscarriage, premature delivery, stillbirth, and fetal mummification. Non-reproductive effects are exhibited as respiratory diseases, immunosuppression, secondary diseases, and increased piglet mortality. These effects combine to cause massive economic losses to the swine industry worldwide.
PRRS outbreaks initially occurred in the United States in 1987, and then spread to Europe and Asia [1]; it was initially called the “mystery pig disease” by the American Animal Health Society in 1990 [2]. In 1991, PRRSV was isolated from diseased animals in the Netherlands and named Lelystad virus (LV) [3]. PRRSV was isolated in the United States in 1992 and named SIRS virus (also known as VR-2332) [4]. According to its genetic and antigenic characteristics, PRRSV can be divided into two genotypes: the European type (PRRSV-1) represented by the LV strain and the American type (PRRSV-2) represented by the VR-2332 strain [5]. The two types share approximately 60% of their nucleotide identity [6]. The first PRRSV strain of China, isolated and identified from aborted fetuses in 1996, was classified as PRRSV-2 [7]. In 2006, an outbreak of highly pathogenic PRRSV (HP-PRRSV) characterized by high mortality, fever, and abortion rates devastated the Chinese swine industry [8].
PRRSV is an RNA virus with an envelope and single positive strand [9], belonging to the order Nidovirales and the family Arteriviridae. It also includes lactate dehydrogenase-elevating virus of mice, equine arteritis virus, and simian hemorrhagic fever virus [10]. The total length of the genome is approximately 15 kb, which forms a cap structure at the 5′ end during mRNA processing, and has a poly-A tail structure at the 3′ end [11]. The PRRSV genome contains 12 open reading frames (ORFs): ORF1a, ORF1a’, TF, ORF1b, ORF2a, ORF2b, ORF3, ORF4, ORF5a, and ORF5–ORF7 [12]. Among them, ORF1a and ORF1b encode polyproteins pp1a and pp1b, which can be processed into 17 nonstructural proteins (NSPs) (NSP1α, NSP1β, NSP2, NSP2N, NSP2TF, NSP3-14) that play a major role in virus replication [13,14]. ORF5 and ORF6 encode the major GP5 and M envelope proteins, respectively, which interact with each other to form heterodimers on the surface of virus particles. GP5 is one of the most variable regions of structural proteins in the PRRSV genome and is, therefore, often used for phylogenetic analyses. ORF2, ORF3, and ORF4 encode the minor GP2a, GP3, and GP4 proteins that form noncovalent heterodimers. Two small non-glycosylated proteins E and GP5a are encoded by ORF2b and ORF5a, respectively. ORF7 encodes the highly conserved nucleocapsid protein (N protein) (Figure 1) [15,16,17,18].
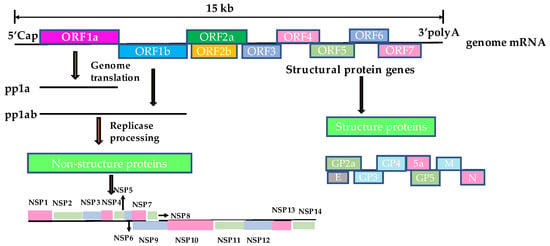
Figure 1.
The structural mode of the PRRSV genome.
GP5 protein is highly variable in PRRSV as it plays an important role in virus infection, as well as simultaneously inducing protective antibodies in the host. Therefore, it is a good target antigen for developing new vaccines [19,20,21]. GP5 protein contains important immune domains related to virus neutralization as well as some peptides or protein motifs, including signal peptides, transmembrane regions, antigenic determinants, and glycosylation sites [22]. As GP5 protein is greatly involved in the invasion, adsorption, and proliferation of PRRSV, it is of great interest to the pathogenicity, diagnosis, prevention, and control of PRRSV [23].
2. Overview of GP5 Protein
GP5 protein is a glycosylated envelope protein encoded by the PRRSV ORF5 gene. Its molecular weight is approximately 25 kDa, and it is composed of approximately 200 amino acids [24]. It is divided into four parts: the N-terminal, which is a signal peptide composed of 32 amino acids; the external functional region, which is composed of 35 amino acids containing varying numbers of glycosylated sites; the hydrophobic region, which is composed of 60 amino acids and three transmembrane domains; and the hydrophilic region, which is at the C-terminal end [5]. The extracellular domain of GP5 protein in the American strain contains four N-glycosylation sites, of which N44 and N51 are of the conservative type, which primarily affect the virus infectivity [25]. Glycosylation of GP5 protein may cause the immune escape of the virus, thus reducing the protection of the host’s immune response [26]. The multiple transmembrane hydrophobic regions contained in GP5 protein can make the translated GP5 protein stay in the endoplasmic reticulum, thus inhibiting its expression [27]. GP5 and M proteins of PRRSV are mainly incorporated into virus particles in the form of disulfide-linked heterodimers or polymers, which are essential for virion formation [28].
3. Genetic Variation Analysis of GP5 Protein
PRRSV GP5 protein has high genetic variation in the PRRSV genome [29]. GP5 protein of the European strain is composed of 201 amino acids, while that of the American strain is composed of 200 amino acids [30]. The amino acid homology deduced from the PRRSV isotype strain of GP5 is between 88% and 99%, while that deduced from GP5 between the European and American strains is between 52% and 55%. The substitution of deduced amino acids of GP5 among strains of the same type mainly occurs in the hypervariable region (26 aa–39 aa) near the outer region of the signal peptide sequence [31,32], whereas, the amino acid variation of GP5 protein of different strains is mainly concentrated in the signal peptide region, neutralizing epitope region, and non-neutralizing epitope region [33].
Zhang et al. [34] amplified the ORF5 of 16 PRRSV strains in different areas of Shandong Province using reverse transcription-polymerase chain reaction (RT-PCR) and performed a sequence analysis. The results showed that there were site mutations in the amino acids of GP5 protein in all 16 strains, from which 12 strains had mutations at the 29th site of the non-neutralizing epitope and the 34th site of quasi-species evolution, from V to A and N to S, respectively. One strain mutated from A to V at the 185th site of the non-neutralizing epitope. Similarly, Li et al. [11] used RT-PCR to analyze the genetic variation of the isolated PRRSV ORF5 gene. The results showed that the 13th and 151st amino acids and neutralizing epitope (36 aa–52 aa) of ORF5 were mutated, which could lead to changes in virulence and immune evasion. In another study, Lu et al. [35] isolated nine Henan strains and reported that the 39th amino acid of all isolates except HeN-3 had changed from F/L to I (Figure 2). In 2011 and 2012, the mutation of the neutralizing epitope Q40, L41, and the new mutation of the decoy epitope L28 in GP5 protein were found in PRRSV strains in Heilongjiang province [36]. In 2014, the amino acid sites of GP5 in South China were analyzed and found to have extensive variations in the signal peptide region, induced epitope, neutralizing epitope, and hypervariable region of the new branch subgroup [37].
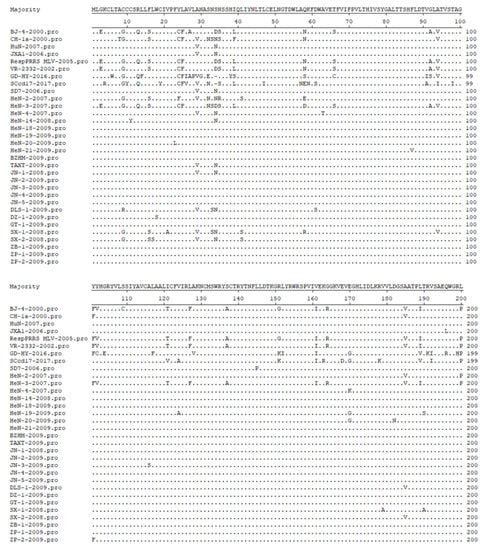
Figure 2.
Alignment analysis of amino acid sequence deduced by GP5.
Besides the mutation of amino acid sites, many studies have shown that the 33rd amino acid of PRRSV GP5 protein is deleted. Jiang et al. [38] reported that the GD-HY strain had an amino acid deletion at the 33rd site. Zhou et al. [39] showed that the ORF5 gene of SCcd17 encodes a 199 aa protein and has a novel 1 aa deletion in hypervariable region 1 (HVR1) at the 33rd site. Zhang [40] reported that the 32nd and 33rd amino acids of the strains were deleted, and the glycosylation sites of GP5 were increased and complexed. In addition, H38→K38 mutation occurred in the epidemic strains. Fan et al. [41] reported that SD7, SD8, and SD9 strains were absent in the 34th amino acid. Via phylogenetic analysis based on GP5 amino acids, Sun et al. [42] revealed that the novel NADC30-like PRRSV with a unique single amino acid deletion at the 34th site has become widespread and evolved into a new subgroup.
The gene sequences of representative PRRSV strains were analyzed. A total of 32 target gene sequences of PRRSV GP5 were obtained. To construct the phylogenetic tree, nucleotide sequences of the target gene were aligned using the ClustalX alignment tool and MEGA software (version 7.0.26, Mega Limited, Auckland, New Zealand). Phylogenetic trees were constructed using the neighbor-joining method with 1000 bootstrap replicates in MEGA7.0 (Figure 3). JN-1 and TAXT are similar to the reference strain JXA1, but far from the strain VR2332, and DLS-1 is in a single branch.
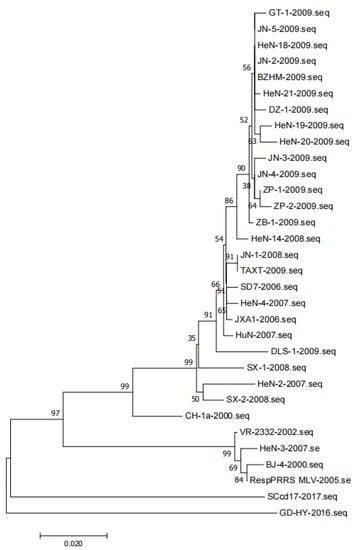
Figure 3.
Phylogenetic tree of PRRSV based on the ORF5 gene.
Furthermore, Zhao et al. [43] reported that a new insertion site (Lys57) appeared at the 57th site of GP5 protein, which was in the hypervariable region (32 aa–35 aa). PRRSV GP5 protein can easily mutate, which leads to the diversity of strains, increases the difficulty of vaccine development, and makes the disease difficult to control. In conclusion, GP5 protein is highly variable in PRRSV and plays an important role in monitoring variations in PRRSV.
4. Immune Function of GP5 Protein
GP5 protein is the main protective antigen targeted by neutralizing antibody induced by PRRSV vaccination or prior infection. The protein has six antigenic determinants. It contains two B cell antigen epitopes: a non-neutralizing epitope A and a neutralizing epitope B. Epitope B induces the body to produce specific neutralizing antibodies against the viral infection. In contrast, epitope A inhibits the recognition of epitope B in the body, thus delaying the production of neutralizing antibodies [30]. Studies show that inserting a Pan-DR T-helper cell epitope (PADRE) between the neutralizing and bait epitopes can reduce or eliminate the bait effect of the non-neutralizing epitope [44]. The reaction between the monoclonal antibody and the expressed product of the deletion mutant shows that GP5 protein has at least two antigenic regions: one extracellular region (27 aa–41 aa) and one at the C-terminal (180 aa–197 aa). The 50 amino acids in the C-terminal of GP5 protein play an important role in maintaining its antigenicity. If these amino acids were absent, GP5 protein would lose its reactivity with the antibody [45].
Akter et al. [46]. showed that the loss of glycan residues in N-linked glycosylation sites (N34, N44, and N51) enhances the immunogenicity of the nearby neutralizing epitope. Leng et al. [47] found that the B antigen region (AR) of HP-PRRSV GP5 did not neutralize AR. The results of indirect enzyme-linked immunosorbent assay (ELISA) and a virus neutralization test showed that there was no correlation between the levels of anti-B AR polypeptide antibody and neutralizing antibodies in the serum of pigs. The specific serum antibody of AR peptide has no neutralizing activity, and glutathione S-transferase-B (GST-B) fusion protein cannot inhibit the neutralizing ability of the antibodies. Three putative N-linked glycosylation sites (N34, N44, and N51) are located in the extracellular domain of GP5, which also include major neutralizing epitopes. Yin et al. [48] indicated that its peptide segments 30–36, 50–55, 140–142, 146–151, and 196–198 may be its dominant B cell epitope regions. Furthermore, T-cell epitopes of PRRSV in GP5 protein have been found (119 aa–127 aa and 151 aa–159 aa) [49]. Zhang et al. [50] used an infectious clone of PRRSV as a vector and replaced Asn with Ala at the 34th or 51st site of GP5 protein. After transfecting the cells, they obtained PRRSV mutants, vVR-N34A and vVR-N51A, which may have been related to the immunogenicity of neutralizing epitope B. The neutralization test results preliminarily suggested that the glycosylation of PRRSV GP5 protein played a role in the host immune function and affected the relationship between the virus and specific neutralizing antibodies. Wang et al. [51] analyzed the carboxyl terminal of PRRSV GP5 protein and predicted and screened the region with a high antigenicity index to construct recombinant phage M13-GP5 168 aa–198 aa. Western blot results showed that the recombinant phage had good reactivity, and the neutralization test confirmed that it could induce piglets to produce high levels of neutralizing antibodies. The latest research shows that a new immune escape mechanism of PRRSV infection has been discovered. Li et al. [52] demonstrated N-terminal acetylation by N-acetyltransferase Nat9 as a novel host defense mechanism that leads to K27-linked-ubiquitination-dependent proteolysis of GP5, which contributes to inhibiting PRRSV infection and proliferation in 3D4/21 cells.
5. Apoptosis Induction by GP5 Protein
GP5 protein not only plays a role in PRRSV infection, cell binding, and virus adsorption, but also in cell apoptosis. In 1996, studies revealed that PRRSV GP5 protein caused strong cytotoxicity, and the apoptosis induced by GP5 was independent of anti-apoptotic protein B-cell lymphoma-2 (Bcl-2) [53]. This apoptosis activity could not be prevented by using cell lines that permanently express Bcl-2 protein, which indicates that GP5 either induces apoptosis downstream of Bcl-2 or employs another unknown apoptosis pathway [54]. In vitro, the constructed cells expressing GP5 exhibit typical apoptosis phenomena, such as gradient fragmentation of genomic DNA, reduction of cell number, and formation of apoptotic bodies [55]. Shen [56] found that mutations at different glycosylation sites of PRRSV GP5 protein had different effects on apoptosis induction. Among them, the mutation of N34 and N35 glycosylation sites led to a significant increase in cell apoptosis, whereas the mutation of other sites did not cause any significant effects. Fernández et al. [57] used recombinant vaccinia virus to express PRRSV GP5 protein in mammalian cells. They found that the first 119 amino acids constituted a region that induced apoptosis due to the resulting strong cytotoxicity. The C-terminal region did not induce apoptosis. Hela cells stably expressing GP5 did not show evidence of apoptotic cell death and they speculated that the difference might be due to a histidine tag and an antigenic tag placed in the N-terminal of GP5 fusion constructs. GP5 protein of the PRRSV SD16 strain inhibits virus replication and leads to G2/M cell cycle arrest, but it does not induce Marc-145 cell apoptosis [58]. Similarly, Ma et al. [59] demonstrated that none of the PRRSV structural proteins, including GP5, had the potential to cause apoptosis in Marc-145 cells. Rather, NSP2 and NSP4 played causative roles in PRRSV-induced apoptosis in Marc-145 cells. Some studies have also found that the expression of GP5, GP5∆84–96, and GP5∆97–119 changes the ratio of Bax/Bcl-2 to different degrees but does not cause the apoptosis of Marc-145 cells [60]. Therefore, the definitive role of GP5 in apoptosis induced by PRRSV is still unknown.
6. Effects of GP5 Protein on Virus Replication
GP5 protein plays an important role in PRRSV replication. Song et al. [61] found that a stable expression of GP5 in Marc-145 cells promoted virus replication at the early stage of PRRSV infection by downregulating interferon (IFN) expression; in contrast, interfering GP5 expression with specific small interfering RNA (siRNA) inhibited PRRSV replication. Wang et al. [62] found that the expression of GP5 D84–119 inhibited PRRSV replication by upregulating IFN expression, especially IFN-β, and that the second extracellular region of GP5 played a regulatory role in PRRSV replication. Song et al. [63,64] successfully constructed three short hairpin RNA (shRNA) expression plasmids targeting the PRRSV GP5, which confirmed that the shRNA could effectively inhibit PRRSV replication in Marc-145 cells. They also screened two deoxyribozymes targeting the PRRSV GP5, which confirmed that the designed deoxyribozymes could effectively inhibit PRRSV replication in Marc-145 cells. Furthermore, a study by Wei et al. [65] evidenced that all N-linked glycans in GP5 are not required for virus viability in vitro but are essential for virus replication in vivo. In a different study, Niu et al. [66] studied the effect of PRRSV GP5 protein on the phosphorylation of interferon regulatory factor-3 (IRF-3) in the IFN signaling pathway. Sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analysis results showed that GP5 protein could inhibit the phosphorylation of IRF-3.
7. Interaction between GP5 Protein and Viral Protein
GP5 and M proteins are the main envelope proteins of PRRSV, which make up more than half of the virus proteins. They can form a heterodimer GP5/M linked by disulfide bonds in virus-infected cells, which is essential for protein transport, assembly, and outflow of progeny virions. Its immunogenicity is strong and conservative, and it allows virus budding to occur on the membrane of the exocytosis pathway [67,68]. The GP5/M protein complex of PRRSV adheres to saliva expressed on cells in order to infect them [69]. When PRRSV was used to infect peptidylgycine α-amidating monooxygenase treated with heparinase, the dimer of M and GP5 proteins was conveniently bound to heparin-like receptors on peptidylgycine α-amidating monooxygenase cells [70]. Zhang et al. [71] found that GP5 and M proteins of PRRSV-1 and PRRSV-2 strains were palmitoylated at cysteine, whether they were expressed alone or in PRRSV-infected cells. Viruses lacking one or two acylation sites in M or GP5 could be saved, but their titers were significantly reduced; in addition, the GP5 and M lacking acylation sites had formed a dimer. Ma et al. [72] used enhanced green fluorescent protein (EGFP) and red fluorescent protein (RFP) as tracers and found that when ORF5-EGFP and ORF6-RFP were co-expressed, GP5 protein could be transported from the endoplasmic reticulum to the high matrix, suggesting that the formation of the GP5/M heterodimer may be related to the post-translational modification, transport, and localization of GP5 protein.
8. Interaction between GP5 Protein and Host Proteins
Xue et al. [73] found that the outer domain (GP5-ecto-1) of the first GP5 directly interacts with non-muscle myosin heavy chain 9 (MYH9) C-terminal domain protein (PRA), and this interaction triggers PRA and endogenous MYH9 to form a silk assembly. MYH9 plays a pivotal role in PRRSV infection by physically interacting with PRRSV GP5 protein through its C-terminal domain, which makes the host cells susceptible to infection [74]. Du et al. [75] screened two types of host proteins using immunoprecipitation, namely, the mitochondrial inner membrane protein and calpain protein, both of which interact with GP5. These proteins can co-locate with GP5 in cells and are related to cell protein glycosylation, cell growth, proliferation, movement, function, and maintenance, as well as the development and function of the nervous system. GP5 contains a T4L (T4 lysozyme)-like domain that locally digests the peptide glycan layer during infection. T4 Spackle protein (encoded by gene 61.3) plays a role in inhibiting GP5 lysozyme activity [76]. Hicks et al. [77] found that GP5 protein interacts with Snapin of the Marc-145 cell line to take advantage of its role in intracellular transport and membrane fusion. Recent studies revealed that glyceraldehyde 3-phosphate dehydrogenase (GAPDH) interacts with GP5 by binding to 13 of its amino acid sequences (93 aa–105 aa), while GP5 interacts with GAPDH at the K277 amino acid. This indicates that during PRRSV infection, GP5 interacts with GAPDH in the cytoplasm to restrict it from entering the nucleus, and PRRSV promotes virus replication by utilizing glycolysis activity of GAPDH [24]. Zhang et al. In [78] cloned porcine ATP synthase subunit alpha (ATP5A) into the vector pFLAG-CMV-2 and transfected human embryonic kidney (HEK) 293 cells with pCI-GP5 along with pFLAG-CMV-2 or pFLAG-pig-ATP5A. Co-immunoprecipitation was performed using anti-FLAG affinity beads, and the immune complexes resolved by SDS-PAGE were probed with either anti-FLAG or anti-GP5 antibodies. The results showed that ATP5A was readily detected only in the presence of GP5 and not in the presence of empty vector. Thus, it is confirmed that GP5 can interact with ATP5A. Understanding the molecular mechanism of the interaction between PRRSV GP5 and host proteins (Figure 4) can lay the foundation for finding new antiviral targets and exploring the mechanism of viral replication.
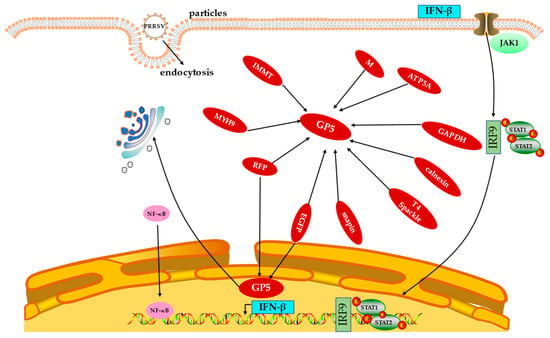
Figure 4.
Interactions between PRRSV GP5 protein and host proteins. Abbreviations: GP5, glycoprotein 5; IFN-β, interferon β; PRRSV, porcine reproductive and respiratory syndrome virus; EGFP, enhanced green fluorescent protein; RFP, red fluorescent protein; MYH9, myosin heavy chain 9; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; ATP5A, ATP synthase subunit alpha; IRF9, interferon regulatory factor 9; NF-κB, nuclear factor κB.
9. Influence of GP5 on Virulence
Wesley et al. [79] compared the amino acid sequences of VR-2332 with its attenuated vaccine. Their study revealed that the mutation of Arg to Asn at the 13th site and Arg to Gly at the 151st site of the American strain was related to the virulence weakening of the vaccine strain. Hence, the 13th amino acid (R13) and the 151st amino acid (R151) in GP5 protein were the sites related to PRRSV virulence. Studies have also shown that the glycosylation sites of certain Chinese strains have changed remarkably since 2006, and the absence of the 33rd glycosylation site and appearance of the 34th and 35th glycosylation sites may be related to the enhancement of PRRSV virulence [33]. Wang et al. [80] found that the 55th site mutations occurred in GP5 of the Guizhou epidemic strain through cloning and sequencing, and the site mutations at the 25th, 26th, 358th, 409th, and 554th nucleotides could lead to the loss or appearance of some enzyme cut-sites. The 55th site mutations in GP5 may play a vital role in the enhancement of PRRSV virulence. Most strains have high arginine-glutamine (RQ) motifs near their carboxyl terminal. Further, the highly conserved RQ motifs overlap with the hypervariable GP5 glycosylation sites, which may influence PRRSV virulence [81].
10. Applications of GP5 Protein in Vaccines
At present, the PRRSV vaccines used in China are mainly inactivated vaccines, such as CH-1a, or attenuated vaccines, such as CH-1R. Some vaccines on the market are listed in Table 1. The inactivated vaccines have poor immune efficacy, short immune protection time, and limited protection against heterologous strains; whereas the attenuated vaccines pose a risk of virulence reversal and have short immune protection time that cannot prevent a strong viral infection. Therefore, the development of efficient, safe, and affordable vaccines has become a trending research topic in recent years. GP5 protein has good immunological properties and induces neutralizing antibodies, making it the preferred protein for developing new vaccines. These vaccines primarily include nucleic acid, subunit, and live vector vaccines.

Table 1.
PRRSV vaccines on the market.
Nucleic acid vaccines, also known as DNA vaccines, induce cellular and humoral immunity simultaneously and have cross-protection to different serotypes of strains. Baroed et al. [82] cloned all the ORFs of PRRSV Danish isolates (DK-111/92) in a DNA vaccine vector, and inoculated pigs. The results showed that the pigs immunized with vaccines prepared using ORF1 and ORF4 quickly developed antibody immune responses against NSP2 and GP4; moreover, neutralizing antibody was detected in all pigs, but those inoculated with ORF5 showed the highest antibody titer. Using a DNA-prime/VACV boost regimen, Cui et al. [83] confirmed that the GP5-Mosaic vaccines conferred protection in pigs against heterologous viruses. Vaccination with the GP5-Mosaic-based vaccines resulted in cellular reactivity and higher levels of neutralizing antibodies to both VR2332 and MN184C PRRSV strains. In contrast, vaccination of animals with the GP5-WT vaccines induced responses only to VR2332. GP5-Mosaic vaccine can induce cross-reactive cellular responses to diverse strains, neutralizing antibodies, and protection in pigs [84]. Jiang et al. [85] obtained the suicide DNA vaccine pSFV-56 co-expressed with ORF5 and ORF6 and reported that it had good immunogenicity and could induce immune animals to produce a greater immune response. Jiang et al. [86] co-expressed GP5 and M proteins of PRRSV and found that the resulting dimer significantly improved DNA immunity. Furthermore, cytokines also improve the body’s immune response levels. Qin et al. [87] developed a nucleic acid vaccine PVIR-IL-18-ORF5 containing porcine interleukin-18 (IL-18) and conducted immune experiments in mice. The percentage of CD4- and CD8-positive cells in the pVIR-IL-18-ORF5 experimental group was significantly higher than that in the pVAX1-ORF5 group, which indicated that IL-18 improved both cellular and humoral immunity in animals, with notable improvement in cellular immunity. Li et al. [88] developed a recombinant plasmid carrying the PRRSV GP5 gene (pVAX-GP5) and porcine interleukin-15 gene (pVAX-IL-15). They inoculated mice with one or both genes and evaluated their humoral and cellular immunity. The proliferation tests showed that the T lymphocyte counts increased in the peripheral blood and spleen of pVAX-GP5-treated mice and were significantly enhanced in combination therapy with pVAX-IL-15. In addition, some studies revealed that adding porcine glutathione peroxidase-1 (GPX1) to PRRSV DNA vaccines produces an adjuvant effect and enhances the humoral and cellular immunity in animals [89]. Recent studies indicate that vaccination with GP5 chimeric DNA vaccine induces cell reaction and high levels of neutralizing antibodies [83]. Although DNA vaccine developments have made great progress, their immune effects are unstable, which warrants further research and improvement.
Recent developments in subunit vaccines have attracted much attention. Plana et al. [90] immunized pregnant sows with insect cell expression products of PRRSV ORF3 and ORF5 of the Spanish isolate (Olot/91) alone or in combination. The results showed that the ORF3 expression product had a protection rate of 68.4%, whereas that of the ORF5 expression product was 50%. Therefore, GP3 and GP5 subunit vaccines have good immunogenicity and are good candidate genes for recombinant subunit vaccine development. Yuan et al. [91] studied the immune-enhancing effects of Taishan Pinus massoniana pollen polysaccharides (TPPPS) and Freund adjuvant on PRRSV GP5 subunit vaccines. The results showed that the recombinant PRRSV GP5 protein induced a significant immune response in animals, which was significantly enhanced by the TPPPS adjuvant, with the middle dose showing the strongest effect. Furthermore, some studies have investigated the use of transgenic Arabidopsis plants expressing codon-optimized and transmembrane deleted antigenic proteins (GP4D and GP5D) as candidate antigens, which produce a good immune response in pigs. Plant-derived GP4D and GP5D proteins provide an alternative platform for producing effective PRRSV subunit vaccines [92]. Guo et al. [93] successfully constructed 10 GP5/M expression vectors. Among the 10 tags, the soluble expression effect of maltose-binding protein (MBP) fused with GP5/M protein was the greatest, and a high-purity recombinant protein of MBP-GP5/M was obtained that laid the foundation for follow-up research and mass preparation of PRRSV subunit vaccines.
Live vector vaccines express the main immunogenic genes of PRRSV through live viruses, such as adenovirus, fowlpox virus, herpes virus, and pseudorabies virus (PRV). Qiu et al. [94] developed recombinant fowlpox virus expressing PRRSV GP5, M, and GP5-M fusion proteins and carried out immunological tests on mice to evaluate its potential in inducing humoral and cellular immune responses. The results showed that, compared with other groups, the recombinant fowlpox virus expressing GP5-M fusion protein stimulated mice to produce high levels of specific neutralizing antibodies, significantly promoted interferon γ (IFN-γ) secretion, and induced the proliferation and expression of specific T lymphocytes, indicating significant enhancement of the humoral and cellular immunity. Wu et al. [95] co-expressed GP5 and M proteins of PRRSV with pseudotyped baculovirus containing hybrid cytomegalovirus (CMV) promoter/SFV (Semliki Forest virus) replicon as vector. The immunogenicity of recombinant baculovirus (BV-SFV-5m6) was compared with that of pseudotype baculovirus vaccine (BV-CMV-5m6), in which the expression of GP5 and M was only driven by CMV promoter. In vitro, BV-SFV-5m6 showed that the expression of foreign protein was enhanced. After immunization in mice, BV-SFV-5m6 induced strong GP5-specific ELISA and neutralizing antibodies against homologous and heterologous viruses. Further, Jiang et al. [96] used live attenuated PRV as the vaccine vector to express GP5 and M proteins in different forms to produce recombinant PRV. The immunized mice subsequently produced PRRSV-specific neutralizing antibodies and had a higher lymphocyte proliferation reaction. Zhao et al. [97] used PRV variant XJ and NADC30-like PRRSV strains (CHSCDJY-2019) as parents to construct a recombinant PRV, rPRV-NC56, with gE/gI/TK deletion and co-expression of NADC30-like PRRSV GP5 and M proteins. The results revealed that inoculation of rPRV-NC56 induced specific humoral and cellular immune responses against PRV and NADC30-like PRRSV in mice and protected them from the PRV XJ strain. Furthermore, Cruz et al. [98] expressed GP5 and M proteins of wild-type PRRSV with porcine transmissible gastroenteritis virus as the vector. The findings showed that the vaccinated animals produced faster and stronger humoral and cellular immune responses than unvaccinated animals.
11. Applications of GP5 Protein in Detection Methods
At present, there are several detection methods for PRRSV, such as immunoperoxidase monolayer assay to detect PRRS serum antibody, indirect fluorescent antibody assay (IFA), serum neutralization test, and ELISA to detect PRRS antibodies. The immunoperoxidase monolayer assay and IFA are not suitable for large-scale testing as they are subjective, laborious, and expensive; therefore, they are limited to laboratory testing. The serum neutralization test is mainly used to detect neutralizing antibodies produced by PRRSV infection in pigs, but it cannot be used to diagnose acute infectious cases [99,100].
ELISA has strong specificity, high sensitivity, and low costs, so it is widely used in the detection of PRRSV antibodies. PRRSV GP5 protein has good immunogenicity and can induce the body to produce neutralizing antibodies. Li et al. [100] amplified PRRSV GP5 by RT-PCR, constructed prokaryotic recombinant expression vector (pGEX-6P-GP5), and transferred it into Escherichia coli BL21 (DE3) for induced expression. The expressed protein was purified by the gradient urea method and then used as coating antigen. Thus, an indirect ELISA method for detecting PRRSV antibodies was established. Hou et al. [101] deleted the transmembrane region of GP5 to match the characteristics of the BB0907 strain of HP-PRRSV GP5 and optimized the remaining gene fragments according to the preference of Escherichia coli. Moreover, they constructed the prokaryotic expression vector of the highly expressed GP5 recombinant protein. The indirect ELISA (GP5-ELISA) method for antibody detection of PRRSV GP5 protein was established by optimizing the reaction conditions. Wang [102] prepared the purified recombinant GP5-3m2 protein, used it as the coating antigen, and optimized the indirect ELISA method using the square array test, thereby establishing the PRRSV GP5 indirect ELISA antibody detection method. Li et al. [103] used the gene sequence of NADC30-like PRRSV strain, which is currently widely used as an amplification template in China; comprehensively analyzed the parameters of secondary structural antigen index, antigenicity, hydrophilicity, surface accessibility, and potential neutralizing epitope of structural PRRSV GP5 and M proteins; and selected the corresponding neutralizing epitopes of GP5 and M proteins as the immunogen. By constructing the fusion expression plasmid pET-32a-GP5/M with linker sequence and expressing soluble GP5/M protein in the Escherichia coli expression system, an indirect ELISA method for detecting PRRSV neutralizing antibody in pigs was established. All the above mentioned studies show that indirect ELISA possesses advantages of high sensitivity, strong specificity, and good repeatability.
In addition, a new method for PRRSV detection was established. Yang et al. [104] used PCR combined with denaturing high-performance liquid chromatography (DHPLC) to detect PRRSV. Specific primers were designed according to the sequence characteristics of PRRSV GP5, and PCR amplification products were rapidly detected by DHPLC technology. The PCR-DHPLC method established in this study had the advantages of specificity, sensitivity, rapidity, and good repeatability, and could be used for early diagnosis of clinical PRRSV infection and in molecular epidemiological investigations.
12. Conclusions
GP5 protein is a highly variable protein in PRRSV, which is the main protective antigenic protein for inducing immunity. GP5 protein promotes virus replication at the early stage of PRRSV infection by downregulating IFN expression; moreover, its N-linked glycan is essential for virus replication in vivo. GP5 and M proteins form the heterodimer GP5-M in virus-infected cells. GP5-M formation may be related to the post-translational modification, transport, and localization of GP5 protein, which is crucial for the assembly, outflow, and virus budding of offspring virions. GP5 protein interacts with PRA and GAPDH in the cytoplasm, thus triggering PRA and endogenous MYH9 to form filament assembly and inhibiting GAPDH from entering the nucleus. In addition, GP5 protein induces apoptosis. For example, GP5 protein expressing PRRSV in mammalian cells using recombinant vaccinia virus provided evidence that the first 119 amino acids form a region that induces apoptosis, whereas the C-terminal region does not. Furthermore, PRRSV detection can be achieved by an indirect ELISA method based on GP5 protein, which possesses high sensitivity, strong specificity, and good repeatability. At present, great progress has been made in the development of new vaccines against PRRSV. However, the immune effect of vaccines is unstable, and the prevention and control of PRRSV are still major concerns that harm the pig industry today. The development of a safer and more effective PRRSV vaccine is needed.
Author Contributions
Q.L.; writing—original draft preparation, Y.Z.; writing—original draft preparation, H.Z.; writing—original draft preparation, Z.Y.; writing—original draft preparation, H.S.: writing—original draft preparation, W.K.; writing—review and editing, M.Z.; funding acquisition, N.W.; funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (31902279, 31902284), Guangdong Science and Technology Program Project (2021A1515012388, 2017A020208079), and the Guangdong Basic and Applied Basic Research Foundation (2021A1515110322).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All datasets are available in the main manuscript. The dataset supporting the conclusions of this article is included within the article.
Acknowledgments
We apologize to all colleagues whose contributions were not discussed and cited owing to space constraints.
Conflicts of Interest
The authors declare that they have no competing interests.
References
- Guo, Z.; Chen, X.X.; Li, R.; Qiao, S.; Zhang, G. The prevalent status and genetic diversity of porcine reproductive and respiratory syndrome virus in China: A molecular epidemiological perspective. Virol. J. 2018, 15, 2. [Google Scholar] [CrossRef]
- Zhang, C. Isolation, Identification, Whole Gene Sequencing, and Genetic Evolution Analysis of PRRSV Shaanxi-2 Strain from Wild Boar. Ph.D. Thesis, Northwest Agricultural and Forestry University, Xianyang, China, 2010. [Google Scholar]
- Karniychuk, U.U.; Nauwynck, H.J. Pathogenesis and prevention of placental and transplacental porcine reproductive and respiratory syndrome virus infection. Vet. Res. 2013, 44, 95. [Google Scholar] [CrossRef] [PubMed]
- Fei, W.T. Prevalence and genetic diversity of porcine reproductive and respiratory syndrome virus (PRRSV) in China: A molecular epidemiological study. Pigs Today 2018, 15, 76–81. [Google Scholar]
- Qin, N. Construction and Preliminary Application of Cell Lines Expressing Major Structural Proteins of PRRSV. Ph.D. Thesis, Guangxi University, Nanning, China, 2019. [Google Scholar]
- Lu, Z.H.; Archibald, A.L.; Ait-Ali, T. Beyond the whole genome consensus: Unravelling of PRRSV phylogenomics using next generation sequencing technologies. Virus Res. 2014, 194, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Wang, C.; Cao, W.; Fang, R.; Zhao, J. Risk Factors and Spatial-Temporal Analysis of Porcine Reproductive and Respiratory Syndrome Seroprevalence in China Before and After African Swine Fever Outbreak. Front. Vet. Sci. 2022, 9, 929596. [Google Scholar] [CrossRef]
- Zhao, H.Z.; Wang, F.X.; Han, X.Y.; Guo, H.; Liu, C.Y.; Hou, L.N.; Wang, Y.X.; Zheng, H.; Wang, L.; Wen, Y.J. Recent advances in the study of NADC34-like porcine reproductive and respiratory syndrome virus in China. Front. Microbiol. 2022, 13, 950402. [Google Scholar] [CrossRef]
- Li, Y.; Fang, L.; Zhou, Y.; Tao, R.; Wang, D.; Xiao, S. Porcine Reproductive and Respiratory Syndrome Virus Infection Induces both eIF2α Phosphorylation-Dependent and-Independent Host Translation Shutoff. J. Virol. 2018, 92, e00600–e00618. [Google Scholar] [CrossRef]
- Dokland, T. The structural biology of PRRSV. Virus Res. 2010, 154, 86–97. [Google Scholar] [CrossRef]
- Li, P.; Shen, Y.; Wang, T.; Li, J.; Li, Y.; Zhao, Y.; Liu, S.; Li, B.; Liu, M.; Meng, F. Epidemiological survey of PRRS and genetic variation analysis of the ORF5 gene in Shandong Province, 2020–2021. Front. Vet. Sci. 2022, 9, 987667. [Google Scholar] [CrossRef]
- Wang, Q.; Peng, J.; Sun, Y.; Chen, J.; An, T.; Leng, C.; Li, L.; Zhao, H.; Guo, X.; Ge, X.; et al. Unique epitopes recognized by monoclonal antibodies against HP-PRRSV: Deep understanding of antigenic structure and virus-antibody interaction. PLoS ONE 2014, 9, e111633. [Google Scholar] [CrossRef]
- Kedkovid, R.; Nuntawan Na Ayudhya, S.; Amonsin, A.; Thanawongnuwech, R. NSP2 gene variation of the North American genotype of the Thai PRRSV in central Thailand. Virol. J. 2010, 7, 340. [Google Scholar] [CrossRef]
- Han, G.; Xu, H.; Wang, K.; He, F. Emergence of Two different recombinant PRRSV strains with low neutralizing antibody susceptibility in China. Sci. Rep. 2019, 9, 2490. [Google Scholar] [CrossRef]
- Chen, N.; Li, S.; Tian, Y.; Li, X.; Li, S.; Li, J.; Qiu, M.; Sun, Z.; Xiao, Y.; Yan, X.; et al. Chimeric HP-PRRSV2 containing an ORF2-6 consensus sequence induces antibodies with broadly neutralizing activity and confers cross protection against virulent NADC30-like isolate. Vet. Res. 2021, 52, 74. [Google Scholar] [CrossRef]
- Evans, A.B.; Loyd, H.; Dunkelberger, J.R.; van Tol, S.; Bolton, M.J.; Dorman, K.S.; Dekkers, J.C.M.; Carpenter, S. Antigenic and Biological Characterization of ORF2-6 Variants at Early Times Following PRRSV Infection. Viruses 2017, 9, 113. [Google Scholar] [CrossRef]
- Guo, J.; Liu, Z.; Tong, X.; Wang, Z.; Xu, S.; Chen, Q.; Zhou, J.; Fang, L.; Wang, D.; Xiao, S. Evolutionary Dynamics of Type 2 Porcine Reproductive and Respiratory Syndrome Virus by Whole-Genome Analysis. Viruses 2021, 13, 2469. [Google Scholar] [CrossRef]
- Zhao, J.; Xu, Z.; Xu, T.; Zhou, Y.; Li, J.; Deng, H.; Li, F.; Xu, L.; Sun, X.; Zhu, L. Molecular Characterization of the Nsp2 and ORF5s of PRRSV Strains in Sichuan China during 2012–2020. Animals 2022, 12, 3309. [Google Scholar] [CrossRef]
- Do, V.T.; Dao, H.T.; Hahn, T.W. Generation of a cold-adapted PRRSV with a nucleotide substitution in the ORF5 and numerous mutations in the hypervariable region of NSP2. J. Vet. Sci. 2020, 21, e85. [Google Scholar] [CrossRef]
- Huang, X.J.; Li, R.; Qiao, S.L.; Yang, Y.Y.; Deng, R.G.; Zhang, G.P. Preparation and identification of monoclonal antibody against GP5 protein of porcine reproductive and respiratory syndrome virus. Chin. Vet. Sci. 2019, 49, 1374–1382. [Google Scholar] [CrossRef]
- Jiang, Z.Y.; Chu, P.P.; Cheng, T.B.; Li, C.L.; Cai, R.J. Genetic Variation Analysis of ORF5 Gene of Porcine Reproductive and Respiratory Syndrome Virus in Guangdong Province in 2021. GD Agr. Sci. 2022, 49, 97–105. [Google Scholar]
- Thuy, N.T.; Thu, N.T.; Son, N.G.; le Ha, T.T.; Hung, V.K.; Nguyen, N.T.; do Khoa, V.A. Genetic analysis of ORF5 porcine reproductive and respiratory syndrome virus isolated in Vietnam. Microbiol. Immunol. 2013, 57, 518–526. [Google Scholar] [CrossRef]
- Wang, Z.H. Preparation and Identification of Monoclonal Antibody against GP5 Protein of Porcine Reproductive and Respiratory Syndrome Virus. Ph.D. Thesis, Zhengzhou University, Luoyang, China, 2012. [Google Scholar]
- Liu, X.; Liu, X.; Bai, J.; Gao, Y.; Song, Z.; Nauwynck, H.; Wang, X.; Yang, Y.; Jiang, P. Glyceraldehyde-3-Phosphate Dehydrogenase Restricted in Cytoplasmic Location by Viral GP5 Facilitates Porcine Reproductive and Respiratory Syndrome Virus Replication via Its Glycolytic Activity. J. Virol. 2021, 95, e0021021. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Screening and Application of Antigenic Epitopes of GP5 Protein and M Protein of Porcine Reproductive and Respiratory Syndrome Virus. Ph.D. Thesis, Zhengzhou University, Luoyang, China, 2021. [Google Scholar]
- Wu, J.S.G.L.; Liu, T.Q.; Ju, M.Y.; Zhang, B.; Han, Y.; Liu, J.Q.; Song, Q.Q.; Guan, P.Y. Prokaryotic expression of GP5 protein of porcine reproductive and respiratory syndrome virus and preparation of polyclonal antibody. China Anim. Health Insp. 2020, 37, 121–126. [Google Scholar]
- Liu, Y. Segmented Expression and Identification of GP5 Protein of Porcine Reproductive and Respiratory Syndrome Virus. Ph.D. Thesis, Shandong Agricultural University, Tai’an, China, 2010. [Google Scholar]
- Shin, G.E.; Park, J.Y.; Lee, K.K.; Ko, M.K.; Ku, B.K.; Park, C.K.; Jeoung, H.Y. Genetic diversity of porcine reproductive and respiratory syndrome virus and evaluation of three one-step real-time RT-PCR assays in Korea. BMC Vet. Res. 2022, 18, 327. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Zhang, Q.; Chen, Y.; Shen, H.; Yang, G.; Jiang, P.; Chen, J.L.; Lin, L. The emergence of a novel recombinant porcine reproductive and respiratory syndrome virus with an amino acid insertion in GP5 protein. Microb. Pathog. 2020, 149, 104573. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.F. Construction of Glycosylated PRRSV GP5 Baculovirus and Study on Its Immunogenicity. Ph.D. Thesis, Shandong Agricultural University, Tai’an, China, 2021. [Google Scholar]
- Cheng, Q. Cloning, Sequence Analysis, and Molecular Epidemiological Investigation of Partial Nsp2 and ORF5~7 Genes of Porcine Reproductive and Respiratory Syndrome Virus in Guangxi. Ph.D. Thesis, Guangxi University, Nanning, China, 2008. [Google Scholar]
- Li, Z.G.; Wu, H. Research progress on genetic variation of porcine reproductive and respiratory syndrome virus genome. Prog. Vet. Med. 2009, 30, 101–105. [Google Scholar] [CrossRef]
- Chen, W.; Li, Y.F.; Wang, X.L.; Jiang, P. Genetic variation and phylogenetic analysis of ORF5 gene of porcine reproductive and respiratory syndrome virus. Anim. Husb. Vet. Med. 2008, 59, 11–14. [Google Scholar]
- Zhang, J.Q.; Wu, J.Q.; Meng, B.; Guo, W.L.; Wang, X.H.; Li, K.; Yu, J.; Liu, S.Y.; Zhang, Y.Y.; Wang, J.B. Genetic variation analysis of PRRSV, CSFV and PRRSV ORF5 genes detected by multiplex RT-PCR. Chin. J. Vet. Sci. 2011, 31, 633–637. [Google Scholar] [CrossRef]
- Lu, X.Y.; Zhou, Y.H.; Li, W.J.; Zhang, Z.Y.; Liu, X.P.; Duan, E.Z.; Xia, P.A.; CUI, B.A. Genetic Variation Analysis of NSP2 and GP5 Genes of Porcine Reproductive and Respiratory Syndrome Virus Isolated from Henan Province. Acta Agric. Boreali-Sin. 2012, 27, 100–104. [Google Scholar]
- Li, G.X.; LI, P.R.; Li, P.C.; Ren, Y.D.; Huang, X.D.; Zhang, R.L.; Wu, X. Genetic variation analysis of GP5 and M genes of northeast strains of porcine reproductive and respiratory syndrome virus from 2006 to 2012. J. Northeast Agric. Univ. 2016, 47, 1–10. [Google Scholar] [CrossRef]
- Deng, S.C.; Feng, J.P.; Cui, J.; Chen, Y.; Cui, T.T.; Bai, X.L.; Shi, Q.W.; Zhang, G.H. Genetic variation analysis of PRRSV ORF5 gene in South China in 2014. Chin. J. Vet. Med. 2016, 52, 3–6. [Google Scholar]
- Jiang, Z.Y.; Cai, R.J.; Li, Y.; Li, C.L. Genetic Variation Analysis of PRRSV GP5 Gene in Guangdong Province from 2014 to 2016. GD Agric. Sci. 2016, 43, 90–95. [Google Scholar] [CrossRef]
- Zhou, L.; Kang, R.; Xie, B.; Tian, Y.; Wu, X.; Lv, X.; Yang, X.; Wang, H. Identification of a Novel Recombinant Type 2 Porcine Reproductive and Respiratory Syndrome Virus in China. Viruses 2018, 10, 151. [Google Scholar] [CrossRef]
- Zhang, J. Genetic Variation of GP5 Gene of Porcine Reproductive and Respiratory Syndrome Virus and Establishment of Double Fluorescence Quantitative PCR Detection Method. Ph.D. Thesis, Nanjing Agricultural University, Nanjing, China, 2020. [Google Scholar]
- Fan, Y.F.; Bai, J.; Jiang, P. Genetic Variation Analysis of GP5 Gene of Porcine Reproductive and Respiratory Syndrome Virus Epidemic Strain in Shandong Province. J. Domest. Anim. Ecol. 2017, 38, 63–67. [Google Scholar]
- Sun, Y.F.; Yu, H.; Jiang, X.; Ma, J.F.; Xu, C.Q.; Yu, X.X.; Li, L.A. Novel ORF5 deletion of NADC30-like porcine reproductive and respiratory syndrome viruses circulating in northern China from 2016 to 2018. J. Vet. Diagn. Investig. 2020, 32, 928–932. [Google Scholar] [CrossRef]
- Zhao, X.Y.; Zhang, G.; Wang, Y.; Liu, H.Y.; Chen, Y.; Zhao, J.; Wang, C.Q.; Chen, L.; Li, Y.T. Molecular epidemiological investigation of porcine reproductive and respiratory syndrome virus in Henan province from 2018 to 2019. Chin. J. Vet. Sci. 2020, 40, 2286–2292. [Google Scholar] [CrossRef]
- Fang, L.; Jiang, Y.; Xiao, S.; Niu, C.; Zhang, H.; Chen, H. Enhanced immunogenicity of the modified GP5 of porcine reproductive and respiratory syndrome virus. Virus Genes 2006, 32, 5–11. [Google Scholar] [CrossRef]
- Wu, J.S.G.L. Prokaryotic Expression of GP5 and M Proteins of PRRSV and Its Effect on Porcine PBMC. Ph.D. Thesis, Inner Mongolia Agricultural University, Huhehaote, China, 2021. [Google Scholar]
- Akter, F.; Roychoudhury, P.; Dutta, T.K.; Subudhi, P.K.; Kumar, S.; Gali, J.M.; Behera, P.; Singh, Y.D. Isolation and molecular characterization of GP5 glycoprotein gene of Betaarterivirus suid 2 from Mizoram, India. Virusdisease 2021, 32, 748–756. [Google Scholar] [CrossRef]
- Leng, C.L.; An, T.Q.; Chen, J.Z.; Gong, D.Q.; Peng, J.M.; Yang, Y.Q.; Wu, J.; Guo, J.J.; Li, D.Y.; Zhang, Y.; et al. Highly pathogenic porcine reproductive and respiratory syndrome virus GP5 B antigenic region is not a neutralizing antigenic region. Vet. Microbiol. 2012, 159, 273–281. [Google Scholar] [CrossRef]
- Yin, G.Y.; Wang, L.S. Cloning of PRRSV-GP5 gene and prediction and analysis of its B cell epitope. J. SC Agric. Univ. 2016, 34, 234–238. [Google Scholar] [CrossRef]
- Vashisht, K.; Goldberg, T.L.; Husmann, R.J.; Schnitzlein, W.; Zuckermann, F.A. Identification of immunodominant T-cell epitopes present in glycoprotein 5 of the North American genotype of porcine reproductive and respiratory syndrome virus. Vaccine 2008, 26, 4747–4753. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, G.L.; Xu, D.Y.; Li, F.; Xu, Z.H. Construction of a mutant with deletion of glycosylation site of GP5 protein of porcine reproductive and respiratory syndrome virus. Chin. Vet. Sci. 2012, 42, 511–516. [Google Scholar] [CrossRef]
- Wang, Y.M.; Gu, J.M.; Lei, L.C.; Feng, X.; Sun, C.J.; Han, W.Y. Phage display of carboxyl terminal of GP5 protein of porcine reproductive and respiratory syndrome virus and its induction of neutralizing antibody level in piglets. Chin. J. Vet. Sci. 2019, 39, 830–834+841. [Google Scholar] [CrossRef]
- Li, X.; Sun, R.; Guo, Y.; Zhang, H.; Xie, R.; Fu, X.; Zhang, L.; Zhang, L.; Li, Z.; Huang, J. N-Acetyltransferase 9 Inhibits Porcine Reproductive and Respiratory Syndrome Virus Proliferation by N-Terminal Acetylation of the Structural Protein GP5. Microbiol. Spectr. 2023, 11, e0244222. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Rogan, D.; Erickson, L.; Zhang, J.; Yoo, D. Characterization of the porcine reproductive and respiratory syndrome virus glycoprotein 5 (GP5) in stably expressing cells. Virus Res. 2004, 104, 33–38. [Google Scholar] [CrossRef]
- Dea, S.; Gagnon, C.A.; Mardassi, H.; Pirzadeh, B.; Rogan, D. Current knowledge on the structural proteins of porcine reproductive and respiratory syndrome (PRRS) virus: Comparison of the North American and European isolates. Arch. Virol. 2000, 145, 659–688. [Google Scholar] [CrossRef]
- Suarez, P.; Diaz-Guerra, M.; Prieto, C.; Esteban, M.; Castro, J.M.; Nieto, A.; Ortin, J. Open reading frame 5 of porcine reproductive and respiratory syndrome virus as a cause of virus-induced apoptosis. J. Virol. 1996, 70, 2876–2882. [Google Scholar] [CrossRef]
- Shen, S. Preliminary Study on Glycosylation Site Mutation and Function of HP-PRRSV Envelope Glycoprotein 5. Ph.D. Thesis, Shandong Agricultural University, Tai’an, China, 2016. [Google Scholar]
- Fernandez, A.; Suarez, P.; Castro, J.M.; Tabares, E.; Diaz-Guerra, M. Characterization of regions in the GP5 protein of porcine reproductive and respiratory syndrome virus required to induce apoptotic cell death. Virus Res. 2002, 83, 103–118. [Google Scholar] [CrossRef]
- Mu, Y.; Li, L.; Zhang, B.; Huang, B.; Gao, J.; Wang, X.; Wang, C.; Xiao, S.; Zhao, Q.; Sun, Y.; et al. Glycoprotein 5 of porcine reproductive and respiratory syndrome virus strain SD16 inhibits viral replication and causes G2/M cell cycle arrest, but does not induce cellular apoptosis in Marc-145 cells. Virology 2015, 484, 136–145. [Google Scholar] [CrossRef]
- Ma, Z.; Wang, Y.; Zhao, H.; Xu, A.T.; Wang, Y.; Tang, J.; Feng, W.H. Correction: Porcine Reproductive and Respiratory Syndrome Virus Nonstructural Protein 4 Induces Apoptosis Dependent on Its 3C-Like Serine Protease Activity. PLoS ONE 2020, 15, e0230086. [Google Scholar] [CrossRef]
- Li, L.L. Preliminary Study on the Relationship between Porcine Reproductive and Respiratory Syndrome Virus GP5, Apoptosis, and Virus Replication. Ph.D. Thesis, Northwest Agricultural and Forestry University, Xianyang, China, 2014. [Google Scholar]
- Song, L.L.; Jia, C.Y.; Han, X.M.; Zhou, E.M.; Mu, Y. Effect of disturbing and stably expressing GP5 on early replication of porcine reproductive and respiratory syndrome virus infection. Chin. J. Anim Vet. Sci. 2018, 49, 1231–1240. [Google Scholar]
- Wang, X.H. Effect of Stable Expression of GP5~(Δ84-119) on Porcine Reproductive and Respiratory Syndrome Virus Replication and Its Mechanism. Ph.D. Thesis, Northwest Agricultural and Forestry University, Xianyang, China, 2016. [Google Scholar]
- Song, D.W.; Ding, Z.; Li, Z.J.; Wang, X.L.; Xuan, H.; Wang, G.P. RNAi targeting GP5 gene inhibits PRRSV replication in MARC-145 cells. In Proceedings of the 2011 Academic Annual Meeting of China Animal Husbandry and Veterinary Society, Chengdu, China, 22 October 2011; p. 419. [Google Scholar]
- Song, D.W.; Yuan, J.; Li, X.; Guo, Y.P.; Yang, J.X.; Xuan, H.; Wang, G.P.; Ding, Z. Inhibition of PRRSV replication in MARC-145 cells by deoxyribozyme targeting GP5 gene. Chin. J. Vet. Sci. 2011, 31, 1695–1701. [Google Scholar] [CrossRef]
- Wei, Z.; Lin, T.; Sun, L.; Li, Y.; Wang, X.; Gao, F.; Liu, R.; Chen, C.; Tong, G.; Yuan, S. N-linked glycosylation of GP5 of porcine reproductive and respiratory syndrome virus is critically important for virus replication in vivo. J. Virol. 2012, 86, 9941–9951. [Google Scholar] [CrossRef]
- Niu, X.Y.; Wang, Y.L.; Xiong, Z.X.; Zhang, X.H.; Li, K.N.; He, D.S. Effect of PRRSV-GP5 protein on phosphorylation of interferon regulatory factor-3. HL Anim. Sci. Vet. Med. 2014, 57, 17–20+205. [Google Scholar] [CrossRef]
- Mardassi, H.; Massie, B.; Dea, S. Intracellular synthesis, processing, and transport of proteins encoded by ORFs 5 to 7 of porcine reproductive and respiratory syndrome virus. Virology 1996, 221, 98–112. [Google Scholar] [CrossRef]
- Yuan, S.L.; Tang, D.Y.; Yang, Z.G.; Han, C.Y.; Yan, R.T.; Chen, C.Z.; Luo, L.; Chen, X.; Liao, Z.B. Research progress on structural protein function of porcine reproductive and respiratory syndrome virus. North. Anim. Husb. 2022, 20, 25. [Google Scholar]
- Cai, J.; Ma, Y.; Li, J.; Yan, C.; Hu, R.; Zhang, J. Construction and characterization of a recombinant canine adenovirus expressing GP5 and M proteins of porcine reproductive and respiratory syndrome virus. J. Vet. Med. Sci. 2010, 72, 1035–1040. [Google Scholar] [CrossRef]
- Xu, L.H.; Wang, L.; Liu, H.L.; Chen, B.Y. Research progress on molecular biology of porcine reproductive and respiratory syndrome virus. J. NX Agric. Coll. 2004, 25, 71–75. [Google Scholar]
- Zhang, M.; Han, X.; Osterrieder, K.; Veit, M. Palmitoylation of the envelope membrane proteins GP5 and M of porcine reproductive and respiratory syndrome virus is essential for virus growth. PLoS Pathog. 2021, 17, e1009554. [Google Scholar] [CrossRef]
- Ma, Y.; Jiang, Y.B.; Xiao, S.B.; Fang, L.R.; Chen, H.C. Heterodimerization of GP5 and M proteins of porcine reproductive and respiratory syndrome virus co expressed in vitro. J. Microbiol. 2006, 54, 639–643. [Google Scholar] [CrossRef]
- Xue, B.; Hou, G.; Zhang, G.; Huang, J.; Li, L.; Nan, Y.; Mu, Y.; Wang, L.; Zhang, L.; Han, X.; et al. MYH9 Aggregation Induced by Direct Interaction With PRRSV GP5 Ectodomain Facilitates Viral Internalization by Permissive Cells. Front. Microbiol. 2019, 10, 2313. [Google Scholar] [CrossRef]
- Gao, J.; Xiao, S.; Xiao, Y.; Wang, X.; Zhang, C.; Zhao, Q.; Nan, Y.; Huang, B.; Liu, H.; Liu, N.; et al. MYH9 is an Essential Factor for Porcine Reproductive and Respiratory Syndrome Virus Infection. Sci. Rep. 2016, 6, 25120. [Google Scholar] [CrossRef] [PubMed]
- Du, J.G.; Wang, Z.; Zhou, L.; Gai, X.N.; Guo, X.; Yang, H.C. Screening and Analysis of Interaction Protein between Porcine Reproductive and Respiratory Syndrome Virus GP5 and Host Cells. In Proceedings of the Chinese Association of Animal Science and Veterinary Medicine, Guangzhou, China, 8 November 2014; p. 352. [Google Scholar]
- Kanamaru, S.; Uchida, K.; Nemoto, M.; Fraser, A.; Arisaka, F.; Leiman, P.G. Structure and Function of the T4 Spackle Protein Gp61.3. Viruses 2020, 12, 1070. [Google Scholar] [CrossRef] [PubMed]
- Hicks, J.A.; Yoo, D.; Liu, H.C. Interaction of porcine reproductive and respiratory syndrome virus major envelope proteins GP5 and M with the cellular protein Snapin. Virus Res. 2018, 249, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zakhartchouk, A. Characterization of the interactome of the porcine reproductive and respiratory syndrome virus glycoprotein-5. Arch. Virol. 2018, 163, 1595–1605. [Google Scholar] [CrossRef] [PubMed]
- Wesley, R.D.; Mengeling, W.L.; Lager, K.M.; Clouser, D.F.; Landgraf, J.G.; Frey, M.L. Differentiation of a porcine reproductive and respiratory syndrome virus vaccine strain from North American field strains by restriction fragment length polymorphism analysis of ORF5. J. Vet. Diagn. Investig. 1998, 10, 140–144. [Google Scholar] [CrossRef]
- Wang, D.S.; Yang, H.F.; Shi, K.Z.; Zhou, B.J.; Wen, M. RLFP Analysis of GP5 Gene of Porcine Reproductive and Respiratory Syndrome Virus Guizhou Epidemic Strain. Anim. Husb. Vet. Med. 2009, 41, 76–78. [Google Scholar]
- Robinson, S.R.; Abrahante, J.E.; Johnson, C.R.; Murtaugh, M.P. Purifying selection in porcine reproductive and respiratory syndrome virus ORF5a protein influences variation in envelope glycoprotein 5 glycosylation. Infect. Genet. Evol. 2013, 20, 362–368. [Google Scholar] [CrossRef]
- Barfoed, A.M.; Blixenkrone-Moller, M.; Jensen, M.H.; Botner, A.; Kamstrup, S. DNA vaccination of pigs with open reading frame 1-7 of PRRS virus. Vaccine 2004, 22, 3628–3641. [Google Scholar] [CrossRef]
- Cui, J.; O’Connell, C.M.; Hagen, C.; Sawicki, K.; Smyth, J.A.; Verardi, P.H.; Kruiningen, H.J.V.; Garmendia, A.E. Broad Protection of Pigs against Heterologous PRRSV Strains by a GP5-Mosaic DNA Vaccine Prime/GP5-Mosaic rVaccinia (VACV) Vaccine Boost. Vaccines 2020, 8, 106. [Google Scholar] [CrossRef]
- Cui, J.; O’Connell, C.M.; Costa, A.; Pan, Y.; Smyth, J.A.; Verardi, P.H.; Burgess, D.J.; Van Kruiningen, H.J.; Garmendia, A.E. A PRRSV GP5-Mosaic vaccine: Protection of pigs from challenge and ex vivo detection of IFNγ responses against several genotype 2 strains. PLoS ONE 2019, 14, e0208801. [Google Scholar] [CrossRef]
- Jiang, Y.B.; Fang, L.R.; Xiao, S.B.; Zhang, H.; Chen, H.C. Construction and Immune Response of Suicide DNA Vaccine Co-expressing GP5 and M Protein of Porcine Reproductive and Respiratory Syndrome Virus. Agric. Sci. China 2006, 47, 1011–1017. [Google Scholar]
- Jiang, Y.; Xiao, S.; Fang, L.; Yu, X.; Song, Y.; Niu, C.; Chen, H. DNA vaccines co-expressing GP5 and M proteins of porcine reproductive and respiratory syndrome virus (PRRSV) display enhanced immunogenicity. Vaccine 2006, 24, 2869–2879. [Google Scholar] [CrossRef]
- Qin, X.G.; Jin, N.Y.; Shen, G.S.; Yin, R.L.; Zhuang, T.Z.; Zheng, M.; Ma, M.X. Construction of experimental nucleic acid vaccine of porcine reproductive and respiratory syndrome virus GP5 protein and preliminary study on its immunogenicity. Chin. J. Virol. 2006, 20, 375–378. [Google Scholar]
- Li, G.; Shi, N.; Suo, S.; Cui, J.; Zarlenga, D.; Ren, X. Vaccination of mice with ORF5 plasmid DNA of PRRSV; enhanced effects by co-immunizing with porcine IL-15. Immunol. Investig. 2012, 41, 231–248. [Google Scholar] [CrossRef]
- Du, L.; Li, B.; Pang, F.; Yu, Z.; Xu, X.; Fan, B.; Tan, Y.; He, K.; Huang, K. Porcine GPX1 enhances GP5-based DNA vaccination against porcine reproductive and respiratory syndrome virus. Vet. Immunol. Immunopathol. 2017, 183, 31–39. [Google Scholar] [CrossRef]
- Plana Duran, J.; Climent, I.; Sarraseca, J.; Urniza, A.; Cortés, E.; Vela, C.; Casal, J.I. Baculovirus expression of proteins of porcine reproductive and respiratory syndrome virus strain Olot/91. Involvement of ORF3 and ORF5 proteins in protection. Virus Genes 1997, 14, 19–29. [Google Scholar] [CrossRef]
- Yuan, Y.M.; Peng, J.; Hu, L.P.; Zhu, L.J.; Wang, S.J.; Dong, W.W.; Ma, N.N.; Zhou, J.B.; Lu, Y.P.; Li, Z.R.; et al. Study on immune enhancement effect of Taishan pine pollen polysaccharide on PRRSV GP5 subunit vaccine. Chin. J. Prev. Vet. Med. 2017, 39, 740–745. [Google Scholar]
- An, C.H.; Nazki, S.; Park, S.C.; Jeong, Y.J.; Lee, J.H.; Park, S.J.; Khatun, A.; Kim, W.I.; Park, Y.I.; Jeong, J.C.; et al. Plant synthetic GP4 and GP5 proteins from porcine reproductive and respiratory syndrome virus elicit immune responses in pigs. Planta 2018, 247, 973–985. [Google Scholar] [CrossRef]
- Guo, Y.K.; Guo, W.Y.; Ming, S.L.; Guo, Y.J.; Yang, G.Y. Soluble Expression and Immunogenicity Analysis of GP5/M Protein of Porcine Reproductive and Respiratory Syndrome Virus. Chin. J. Vet. Sci. 2018, 38, 609–617. [Google Scholar] [CrossRef]
- Qiu, Y.; Wang, M.Z. Study on the Immunity of Three Different Live Vector Vaccines of Porcine Reproductive and Respiratory Syndrome Virus. GD J. Anim. Vet. Sci. 2011, 36, 28–31. [Google Scholar]
- Wu, Q.; Xu, F.; Fang, L.; Xu, J.; Li, B.; Jiang, Y.; Chen, H.; Xiao, S. Enhanced immunogenicity induced by an alphavirus replicon-based pseudotyped baculovirus vaccine against porcine reproductive and respiratory syndrome virus. J. Virol. Methods 2013, 187, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Fang, L.; Xiao, S.; Zhang, H.; Pan, Y.; Luo, R.; Li, B.; Chen, H. Immunogenicity and protective efficacy of recombinant pseudorabies virus expressing the two major membrane-associated proteins of porcine reproductive and respiratory syndrome virus. Vaccine 2007, 25, 547–560. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhu, L.; Xu, L.; Li, F.; Deng, H.; Huang, Y.; Gu, S.; Sun, X.; Zhou, Y.; Xu, Z. The Construction and Immunogenicity Analyses of Recombinant Pseudorabies Virus with NADC30-Like Porcine Reproductive and Respiratory Syndrome Virus-Like Particles Co-expression. Front. Microbiol. 2022, 13, 846079. [Google Scholar] [CrossRef] [PubMed]
- Cruz, J.L.; Zuniga, S.; Becares, M.; Sola, I.; Ceriani, J.E.; Juanola, S.; Plana, J.; Enjuanes, L. Vectored vaccines to protect against PRRSV. Virus Res. 2010, 154, 150–160. [Google Scholar] [CrossRef]
- Deng, Z.T. Establishment of Indirect ELISA Method for Detection of PRRSV GP5 Protein and Development of Monoclonal Antibody. Ph.D. Thesis, Yangzhou University, Yangzhou, China, 2020. [Google Scholar]
- Li, Z.J.; Ti, J.F.; Li, F.; Wang, Z.Y.; Diao, Y.R. Expression of GP5 protein of porcine reproductive and respiratory syndrome virus and establishment of indirect ELISA for antibody detection. Chin. J. Prev. Vet. Med. 2018, 40, 24–28. [Google Scholar]
- Hou, H.F.; Liu, X.W.; Bai, J.; Song, Z.B.; Jiang, P. Establishment and application of indirect ELISA for detection of GP5 protein antibody of porcine reproductive and respiratory syndrome virus. Chin. Vet. Sci. 2018, 48, 1086–1092. [Google Scholar] [CrossRef]
- Wang, Y. Prokaryotic Expression of GP5 Protein of Porcine Reproductive and Respiratory Syndrome Virus and Establishment of Indirect ELISA Antibody Detection Method. Ph.D. Thesis, Nanjing Agricultural University, Nanjing, China, 2019. [Google Scholar]
- Li, L.B.; Li, X.X.; Wang, R.N.; Zhou, Z.; Liu, Y.L.; Zhao, B.L.; Sun, H.; Feng, B.; Chen, L.; Wang, C.B.; et al. Fusion expression and purification of neutralizing epitope of PRRSV GP5 and M antigen and establishment of indirect ELISA detection method. China Anim. Health Insp. 2022, 39, 71–79. [Google Scholar]
- Yang, C.H.; Zhu, J.X.; Zhong, Y.; Sun, S.Y.; Shen, Y.; Chen, Q.F.; Wang, C.Q. Establishment and application of a new PCR-DHPLC detection technique for porcine reproductive and respiratory syndrome virus. Chin. J. Vet. Sci. 2012, 32, 167–171. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).