A Comparative Study on the Effect of Euthanasia Methods and Sample Storage Conditions on RNA Yield and Quality in Porcine Tissues
Abstract
Simple Summary
Abstract
1. Introduction
2. Material and Methods
2.1. Experimental Set Up
2.2. Euthanasia Method
2.3. Sampling and Storage
2.4. RNA Extraction
2.5. Statistical Analysis
3. Results
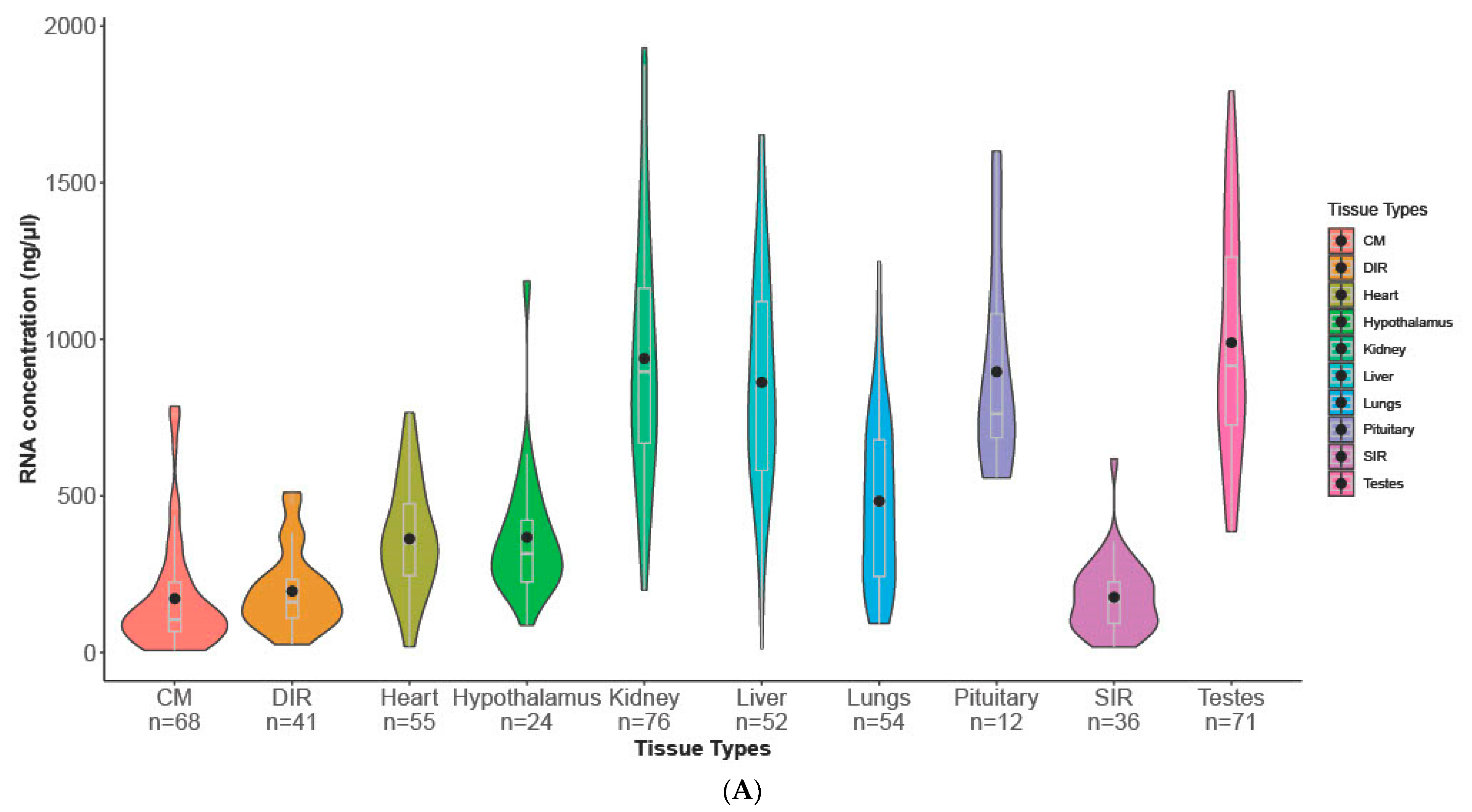
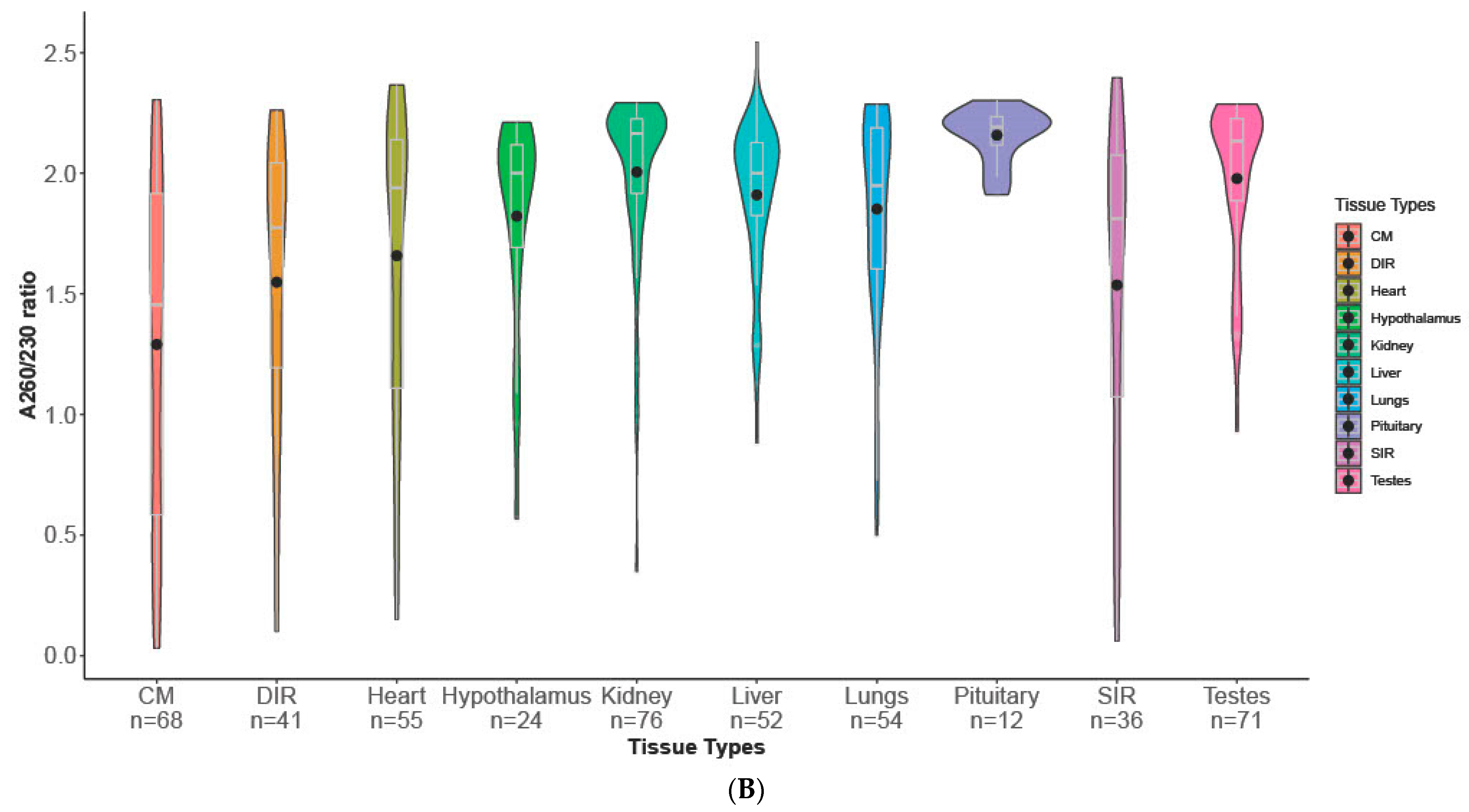
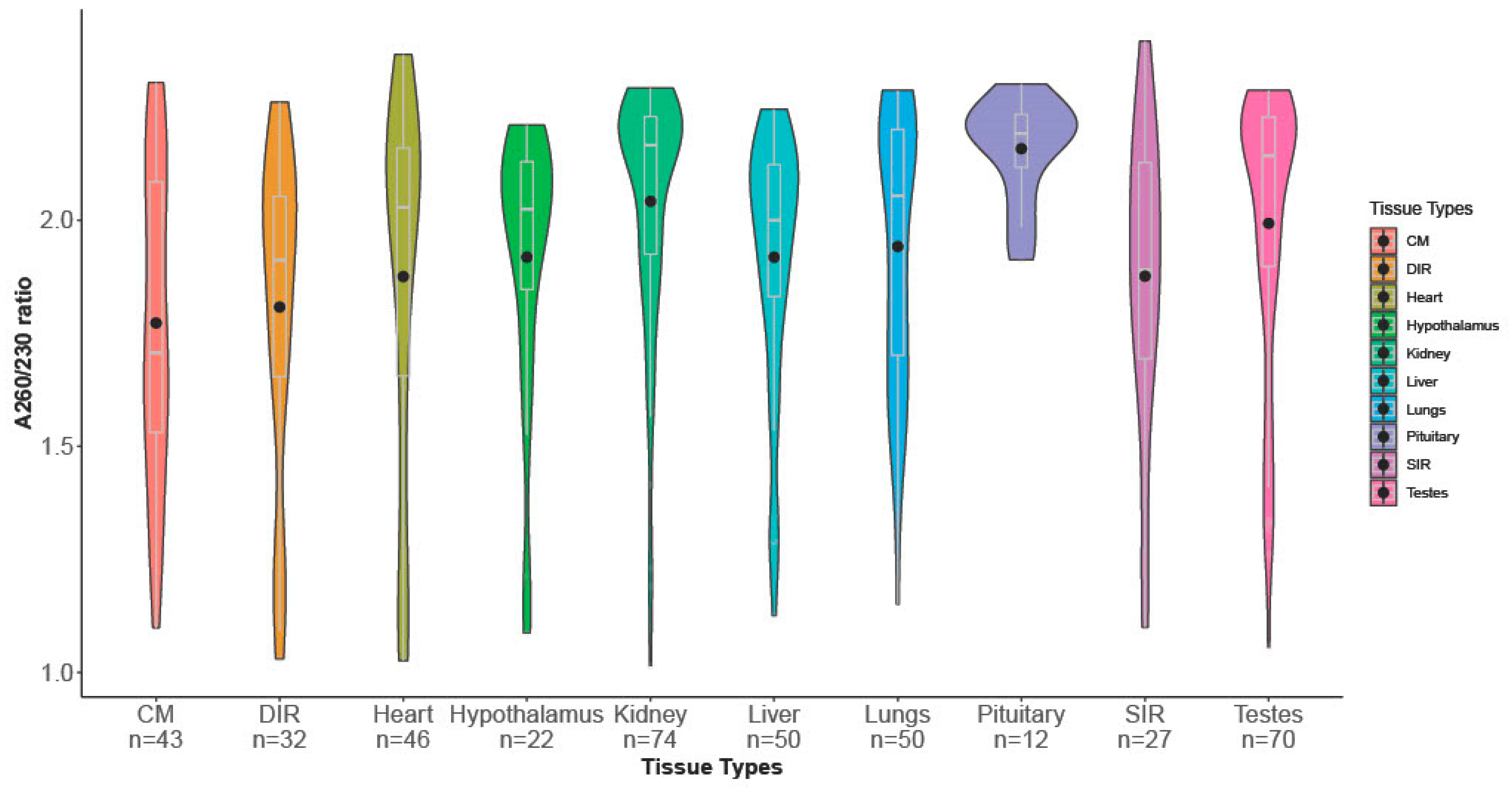
3.1. Descriptive Statistics
3.2. Data Filtering Based on Thresholds Set for RNA Measurements
3.3. Linear Model to Determine Factors Influencing Dependent Variables
3.3.1. RNA Concentration
3.3.2. RNA Quality—A260/230 Ratio
3.3.3. RNA Quality—A260/280 Ratio
3.3.4. RNA Integrity Number (RIN)
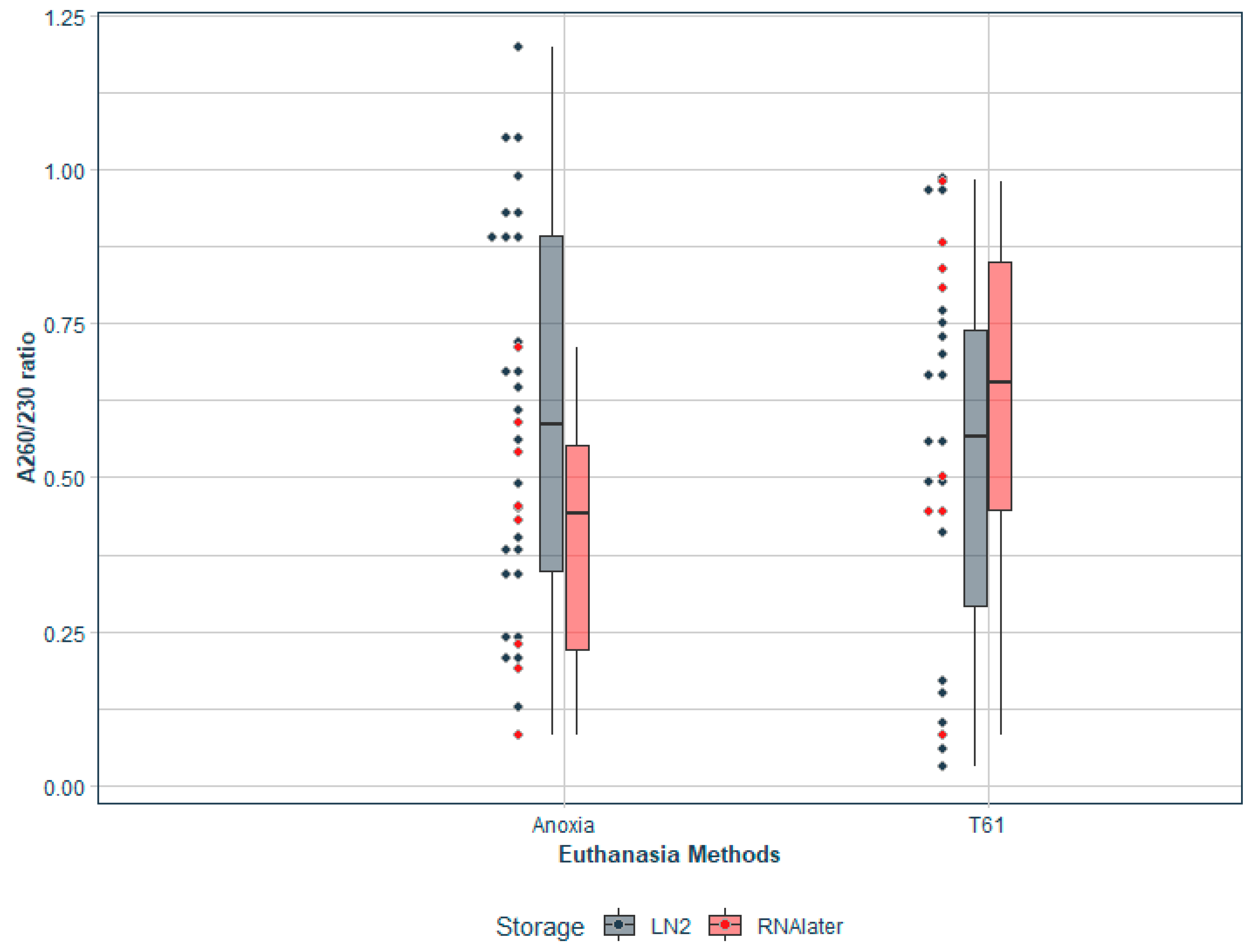
3.4. Test of Equivalence
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Close, B.; Croft, B.; Lane, B.; Knoll, B. Recommendations for Euthanasia of Experimental Animals: Part 1. Lab. Anim. 1996, 30, 293–316. [Google Scholar] [CrossRef] [PubMed]
- AVMA. AVMA Guidelines for the Euthanasia of Animals: 2020 Edition; AVMA: Denver, CO, USA, 2020; ISBN 9781882691548. Available online: www.avma.org/KB/Policies/Documents/euthanasia.pdf (accessed on 11 May 2022).
- Council Regulation (EC) No. 1099/2009 on the Protection of Animals at the Time of Killing. Available online: https://www.ecolex.org/details/legislation/council-regulation-ec-no-10992009-on-the-protection-of-animals-at-the-time-of-killing-lex-faoc090989/ (accessed on 27 March 2022).
- Close, B.; Banister, K.; Baumans, V.; Bernoth, E.M.; Bromage, N.; Bunyan, J.; Erhardt, W.; Flecknell, P.; Gregory, N.; Hackbarth, H.; et al. Recommendations for Euthanasia of Experimental Animals: Part 2. DGXT of the European Commission. Lab. Anim. 1997, 31, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Sindhøj, E.; Lindahl, C.; Bark, L. Review: Potential alternatives to high-concentration carbon dioxide stunning of pigs at slaughter. Animal 2021, 15, 100164. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, C.; Sindhøj, E.; Hellgren, R.B.; Berg, C.; Wallenbeck, A. Responses of Pigs to Stunning with Nitrogen Filled High-Expansion Foam. Animals 2020, 10, 2210. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.S.; Song, D.-H.; Lee, J.-A.; Hoa, V.-B.; Hwang, I.; Kim, H.-W.; Kang, S.-M.; Cho, S.-H.; Seol, K.-H. Effects of High Concentration Nitrogen Gas Stunning of Pigs on the Quality Traits of Meat and Small Intestine. Animals 2022, 12, 2249. [Google Scholar] [CrossRef] [PubMed]
- Nouvel, A.; Laget, J.; Duranton, F.; Leroy, J.; Desmetz, C.; Servais, M.-D.; de Préville, N.; Galtier, F.; Nocca, D.; Builles, N.; et al. Optimization of RNA extraction methods from human metabolic tissue samples of the COMET biobank. Sci. Rep. 2021, 11, 20975. [Google Scholar] [CrossRef]
- Kap, M.; Sieuwerts, A.M.; Kubista, M.; Oomen, M.; Arshad, S.; Riegman, P. The Influence of Tissue Procurement Procedures on RNA Integrity, Gene Expression, and Morphology in Porcine and Human Liver Tissue. Biopreserv. Biobank. 2015, 13, 200–206. [Google Scholar] [CrossRef]
- Wong, P.B.; Wiley, E.O.; Johnson, W.E.; Ryder, O.A.; O’Brien, S.J.; Haussler, D.; Koepfli, K.P.; Houck, M.L.; Perelman, P.; Mastromonaco, G.; et al. Tissue sampling methods and standards for vertebrate genomics. Gigascience 2012, 1, 2047–2217X. [Google Scholar] [CrossRef]
- Nagy, Z.T. A hands-on overview of tissue preservation methods for molecular genetic analyses. Org. Divers. Evol. 2010, 10, 91–105. [Google Scholar] [CrossRef]
- Fleige, S.; Pfaffl, M.W. RNA integrity and the effect on the real-time qRT-PCR performance. Mol. Asp. Med. 2006, 27, 126–139. [Google Scholar] [CrossRef]
- Imbeaud, S. Towards standardization of RNA quality assessment using user-independent classifiers of microcapillary electrophoresis traces. Nucleic Acids Res. 2005, 33, e56. [Google Scholar] [CrossRef]
- Auer, H.; Mobley, J.; Ayers, L.; Bowen, J.; Chuaqui, R.; Johnson, L.; Livolsi, V.; Lubensky, I.; McGarvey, D.; Monovich, L.; et al. The effects of frozen tissue storage conditions on the integrity of RNA and protein. Biotech. Histochem. 2014, 89, 518–528. [Google Scholar] [CrossRef]
- Van Eijsden, R.G.E.; Stassen, C.; Daenen, L.; Van Mulders, S.E.; Bapat, P.M.; Siewers, V.; Goossens, K.V.Y.; Nielsen, J.; Delvaux, F.R.; Van Hummelen, P.; et al. A universal fixation method based on quaternary ammonium salts (RNAlater) for omics-technologies: Saccharomyces cerevisiae as a case study. Biotechnol. Lett. 2013, 35, 891–900. [Google Scholar] [CrossRef]
- Williams, M.A. Article Commentary: Stabilizing the Code—Methods to Preserve RNA Prove Their Worth. Biomark. Insights 2010, 5, BMI.S6094. [Google Scholar] [CrossRef]
- Qiagen Application Note: Effects of Low A260/A230 Ratios in RNA Preparations on Downstream Applications. 2018. Available online: https://www.qiagen.com/de/resources/download.aspx?id=11226191-0a82-4a9b-ba4a-99800b6f8595&lang=en (accessed on 14 October 2022).
- Thermo Scientific Technical Bulletin NanoDrop Spectrophotometers, 260/280 and 260/230 Ratios. Available online: https://tools.thermofisher.com/content/sfs/manuals/ts-nanodrop-nucleicacid-olv-r2.pdf (accessed on 26 April 2022).
- Schroeder, A.; Mueller, O.; Stocker, S.; Salowsky, R.; Leiber, M.; Gassmann, M.; Lightfoot, S.; Menzel, W.; Granzow, M.; Ragg, T. The RIN: An RNA Integrity Number for Assigning Integrity Values to RNA Measurements. BMC Mol. Biol. 2006, 7, 3. [Google Scholar] [CrossRef]
- A Microfluidic System for High-Speed Reproducible DNA Sizing and Quantitation. Available online: https://analyticalsciencejournals.onlinelibrary.wiley.com/doi/epdf/10.1002/%28SICI%291522-2683%2820000101%2921%3A1%3C128%3A%3AAID-ELPS128%3E3.0.CO%3B2-M (accessed on 13 December 2022).
- Qiagen All Insights Start with the Sample Your Comprehensive Guide for Isolating Top-Quality RNA Sample to Insight. Available online: https://www.qiagen.com/us/resources/resourcedetail?id=19988eb0-6df5-4d26-8435-33efe98197da&lang=en (accessed on 12 September 2022).
- Sheng, Q.; Vickers, K.; Zhao, S.; Wang, J.; Samuels, D.C.; Koues, O.; Shyr, Y.; Guo, Y. Multi-perspective quality control of Illumina RNA sequencing data analysis. Briefings Funct. Genom. 2016, 16, 194–204. [Google Scholar] [CrossRef]
- Weber, D.G.; Casjens, S.; Rozynek, P.; Lehnert, M.; Zilch-Schöneweis, S.; Bryk, O.; Taeger, D.; Gomolka, M.; Kreuzer, M.; Otten, H.; et al. Assessment of mRNA and microRNA Stabilization in Peripheral Human Blood for Multicenter Studies and Biobanks. Biomark. Insights 2010, 5, BMI.S5522. [Google Scholar] [CrossRef]
- Hatzis, C.; Sun, H.; Yao, H.; Hubbard, R.E.; Meric-Bernstam, F.; Babiera, G.V.; Wu, Y.; Pusztai, L.; Symmans, W.F. Effects of Tissue Handling on RNA Integrity and Microarray Measurements from Resected Breast Cancers. Gynecol. Oncol. 2011, 103, 1871–1883. [Google Scholar] [CrossRef]
- Mutter, G.L.; Zahrieh, D.; Liu, C.; Neuberg, D.; Finkelstein, D.; Baker, H.E.; A Warrington, J. Comparison of frozen and RNALater solid tissue storage methods for use in RNA expression microarrays. BMC Genom. 2004, 5, 88. [Google Scholar] [CrossRef]
- Ko, M.J.; Mulia, G.E.; Van Rijn, R.M. Commonly Used Anesthesia/Euthanasia Methods for Brain Collection Differentially Impact MAPK Activity in Male and Female C57BL/6 Mice. Front. Cell. Neurosci. 2019, 13, 96. [Google Scholar] [CrossRef]
- Overmyer, K.A.; Thonusin, C.; Qi, N.R.; Burant, C.F.; Evans, C.R. Impact of Anesthesia and Euthanasia on Metabolomics of Mammalian Tissues: Studies in a C57BL/6J Mouse Model. PLoS ONE 2015, 10, e0117232. [Google Scholar] [CrossRef] [PubMed]
- Barocio, L.D. Review of Literature on Use of T-61 as an Euthanasic Agent. Int. J. Study Anim. Probl. 1983, 4, 336–342. [Google Scholar]
- About Us—Heft International. Available online: https://heftinternational.com/about-heft/ (accessed on 12 January 2023).
- Tukey, J.W. Comparing Individual Means in the Analysis of Variance. Biometrics 1949, 5, 99–114. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2018. [Google Scholar]
- The Jamovi Project [Computer Software]. 2021. Available online: http://www.jamovi.org (accessed on 1 January 2020).
- Dixon, P.M.; Saint-Maurice, P.F.; Kim, Y.; Hibbing, P.; Bai, Y.; Welk, G.J. A Primer on the Use of Equivalence Testing for Evaluating Measurement Agreement. Med. Sci. Sports Exerc. 2018, 50, 837–845. [Google Scholar] [CrossRef] [PubMed]
- Matos, B.; Publicover, S.J.; Castro, L.F.C.; Esteves, P.J.; Fardilha, M. Brain and testis: More alike than previously thought? Open Biol. 2021, 11, 200322. [Google Scholar] [CrossRef]
- Wang, Z.; Ying, Z.; Bosy-Westphal, A.; Zhang, J.; Schautz, B.; Later, W.; Heymsfield, S.B.; Müller, M.J. Specific metabolic rates of major organs and tissues across adulthood: Evaluation by mechanistic model of resting energy expenditure. Am. J. Clin. Nutr. 2010, 92, 1369–1377. [Google Scholar] [CrossRef]
- Viñuela, A.; Brown, A.; Buil, A.; Tsai, P.-C.; Davies, M.N.; Bell, J.; Dermitzakis, E.; Spector, T.D.; Small, K.S. Age-dependent changes in mean and variance of gene expression across tissues in a twin cohort. Hum. Mol. Genet. 2017, 27, 732–741. [Google Scholar] [CrossRef]
- Oliva, M.; Muñoz-Aguirre, M.; Kim-Hellmuth, S.; Wucher, V.; Gewirtz, A.D.H.; Cotter, D.J.; Parsana, P.; Kasela, S.; Balliu, B.; Viñuela, A.; et al. The impact of sex on gene expression across human tissues. Science 2020, 369, eaba3066. [Google Scholar] [CrossRef]
- Kukurba, K.R.; Montgomery, S.B. RNA Sequencing and Analysis. Cold Spring Harb. Protoc. 2015, 2015, 951–969. [Google Scholar] [CrossRef]
- Kruse, C.P.S.; Basu, P.; Luesse, D.R.; Wyatt, S.E. Transcriptome and proteome responses in RNAlater preserved tissue of Arabidopsis thaliana. PLoS ONE 2017, 12, e0175943. [Google Scholar] [CrossRef]
- Ozkan, H.; Kerman, E. Comparative Evaluation of RNAlater Solution and Snap Frozen Methods for Gene Expression Studies in Different Tissues. Rom. Rev. Lab. Med. 2020, 28, 287–297. [Google Scholar] [CrossRef]


| RNA Concentration (SD) | A260/230 Ratio (SD) | A260/280 Ratio (SD) | RIN (SD) | |
|---|---|---|---|---|
| ANOXIATM | 604.56 (462.61) | 1.77 (0.57) | 2.13 (0.04) | 8.85 (1.00) |
| T-61® | 525.45 (404.30) | 1.75 (0.56) | 2.11 (0.04) | 8.77 (1.32) |
| RNAlaterTM | 611.99 (433.88) | 1.86 (0.47) | 2.12 (0.03) | 9.26 (0.66) |
| LN2 | 525.12 (439.57) | 1.66 (0.63) | 2.12 (0.05) | 7.99 (1.40) |
| RNA Concentration | A260/230 Ratio | A260/280 Ratio | RIN | |
|---|---|---|---|---|
| Final Model | Final Model | Final Model | Final Model | |
| N | 239 | 426 | 426 | 55 |
| Overall mean | 633.3 | 1.93 | 2.116 | 8.79 |
| RMSE | 248.18 | 0.31 | 0.02 | 0.63 |
| pvalues of F-test | ||||
| FACTORS | ||||
| Euthanasia Method | *** | 0.25 | *** | 0.51 |
| Storage | 0.49 | 0.18 | 0.47 | *** |
| Tissue Type | *** | *** | *** | *** |
| Time of Sampling | 0.41 | 0.18 | * | 0.56 |
| Tissue Weight | *** | |||
| LSmeans for treatment levels (std. error in brackets) | ||||
| Euthanasia Method | ||||
| ANOXIATM | 761.7 (39.5) a | 1.95 (0.02) a | 2.12 (0.002) a | 8.85 (0.18) a |
| T-61® | 647.7 (42.9) b | 1.91 (0.02) a | 2.11 (0.002) b | 8.73 (0.26) a |
| Storage Condition | ||||
| RNAlater | 729.4(40.0) a | 1.95 (0.02) a | 2.117 (0.002) a | 9.25 (0.11) a |
| LN2 | 703.7(44.0) a | 1.90 (0.03) a | 2.114 (0.002) a | 7.99 (0.31) b |
| Estimates for Time of Sampling and Tissue Weight (std. error in brackets) | ||||
| Time of Sampling (minutes) | 0.94 (0.72) | −0.001 (0.001) | 0.13 (0.05) | 0.04 (0.07) |
| Tissue Weight (milligram) | 21.02 (3.37) | |||
| t | df | p | |||
|---|---|---|---|---|---|
| A260/230 Ratio | t-test | 1.109 | 424 | 0.268 | |
| TOST Upper | −4.03 | 424 | <0.001 | ||
| TOST Lower | 6.25 | 424 | <0.001 | ||
| RIN | t-test | 0.385 | 53 | 0.701 | |
| TOST Upper | −1.47 | 53 | 0.074 | ||
| TOST Lower | 2.24 | 53 | 0.015 | ||
| Equivalence Bounds | |||||
| 90% Confidence interval | |||||
| Low | High | Lower | Upper | ||
| A260/230 Ratio | Cohen’s d | −0.5 | 0.5 | ||
| Raw | −0.164 | 0.164 | −0.0172 | 0.0881 | |
| RIN | Cohen’s d | −0.5 | 0.5 | ||
| Raw | −0.583 | 0.583 | −0.4057 | 0.6484 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chakkingal Bhaskaran, B.; Meyermans, R.; Gorssen, W.; Maes, G.E.; Janssens, S.; Buys, N. A Comparative Study on the Effect of Euthanasia Methods and Sample Storage Conditions on RNA Yield and Quality in Porcine Tissues. Animals 2023, 13, 698. https://doi.org/10.3390/ani13040698
Chakkingal Bhaskaran B, Meyermans R, Gorssen W, Maes GE, Janssens S, Buys N. A Comparative Study on the Effect of Euthanasia Methods and Sample Storage Conditions on RNA Yield and Quality in Porcine Tissues. Animals. 2023; 13(4):698. https://doi.org/10.3390/ani13040698
Chicago/Turabian StyleChakkingal Bhaskaran, Bimal, Roel Meyermans, Wim Gorssen, Gregory Erich Maes, Steven Janssens, and Nadine Buys. 2023. "A Comparative Study on the Effect of Euthanasia Methods and Sample Storage Conditions on RNA Yield and Quality in Porcine Tissues" Animals 13, no. 4: 698. https://doi.org/10.3390/ani13040698
APA StyleChakkingal Bhaskaran, B., Meyermans, R., Gorssen, W., Maes, G. E., Janssens, S., & Buys, N. (2023). A Comparative Study on the Effect of Euthanasia Methods and Sample Storage Conditions on RNA Yield and Quality in Porcine Tissues. Animals, 13(4), 698. https://doi.org/10.3390/ani13040698







