Insights into Transcriptomic Differences in Ovaries between Lambs and Adult Sheep after Superovulation Treatment
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Research Ethics
2.2. Experimental Design, Superovulation Techniques, and Sample Collection
2.3. RNA Extraction, Library Construction, and Sequencing
2.4. Sequencing Data Analysis
2.5. Identification of Candidate lncRNAs and mRNAs
2.6. Functional Analysis
2.7. Real-Time qPCR Validation
2.8. Statistical Analysis
3. Result
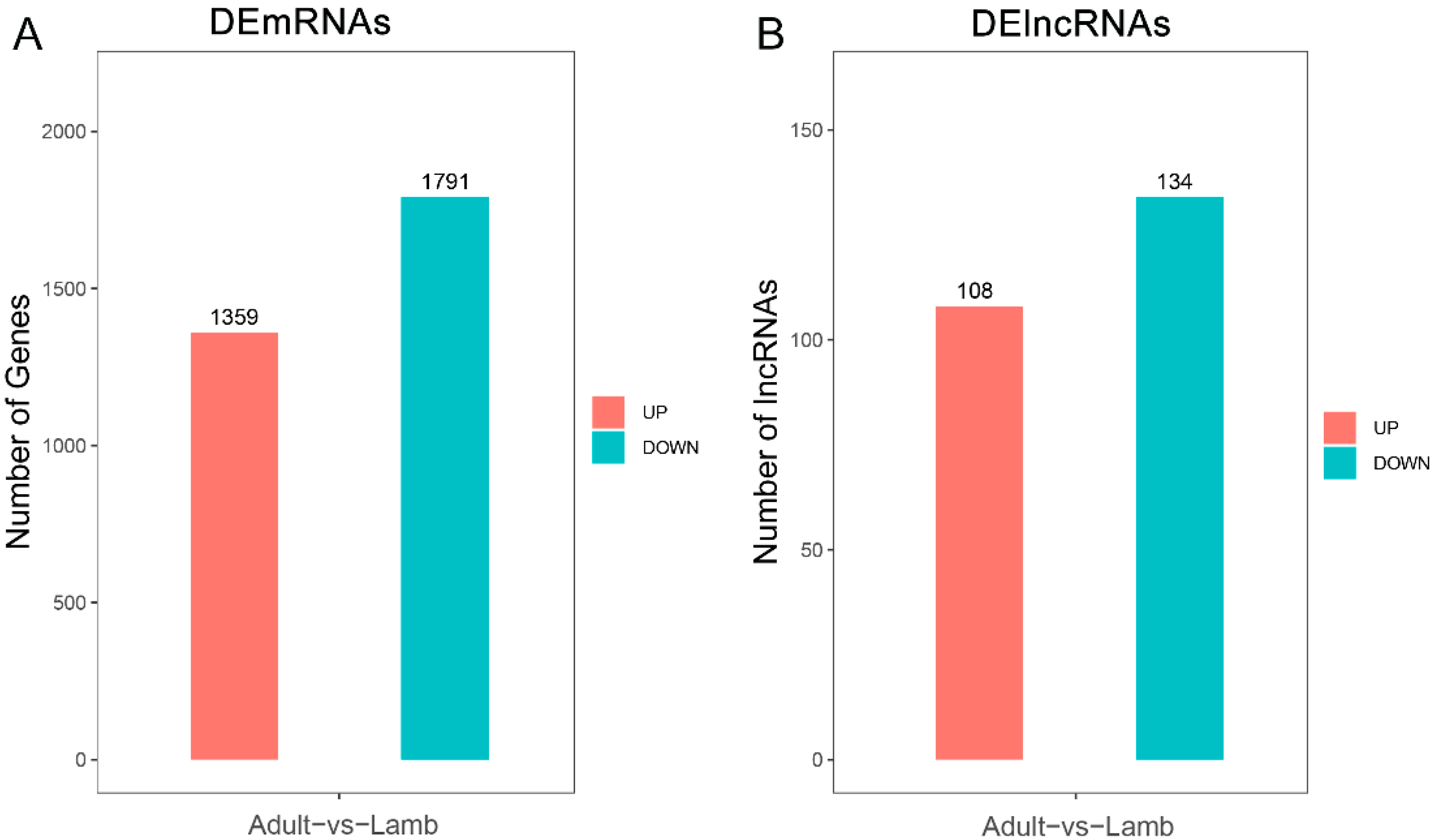
3.1. Sequence Data Summary
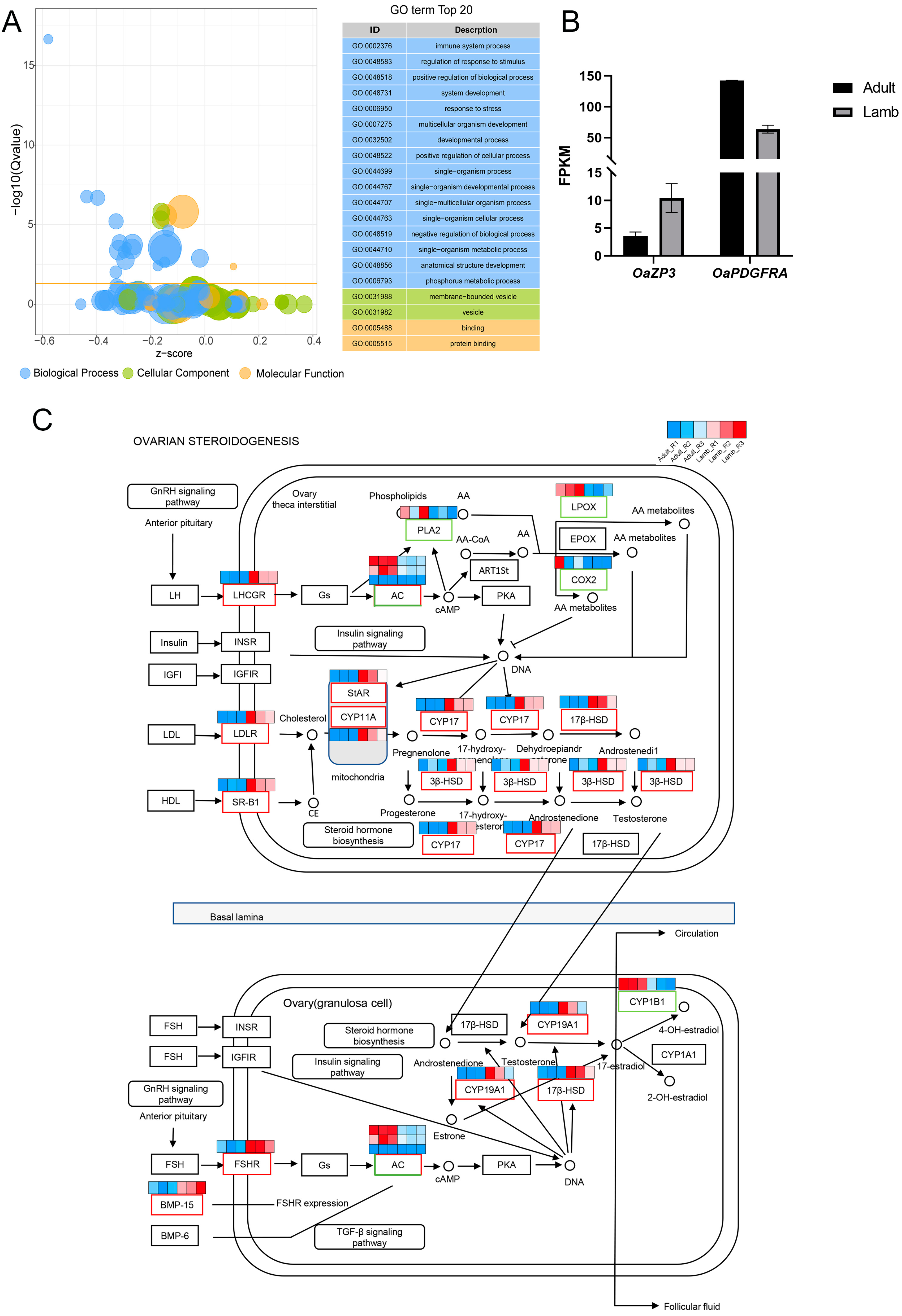
3.2. Functional Analysis
3.3. LncRNA-mRNA Interaction Analysis
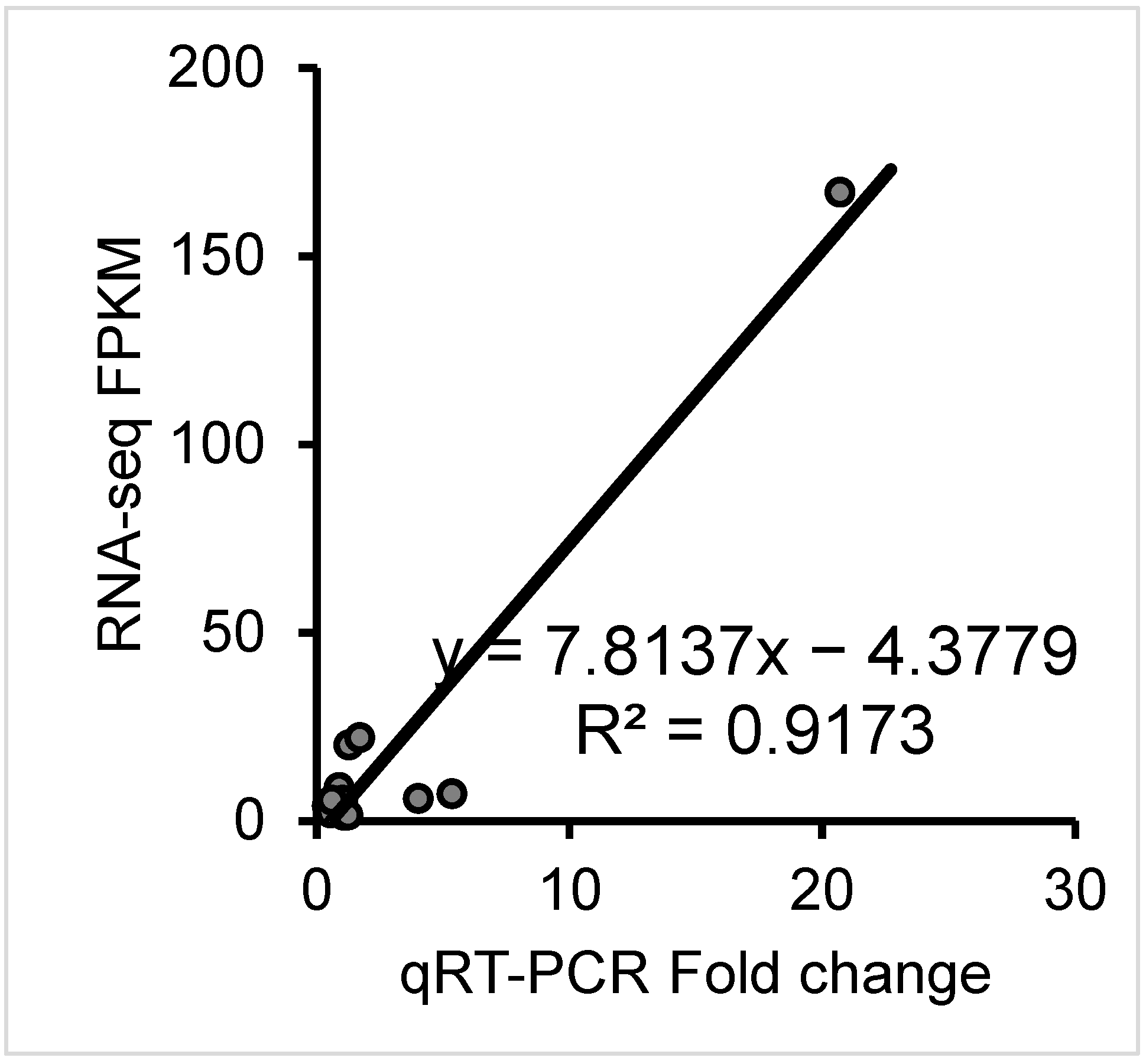
3.4. Quantitative Real-Time PCR (qRT-PCR) Confirmation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Morton, K.M.; Catt, S.L.; Maxwell, W.M.C.; Evans, G. An efficient method of ovarian stimulation and in vitro embryo production from prepubertal lambs. Reprod. Fertil. Dev. 2005, 17, 701–706. [Google Scholar] [CrossRef]
- Daly, J.; Smith, H.; McGrice, H.A.; Kind, K.L.; van Wettere, W.H. Towards Improving the Outcomes of Assisted Reproductive Technologies of Cattle and Sheep, with Particular Focus on Recipient Management. Animals 2020, 10, 293. [Google Scholar] [CrossRef] [PubMed]
- Ptak, G.; Loi, P.; Dattena, M.; Tischner, M.; Cappai, P. Offspring from One-Month-Old Lambs: Studies on the Developmental Capability of Prepubertal Oocytes1. Biol. Reprod. 1999, 61, 1568–1574. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [PubMed]
- Youngquist, R.S.; Threlfall, W.R. Artificial Insemination and Embryo Transfer in Sheep. In Current Therapy in Large Animal Theriogenology, 2nd ed.; Elsevier: Amsterdam, The Netherlands, 2007; pp. 629–641. [Google Scholar]
- Bartlewski, P.M.; Seaton, P.; Szpila, P.; Oliveira, M.E.F.; Murawski, M.; Schwarz, T.; Kridli, R.T.; Zieba, D.A. Comparison of the effects of pretreatment with Veramix sponge (medroxyprogesterone acetate) or CIDR (natural progesterone) in combination with an injection of estradiol-17β on ovarian activity, endocrine profiles, and embryo yields in cyclic ewes superovulated in the multiple-dose Folltropin-V (porcine FSH) regimen. Theriogenology 2015, 84, 1225–1237. [Google Scholar]
- Bartlewski, P.M.; Alexander, B.D.; Rawlings, N.C.; Barrett, D.M.W.; King, W.A. Ovarian responses, hormonal profiles and embryo yields in anoestrous ewes superovulated with Folltropin-V after pretreatment with medroxyprogesterone acetate-releasing vaginal sponges and a single dose of oestradiol-17beta. Reprod. Domest. Anim. 2008, 43, 299–307. [Google Scholar] [CrossRef] [PubMed]
- Gherardi, P.B.; Lindsay, D.R. The effect of season on the ovulatory response of Merino ewes to serum from pregnant mares. Reproduction 1980, 60, 425–429. [Google Scholar] [CrossRef]
- Zhang, L.; Chai, M.; Tian, X.; Wang, F.; Fu, Y.; He, C.; Deng, S.-L.; Lian, Z.; Feng, J.; Tan, D.-X.; et al. Effects of melatonin on superovulation and transgenic embryo transplantation in small-tailed han sheep (Ovis aries). Neuro Endocrinol. Lett. 2013, 34, 294–301. [Google Scholar] [PubMed]
- Amiridis, G.; Cseh, S. Assisted reproductive technologies in the reproductive management of small ruminants. Anim. Reprod. Sci. 2012, 130, 152–161. [Google Scholar] [CrossRef]
- O’Callaghan, D.; Yaakub, H.; Hyttel, P.; Spicer, L.J.; Boland, M.P. Effect of nutrition and superovulation on oocyte morphology, follicular fluid composition and systemic hormone concentrations in ewes. J. Reprod. Fertil. 2000, 118, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Jabbour, H.; Ryan, J.; Evans, G.; Maxwell, W. Effects of season, GnRH administration and lupin supplementation on the ovarian and endocrine responses of merino ewes treated with PMSG and FSH-P to induce superovulation. Reprod. Fertil. Dev. 1991, 3, 699–707. [Google Scholar] [CrossRef]
- Abecia, J.; Lozano, J.; Forcada, F.; Zarazaga, L. Effect of level of dietary energy and protein on embryo survival and progesterone production on day eight of pregnancy in Rasa Aragonesa ewes. Anim. Reprod. Sci. 1997, 48, 209–218. [Google Scholar] [CrossRef]
- Abecia, J.; Forcada, F.; Palacín, I.; Sánchez-Prieto, L.; Sosa, C.; Fernández-Foren, A.; Meikle, A. Undernutrition affects embryo quality of superovulated ewes. Zygote 2015, 23, 116–124. [Google Scholar] [CrossRef] [PubMed]
- Lozano, J.M.; Lonergan, P.; Boland, M.P.; Callaghan, D.O. Influence of nutrition on the effectiveness of superovulation programmes in ewes: Effect on oocyte quality and post-fertilization development. Reproduction 2003, 125, 543–553. [Google Scholar] [CrossRef] [PubMed]
- Veiga-Lopez, A.; Gonzalez-Bulnes, A.; Garcia, R.; Dominguez, V.; Cocero, M.J. The effects of previous ovarian status on ovulation rate and early embryo development in response to superovulatory FSH treatments in sheep. Theriogenology 2005, 63, 1973–1983. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Li, X.; Ma, Q.; Zhang, X.; Cao, Y.; Yao, Y.; You, S.; Wang, D.; Quan, R.; Hou, X.; et al. Genome-wide analysis of circular RNAs in prenatal and postnatal pituitary glands of sheep. Sci. Rep. 2017, 7, 16143. [Google Scholar] [CrossRef]
- Wang, H.; Feng, X.; Muhatai, G.; Wang, L. Expression profile analysis of sheep ovary after superovulation and estrus synchronisation treatment. Veter- Med. Sci. 2022, 8, 1276–1287. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Dong, C.; Yang, J.; Li, Y.; Feng, J.; Wang, B.; Zhang, J.; Guo, X. The Roles of the miRNAome and Transcriptome in the Ovine Ovary Reveal Poor Efficiency in Juvenile Superovulation. Animals 2021, 11, 239. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Hooper, J.E. A survey of software for genome-wide discovery of differential splicing in RNA-Seq data. Hum. Genom. 2014, 8, 3. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, D.T.; Kotaras, P.J.; Earl, C.R. Advances in production of embryos in vitro from juvenile and prepubertal oocytes from the calf and lamb. Reprod. Fertil. Dev. 1997, 9, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Reader, K.L.; Cox, N.R.; Stanton, J.-A.; Juengel, J. Mitochondria and vesicles differ between adult and prepubertal sheep oocytes during IVM. Reprod. Fertil. Dev. 2015, 27, 513–522. [Google Scholar] [CrossRef] [PubMed]
- Gou, K.-M.; Guan, H.; Bai, J.-H.; Cui, X.-H.; Wu, Z.-F.; Yan, F.-X.; An, X.-R. Field evaluation of juvenile in vitro embryo transfer (JIVET) in sheep. Anim. Reprod. Sci. 2009, 112, 316–324. [Google Scholar] [CrossRef]
- Earl, C.R.; Fry, R.C.; Maclellan, L.J.; Kelly, J.; MandArmstrong, D.T. In vitro fertilisation and developmental potential of prepu bertal calf oocytes. In Gametes: Development and Function; Giannaroli, I., Ed.; Serono Symposia: Rome, Italy, 1998; pp. 115–137. [Google Scholar]
- Zhao, Y.; Pan, S.; Li, Y.; Wu, X. Exosomal miR-143-3p derived from follicular fluid promotes granulosa cell apoptosis by targeting BMPR1A in polycystic ovary syndrome. Sci. Rep. 2022, 12, 1–12. [Google Scholar] [CrossRef]
- Martins, I.J.; Hone, E.; Chi, C.; Seydel, U.; Martins, R.N.; Redgrave, T.G. Relative roles of LDLr and LRP in the metabolism of chylomicron remnants in genetically manipulated mice. J. Lipid Res. 2000, 41, 205–213. [Google Scholar] [CrossRef]
- Yoon, S.-J.; Kim, K.-H.; Chung, H.-M.; Choi, D.-H.; Lee, W.-S.; Cha, K.-Y.; Lee, K.-A. Gene expression profiling of early follicular development in primordial, primary, and secondary follicles. Fertil. Steril. 2006, 85, 193–203. [Google Scholar] [CrossRef] [PubMed]
- Larqué, E.; Ruiz-Palacios, M.; Koletzko, B. Placental regulation of fetal nutrient supply. Curr. Opin. Clin. Nutr. Metab. Care 2013, 16, 292–297. [Google Scholar] [CrossRef]
- Azhar, S.; Medicherla, S.; Shen, W.-J.; Fujioka, Y.; Fong, L.G.; Reaven, E.; Cooper, A.D. LDL and cAMP cooperate to regulate the functional expression of the LRP in rat ovarian granulosa cells. J. Lipid Res. 2006, 47, 2538–2550. [Google Scholar] [CrossRef]
- Cheung, J.; Lokman, N.A.; Abraham, R.D.; MacPherson, A.M.; Lee, E.; Grutzner, F.; Ghinea, N.; Oehler, M.K.; Ricciardelli, C. Reduced Gonadotrophin Receptor Expression Is Associated with a More Aggressive Ovarian Cancer Phenotype. Int. J. Mol. Sci. 2020, 22, 71. [Google Scholar] [CrossRef]
- Zhang, Z.; Lau, S.-W.; Zhang, L.; Ge, W. Disruption of Zebrafish Follicle-Stimulating Hormone Receptor (fshr) But Not Luteinizing Hormone Receptor (lhcgr) Gene by TALEN Leads to Failed Follicle Activation in Females Followed by Sexual Reversal to Males. Endocrinology 2015, 156, 3747–3762. [Google Scholar] [CrossRef]
- El-Hayek, S.; Demeestere, I.; Clarke, H.J. Follicle-stimulating hormone regulates expression and activity of epidermal growth factor receptor in the murine ovarian follicle. Proc. Natl. Acad. Sci. USA 2014, 111, 16778–16783. [Google Scholar] [CrossRef] [PubMed]
- Richards, J.S.; Pangas, S.A. The ovary: Basic biology and clinical implications. J. Clin. Investig. 2010, 120, 963–972. [Google Scholar] [CrossRef] [PubMed]
- Kumar, T.R.; Wang, Y.; Lu, N.; Matzuk, M.M. Follicle stimulating hormone is required for ovarian follicle maturation but not male fertility. Nat. Genet. 1997, 15, 201–204. [Google Scholar] [CrossRef]
- Balla, A.; Danilovich, N.; Yang, Y.; Sairam, M.R. Dynamics of Ovarian Development in the FORKO Immature Mouse: Structural and Functional Implications for Ovarian Reserve. Biol. Reprod. 2003, 69, 1281–1293. [Google Scholar] [CrossRef]
- Hardy, K.; Fenwick, M.; Mora, J.; Laird, M.; Thomson, K.; Franks, S. Onset and Heterogeneity of Responsiveness to FSH in Mouse Preantral Follicles in Culture. Endocrinology 2016, 158, 134–147. [Google Scholar] [CrossRef]
- McGee, E.A.; Perlas, E.; LaPolt, P.S.; Tsafriri, A.; Hsueh, A.J. Follicle-Stimulating Hormone Enhances the Development of Preantral Follicles in Juvenile Rats1. Biol. Reprod. 1997, 57, 990–998. [Google Scholar] [CrossRef] [PubMed]
- Birt, J.A.; Nabli, H.; Stilley, J.A.; Windham, E.A.; Frazier, S.R.; Sharpe-Timms, K.L. Elevated Peritoneal Fluid TNF-α Incites Ovarian Early Growth Response Factor 1 Expression and Downstream Protease Mediators: A Correlation with Ovulatory Dysfunction in Endometriosis. Reprod. Sci. 2013, 20, 514–523. [Google Scholar] [CrossRef]
- Torres-Rovira, L.; Gonzalez-Bulnes, A.; Succu, S.; Spezzigu, A.; Manca, M.E.; Leoni, G.G.; Sanna, M.; Pirino, S.; Gallus, M.; Naitana, S.; et al. Predictive value of antral follicle count and anti-Müllerian hormone for follicle and oocyte developmental competence during the early prepubertal period in a sheep model. Reprod. Fertil. Dev. 2014, 26, 1094–1106. [Google Scholar] [CrossRef]
- Sleer, L.S.; Taylor, C.C. Cell-Type Localization of Platelet-Derived Growth Factors and Receptors in the Postnatal Rat Ovary and Follicle1. Biol. Reprod. 2007, 76, 379–390. [Google Scholar] [CrossRef]
- Chen, J.; Su, Y.; Pi, S.; Hu, B.; Mao, L. The Dual Role of Low-Density Lipoprotein Receptor-Related Protein 1 in Atherosclerosis. Front. Cardiovasc. Med. 2021, 8, 682389. [Google Scholar] [CrossRef] [PubMed]
| Sample | Raw Base (bp) | Clean Base (%) | Total Mapped (%) | Unique Mapped (%) | Q30 (%) | Q20 (%) | GC (%) |
|---|---|---|---|---|---|---|---|
| Lamb_1 | 96499362 | 99.80 | 96.70 | 93.46 | 92.33 | 97.25 | 44.60 |
| Lamb_2 | 67309706 | 99.81 | 97.28 | 94.02 | 93.66 | 97.78 | 44.36 |
| Lamb_3 | 76906026 | 99.82 | 97.12 | 93.99 | 93.47 | 97.71 | 44.24 |
| Adult_1 | 73783056 | 99.82 | 97.12 | 93.99 | 93.65 | 97.78 | 43.76 |
| Adult_2 | 78182148 | 99.85 | 97.28 | 94.11 | 93.95 | 97.93 | 44.82 |
| Adult_3 | 76149668 | 99.83 | 97.13 | 93.73 | 93.83 | 97.85 | 44.60 |
| Antisense | Cis | |||||
|---|---|---|---|---|---|---|
| lncRNA | mRNA | Pair | lncRNA | mRNA | Pair | |
| All | 539 | 402 | 561 | 1973 | 1840 | 2625 |
| Differently expressed | 5 | 4 | 5 | 30 | 27 | 30 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Q.; Guo, X.; Yao, D.; Wang, B.; Li, Y.; Zhang, J.; Zhang, X. Insights into Transcriptomic Differences in Ovaries between Lambs and Adult Sheep after Superovulation Treatment. Animals 2023, 13, 665. https://doi.org/10.3390/ani13040665
Wang Q, Guo X, Yao D, Wang B, Li Y, Zhang J, Zhang X. Insights into Transcriptomic Differences in Ovaries between Lambs and Adult Sheep after Superovulation Treatment. Animals. 2023; 13(4):665. https://doi.org/10.3390/ani13040665
Chicago/Turabian StyleWang, Qingwei, Xiaofei Guo, Dawei Yao, Biao Wang, Yupeng Li, Jinlong Zhang, and Xiaosheng Zhang. 2023. "Insights into Transcriptomic Differences in Ovaries between Lambs and Adult Sheep after Superovulation Treatment" Animals 13, no. 4: 665. https://doi.org/10.3390/ani13040665
APA StyleWang, Q., Guo, X., Yao, D., Wang, B., Li, Y., Zhang, J., & Zhang, X. (2023). Insights into Transcriptomic Differences in Ovaries between Lambs and Adult Sheep after Superovulation Treatment. Animals, 13(4), 665. https://doi.org/10.3390/ani13040665






