Comprehensive Comparative Analysis Sheds Light on the Patterns of Microsatellite Distribution across Birds Based on the Chromosome-Level Genomes
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sources of Genomic Dataset
2.2. SSRs Identification and Characterization
2.3. Variation Analysis of P-SSRs
2.4. Functional Analysis
3. Result
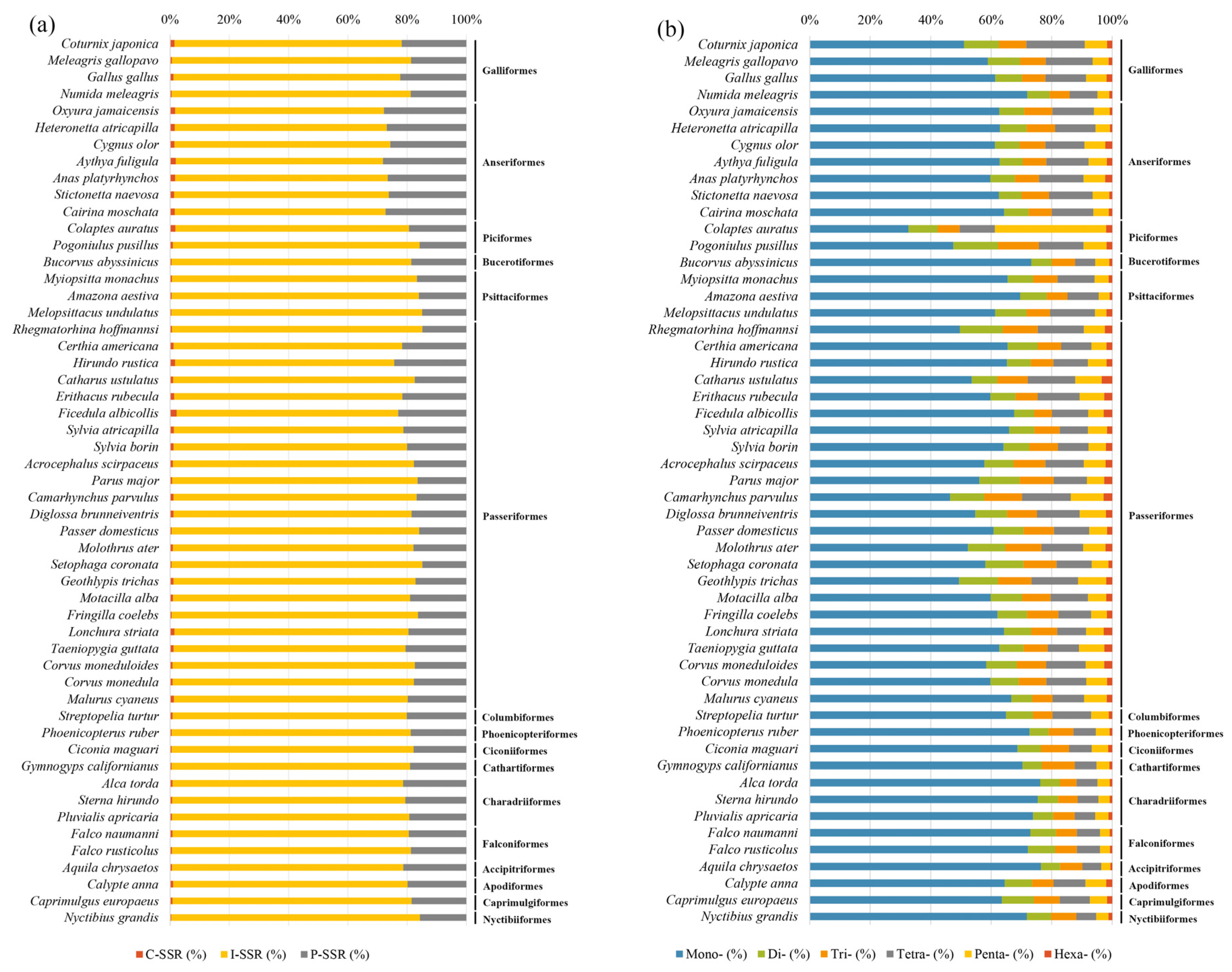
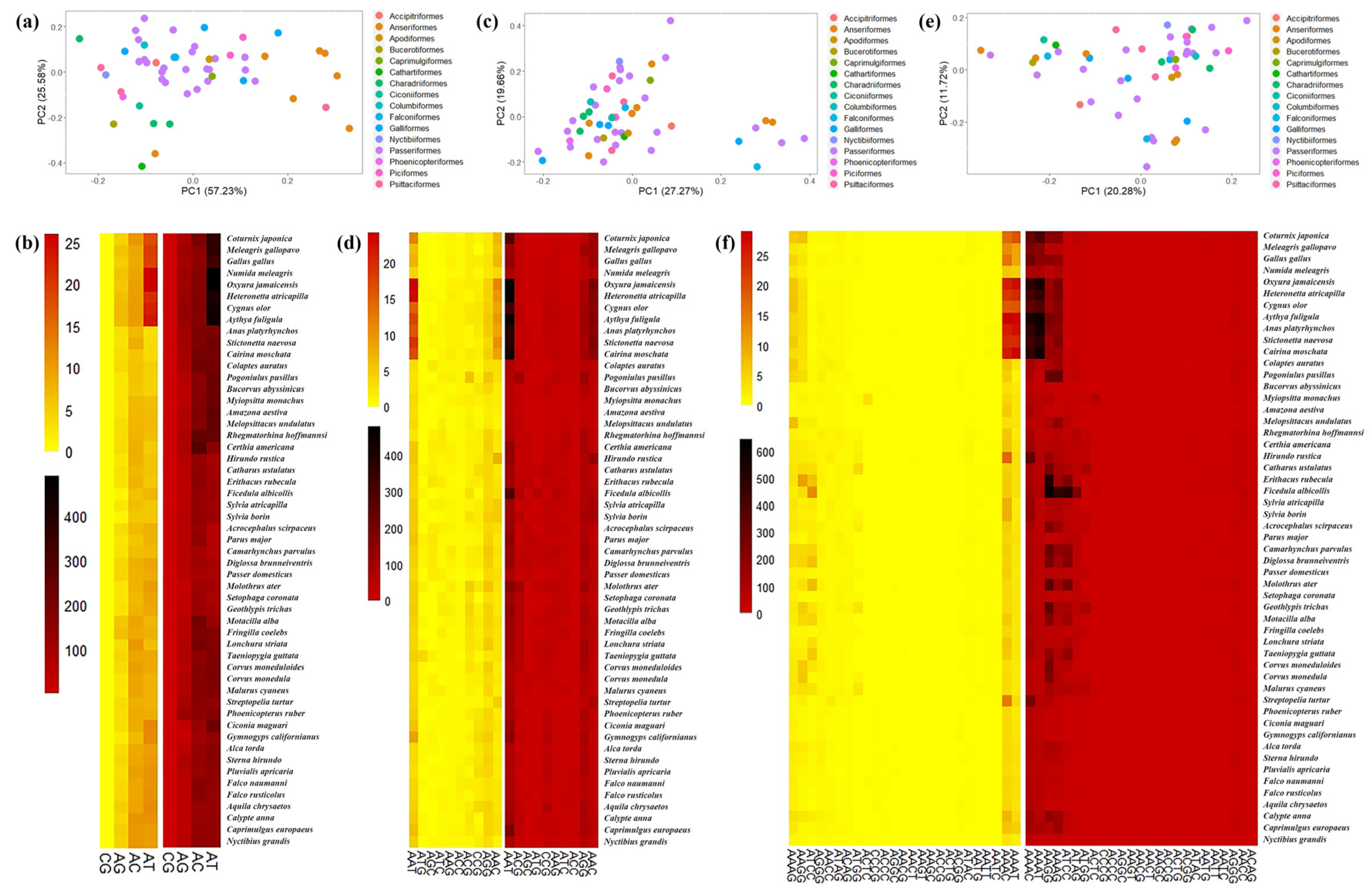
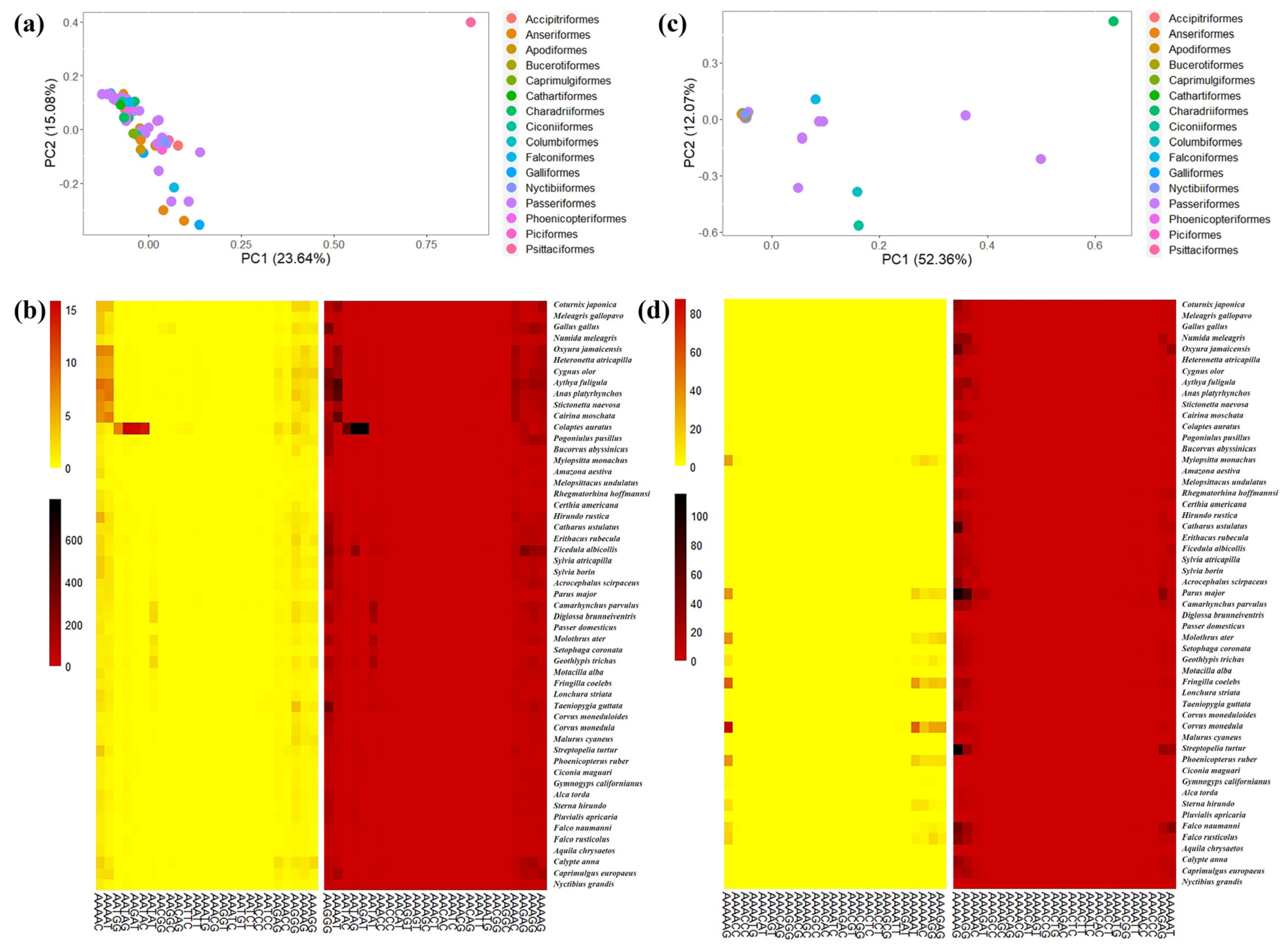
3.1. Characteristics of Avian SSRs
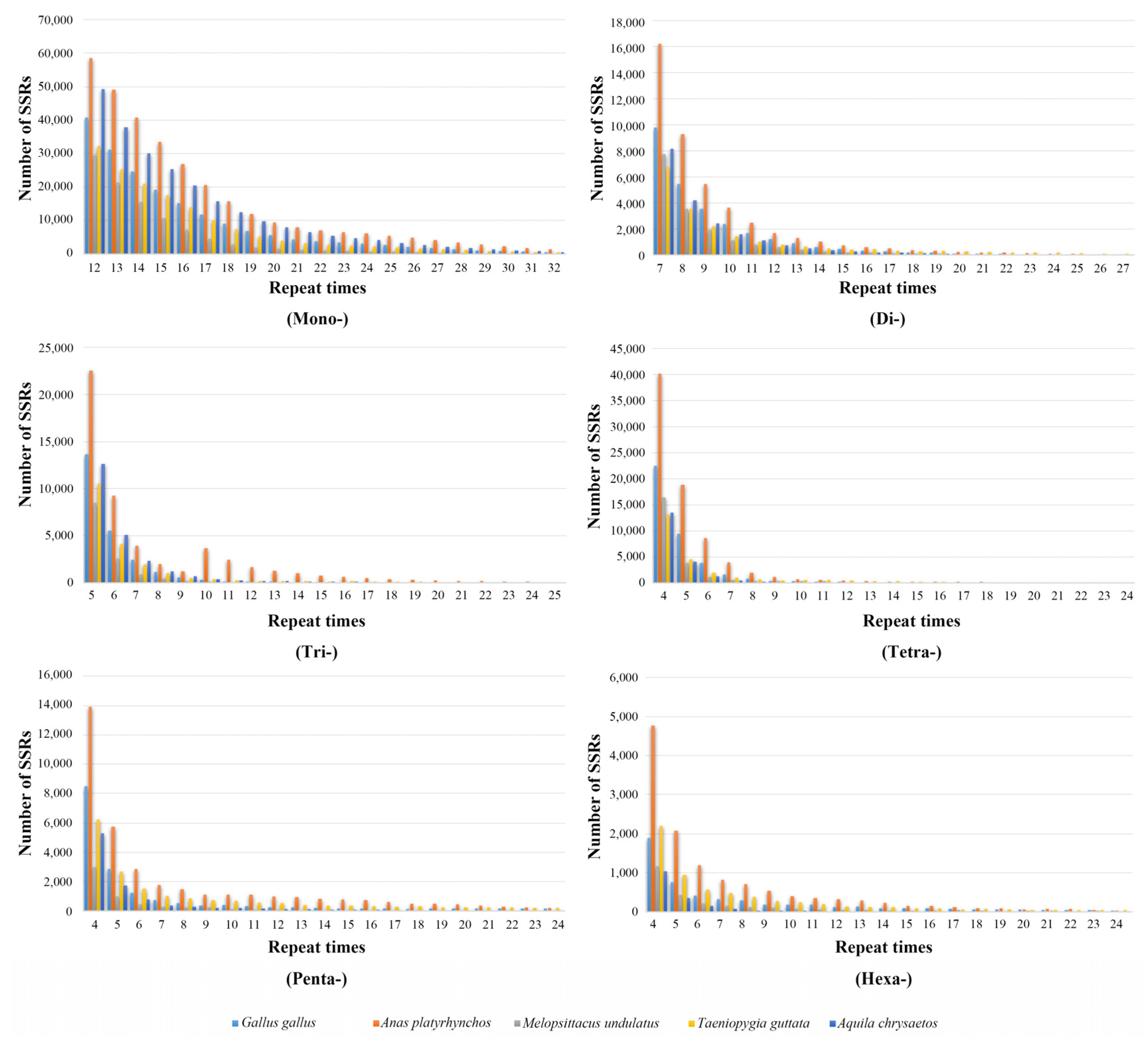
3.2. The Variability of Repeat Copy Number
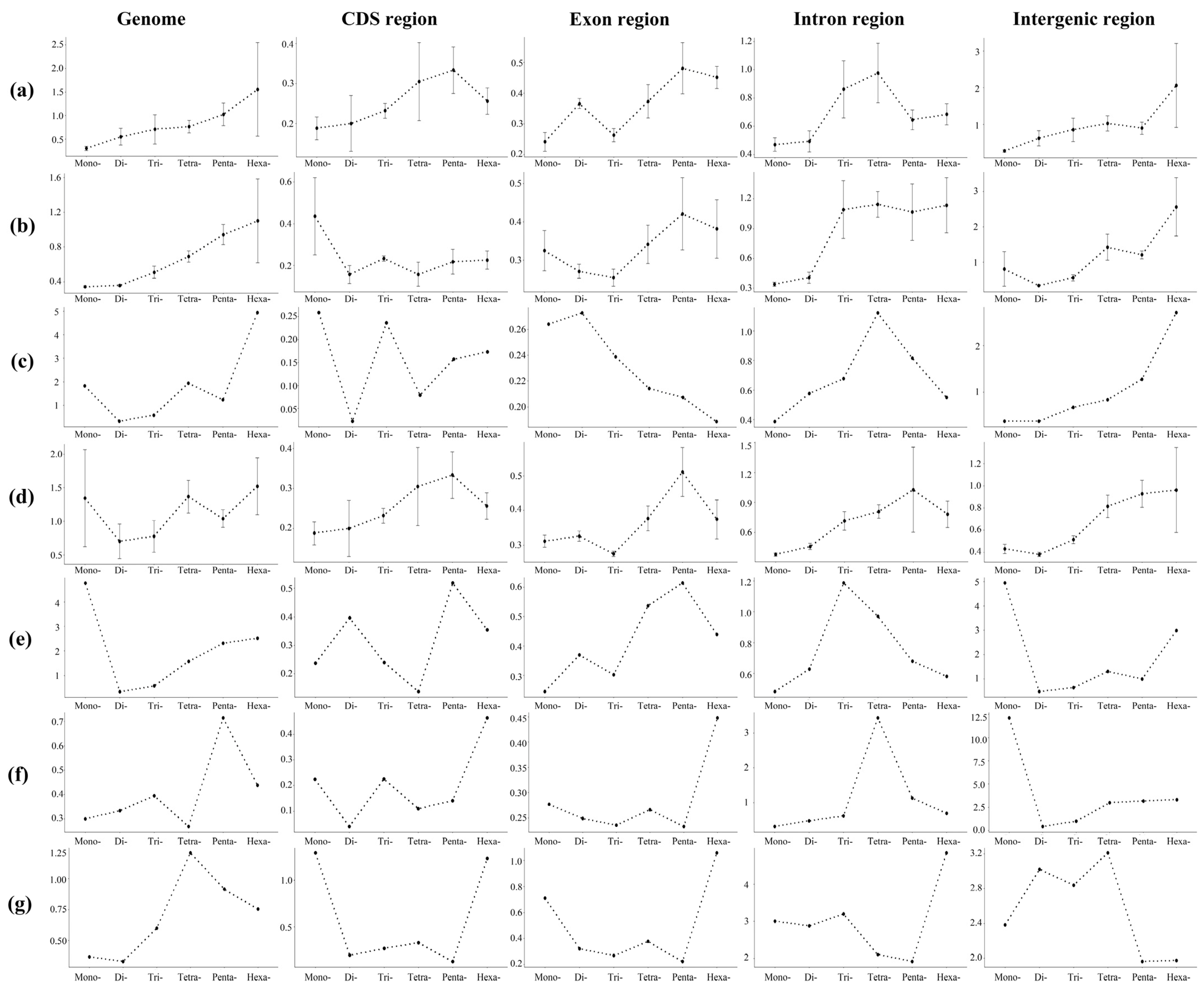
3.3. GC Content, and Functional Analysis
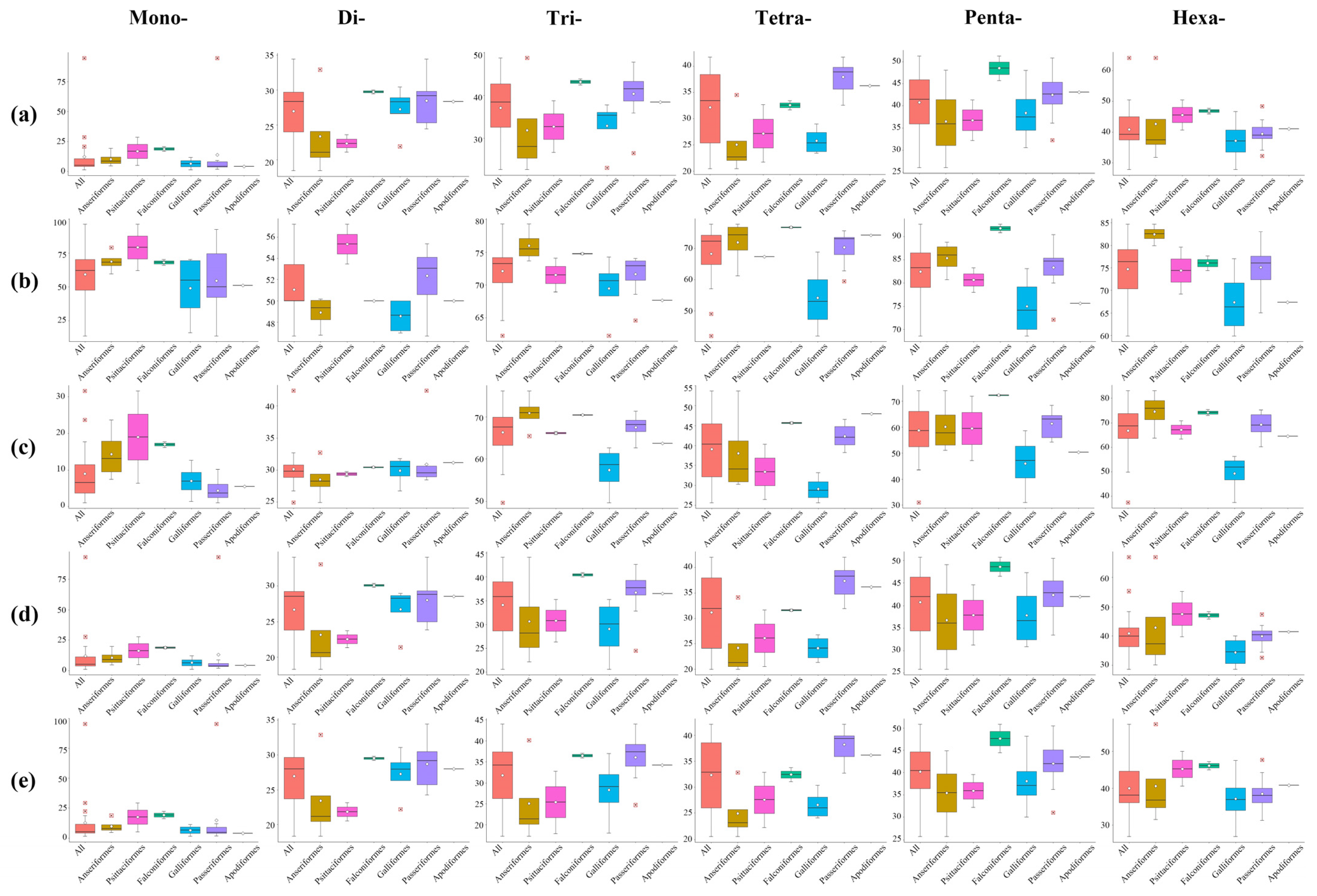
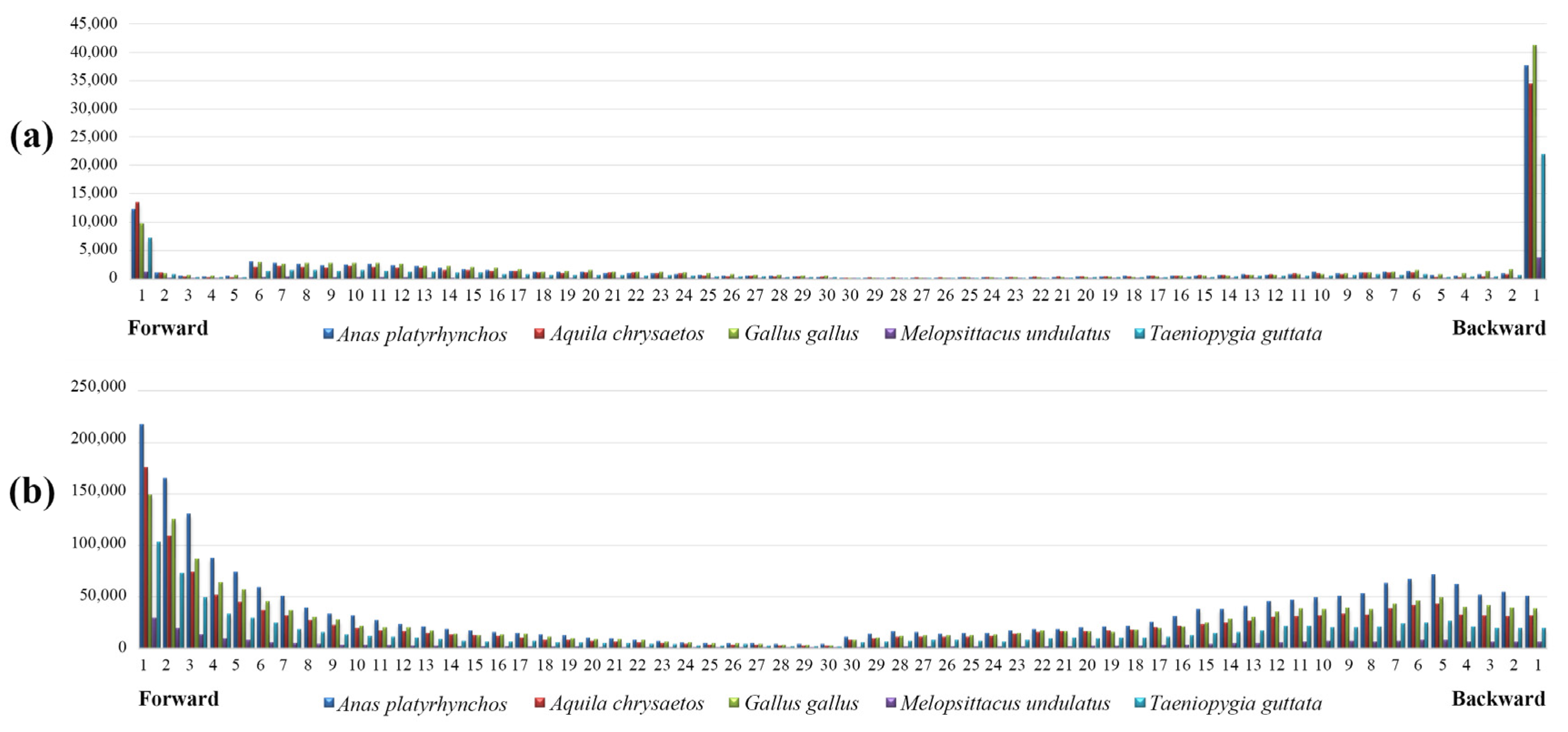
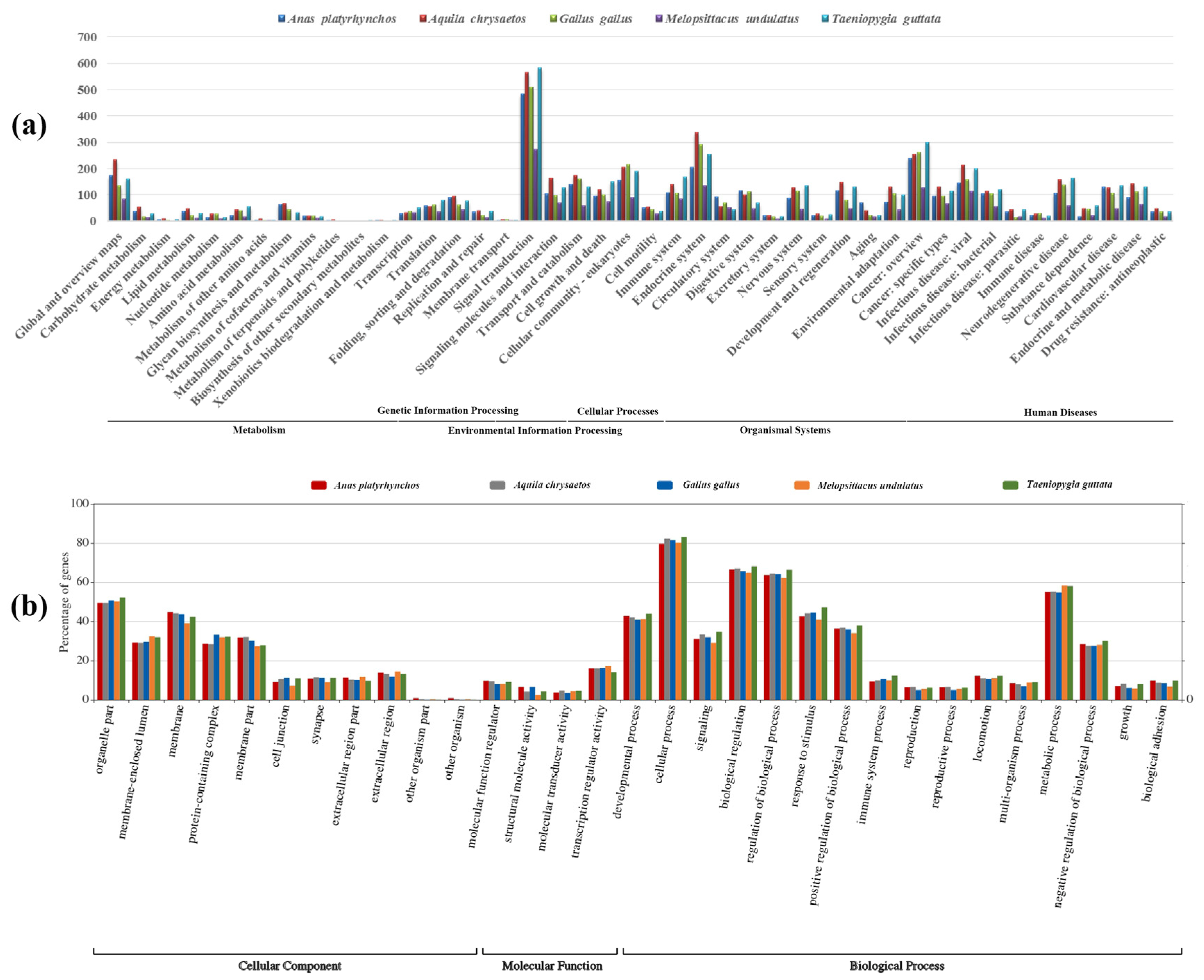
4. Discussion
4.1. Abundance of SSRs in Avian Genomes
4.2. Distribution of SSRs across Avian Lineages
4.3. Length Variation of P-SSRs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Toth, G.; Gaspari, Z.; Jurka, J. Microsatellites in Different Eukaryotic Genomes: Survey and Analysis. Genome Res. 2000, 10, 967–981. [Google Scholar] [CrossRef]
- Li, Y.-C.; Korol, A.B.; Fahima, T.; Beiles, A.; Nevo, E. Microsatellites: Genomic distribution, putative functions and mutational mechanisms: A review. Mol. Ecol. 2002, 11, 2453–2465. [Google Scholar] [CrossRef]
- Sharma, P.C.; Grover, A.; Kahl, G. Mining microsatellites in eukaryotic genomes. Trends Biotechnol. 2007, 25, 490–498. [Google Scholar] [CrossRef]
- Song, X.; Yang, T.; Yan, X.; Zheng, F.; Xu, X.; Zhou, C. Comparison of microsatellite distribution patterns in twenty-nine beetle genomes. Gene 2020, 757, 144919. [Google Scholar] [CrossRef]
- Qi, W.-H.; Lu, T.; Zheng, C.-L.; Jiang, X.-M.; Jie, H.; Zhang, X.-Y.; Yue, B.-S.; Zhao, G.-J. Distribution patterns of microsatellites and development of its marker in different genomic regions of forest musk deer genome based on high throughput sequencing. Aging 2020, 12, 4445–4462. [Google Scholar] [CrossRef] [PubMed]
- Kashi, Y.; King, D.G. Simple sequence repeats as advantageous mutators in evolution. Trends Genet. 2006, 22, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Gemayel, R.; Vinces, M.D.; Legendre, M.; Verstrepen, K.J. Variable Tandem Repeats Accelerate Evolution of Coding and Regulatory Sequences. Annu. Rev. Genet. 2010, 44, 445–477. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira Furo, I.; Kretschmer, R.; dos Santos, M.S.; de Lima Carvalho, C.A.; Gunski, R.J.; O’Brien, P.C.M.; Ferguson-Smith, M.A.; Cioffi, M.B.; de Oliveira, E.H.C. Chromosomal Mapping of Repetitive DNAs in Myiopsitta monachus and Amazona aestiva (Psittaciformes, Psittacidae) with Emphasis on the Sex Chromosomes. Cytogenet. Genome Res. 2017, 151, 151–160. [Google Scholar] [CrossRef]
- Barcellos, S.A.; Kretschmer, R.; de Souza, M.S.; Costa, A.L.; Degrandi, T.M.; dos Santos, M.S.; de Oliveira, E.H.C.; Cioffi, M.B.; Gunski, R.J.; Garnero, A.D.V. Karyotype Evolution and Distinct Evolutionary History of the W Chromosomes in Swallows (Aves, Passeriformes). Cytogenet. Genome Res. 2019, 158, 98–105. [Google Scholar] [CrossRef] [PubMed]
- de Souza, M.S.; Kretschmer, R.; Barcellos, S.A.; Costa, A.L.; Cioffi, M.D.; de Oliveira, E.H.; Garnero, A.D.; Gunski, R.J. Repeat Sequence Mapping Shows Different W Chromosome Evolutionary Pathways in Two Caprimulgiformes Families. Birds 2020, 1, 19–34. [Google Scholar] [CrossRef]
- Gunski, R.J.; Kretschmer, R.; Santos de Souza, M.; de Oliveira Furo, I.; Barcellos, S.A.; Costa, A.L.; Cioffi, M.B.; de Oliveira, E.H.C.; del Valle Garnero, A. Evolution of Bird Sex Chromosomes Narrated by Repetitive Sequences: Unusual W Chromosome Enlargement in Gallinula melanops (Aves: Gruiformes: Rallidae). Cytogenet. Genome Res. 2019, 158, 152–159. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, T.D.; Kretschmer, R.; Bertocchi, N.A.; Degrandi, T.M.; de Oliveira, E.H.C.; Cioffi, M.d.B.; Garnero, A.d.V.; Gunski, R.J. Genomic Organization of Repetitive DNA in Woodpeckers (Aves, Piciformes): Implications for Karyotype and ZW Sex Chromosome Differentiation. PLoS ONE 2017, 12, e0169987. [Google Scholar] [CrossRef]
- Matsubara, K.; O’Meally, D.; Azad, B.; Georges, A.; Sarre, S.D.; Graves, J.A.M.; Matsuda, Y.; Ezaz, T. Amplification of microsatellite repeat motifs is associated with the evolutionary differentiation and heterochromatinization of sex chromosomes in Sauropsida. Chromosoma 2016, 125, 111–123. [Google Scholar] [CrossRef]
- Kretschmer, R.; Rodrigues, B.S.; Barcellos, S.A.; Costa, A.L.; Cioffi, M.D.; Garnero, A.D.; Gunski, R.J.; de Oliveira, E.H.; Griffin, D.K. Karyotype Evolution and Genomic Organization of Repetitive DNAs in the Saffron Finch, Sicalis flaveola (Passeriformes, Aves). Animals 2021, 11, 1456. [Google Scholar] [CrossRef]
- Kretschmer, R.; de Oliveira, T.; Furo, I.; Silva, F.; Gunski, R.; Garnero, A.; Cioffi, M.; De Oliveira, E.; Freitas, T. Repetitive DNAs and shrink genomes: A chromosomal analysis in nine Columbidae species (Aves, Columbiformes). Genet. Mol. Biol. 2018, 41, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.; Avvaru, A.K.; Sowpati, D.T.; Mishra, R.K. Patterns of microsatellite distribution across eukaryotic genomes. BMC Genom. 2019, 20, 153. [Google Scholar] [CrossRef]
- Lim, K.G.; Kwoh, C.K.; Hsu, L.Y.; Wirawan, A. Review of tandem repeat search tools: A systematic approach to evaluating algorithmic performance. Brief. Bioinform. 2013, 14, 67–81. [Google Scholar] [CrossRef]
- Qi, W.-H.; Jiang, X.-M.; Du, L.-M.; Xiao, G.-S.; Hu, T.-Z.; Yue, B.-S.; Quan, Q.-M. Genome-Wide Survey and Analysis of Microsatellite Sequences in Bovid Species. PLoS ONE 2015, 10, e0133667. [Google Scholar] [CrossRef]
- Xu, Y.; Hu, Z.; Wang, C.; Zhang, X.; Li, J.; Yue, B. Characterization of perfect microsatellite based on genome-wide and chromosome level in Rhesus monkey (Macaca mulatta). Gene 2016, 592, 269–275. [Google Scholar] [CrossRef]
- Pigot, A.L.; Sheard, C.; Miller, E.T.; Bregman, T.P.; Freeman, B.G.; Roll, U.; Seddon, N.; Trisos, C.H.; Weeks, B.C.; Tobias, J.A. Macroevolutionary convergence connects morphological form to ecological function in birds. Nat. Ecol. Evol. 2020, 4, 230–239. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Li, C.; Li, Q.; Li, B.; Larkin, D.M.; Lee, C.; Storz, J.F.; Antunes, A.; Greenwold, M.J.; Meredith, R.W.; et al. Comparative genomics reveals insights into avian genome evolution and adaptation. Science 2014, 346, 1311–1320. [Google Scholar] [CrossRef]
- Hughes, A.L.; Piontkivska, H. DNA repeat arrays in chicken and human genomes and the adaptive evolution of avian genome size. BMC Evol. Biol. 2005, 5, 12. [Google Scholar] [CrossRef]
- Hughes, A.L.; Hughes, M.K. Small genomes for better flyers. Nature 1995, 377, 391. [Google Scholar] [CrossRef]
- Huang, J.; Li, W.; Jian, Z.; Yue, B.; Yan, Y. Genome-wide distribution and organization of microsatellites in six species of birds. Biochem. Syst. Ecol. 2016, 67, 95–102. [Google Scholar] [CrossRef]
- Adams, R.; Blackmon, H.; Reyes Velasco, J.; Schield, D.; Card, D.; Andrew, A.; Waynewood, N.; Castoe, T. Microsatellite landscape evolutionary dynamics across 450 million years of vertebrate genome evolution. Genome 2016, 59, 295–310. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Suleski, M.; Hedges, S.B. TimeTree: A Resource for Timelines, Timetrees, and Divergence Times. Mol. Biol. Evol. 2017, 34, 1812–1819. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Zhang, C.; Liu, Q.; Zhang, X.; Yue, B. Krait: An ultrafast tool for genome-wide survey of microsatellites and primer design. Bioinformatics 2018, 34, 681–683. [Google Scholar] [CrossRef]
- Zhou, C.; Li, F.; Wen, Q.; Price, M.; Yang, N.; Yue, B. Characterization of microsatellites in the endangered snow leopard based on the chromosome-level genome. Mammal Res. 2021, 66, 385–398. [Google Scholar] [CrossRef]
- Revell, L.J. phytools: An R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 2012, 3, 217–223. [Google Scholar] [CrossRef]
- Qi, W.-H.; Jiang, X.-M.; Yan, C.-C.; Zhang, W.-Q.; Xiao, G.-S.; Yue, B.-S.; Zhou, C.-Q. Distribution patterns and variation analysis of simple sequence repeats in different genomic regions of bovid genomes. Sci. Rep. 2018, 8, 14407. [Google Scholar] [CrossRef]
- Song, X.; Yang, T.; Zhang, X.; Yuan, Y.; Yan, X.; Wei, Y.; Zhang, J.; Zhou, C. Comparison of the Microsatellite Distribution Patterns in the Genomes of Euarchontoglires at the Taxonomic Level. Front. Genet. 2021, 12, 622724. [Google Scholar] [CrossRef]
- Ledenyova, M.L.; Tkachenko, G.A.; Shpak, I.M. Imperfect and Compound Microsatellites in the Genomes of Burkholderia pseudomallei Strains. Mol. Biol. 2019, 53, 127–137. [Google Scholar] [CrossRef]
- Gáspári, Z.; Ortutay, C.; Tóth, G. Divergent microsatellite evolution in the human and chimpanzee lineages. FEBS Lett. 2007, 581, 2523–2526. [Google Scholar] [CrossRef]
- Boeva, V.; Regnier, M.; Papatsenko, D.; Makeev, V. Short fuzzy tandem repeats in genomic sequences, identification, and possible role in regulation of gene expression. Bioinformatics 2006, 22, 676–684. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Shen, F.; Huang, J.; Huang, Y.; Du, L.; Wang, C.; Fan, Z.; Hou, R.; Yue, B.; Zhang, X. Transcriptome-Derived Tetranucleotide Microsatellites and Their Associated Genes from the Giant Panda (Ailuropoda melanoleuca). J. Hered. 2016, 107, 423–430. [Google Scholar] [CrossRef]
- Campregher, C.; Scharl, T.; Nemeth, M.; Honeder, C.; Jascur, T.; Boland, C.R.; Gasche, C. The nucleotide composition of microsatellites impacts both replication fidelity and mismatch repair in human colorectal cells. Hum. Mol. Genet. 2010, 19, 2648–2657. [Google Scholar] [CrossRef] [PubMed]
- Alam, C.M.; George, B.; Sharfuddin, C.; Jain, S.K.; Chakraborty, S. Occurrence and analysis of imperfect microsatellites in diverse potyvirus genomes. Gene 2013, 521, 238–244. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Hou, W.; Sun, T.; Xu, Y.; Li, P.; Yue, B.; Fan, Z.; Li, J. Genome-wide mining and comparative analysis of microsatellites in three macaque species. Mol. Genet. Genom. 2017, 292, 537–550. [Google Scholar] [CrossRef] [PubMed]
- Kaessmann, H.; Vinckenbosch, N.; Long, M. RNA-based gene duplication: Mechanistic and evolutionary insights. Nat. Rev. Genet. 2009, 10, 19–31. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Tan, Z.; Feng, H.; Yang, R.; Li, M.; Jiang, J.; Shen, G.; Yu, R. Microsatellites in different Potyvirus genomes: Survey and analysis. Gene 2011, 488, 52–56. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A.; Takai, D. The Role of DNA Methylation in Mammalian Epigenetics. Science 2001, 293, 1068–1070. [Google Scholar] [CrossRef]
- Ren, L.; Gao, G.; Zhao, D.; Ding, M.; Luo, J.; Deng, H. Developmental stage related patterns of codon usage and genomic GC content: Searching for evolutionary fingerprints with models of stem cell differentiation. Genome Biol. 2007, 8, R35. [Google Scholar] [CrossRef]
- Ellegren, H. Microsatellites: Simple sequences with complex evolution. Nat. Rev. Genet. 2004, 5, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Payseur, B.A.; Jing, P.; Haasl, R.J. A Genomic Portrait of Human Microsatellite Variation. Mol. Biol. Evol. 2011, 28, 303–312. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.-H.; Kussell, E. Evolutionary pressures on simple sequence repeats in prokaryotic coding regions. Nucleic Acids Res. 2012, 40, 2399–2413. [Google Scholar] [CrossRef]
- Castagnone-Sereno, P.; Danchin, E.G.J.; Deleury, E.; Guillemaud, T.; Malausa, T.; Abad, P. Genome-wide survey and analysis of microsatellites in nematodes, with a focus on the plant-parasitic species Meloidogyne incognita. BMC Genom. 2010, 11, 598. [Google Scholar] [CrossRef]
- Katti, M.V.; Ranjekar, P.K.; Gupta, V.S. Differential Distribution of Simple Sequence Repeats in Eukaryotic Genome Sequences. Mol. Biol. Evol. 2001, 18, 1161–1167. [Google Scholar] [CrossRef]
- Bell, G.I.; Jurka, J. The Length Distribution of Perfect Dimer Repetitive DNA Is Consistent with Its Evolution by an Unbiased Single-Step Mutation Process. J. Mol. Evol. 1997, 44, 414–421. [Google Scholar] [CrossRef] [PubMed]
- Kruglyak, S.; Durrett, R.T.; Schug, M.D.; Aquadro, C.F. Equilibrium distributions of microsatellite repeat length resulting from a balance between slippage events and point mutations. Proc. Natl. Acad. Sci. USA 1998, 95, 10774–10778. [Google Scholar] [CrossRef]
- Leopoldino, A.M.; Pena, S.D.J. The mutational spectrum of human autosomal tetranucleotide microsatellites. Hum. Mutat. 2003, 21, 71–79. [Google Scholar] [CrossRef]
- Vowles, E.J.; Amos, W. Quantifying Ascertainment Bias and Species-Specific Length Differences in Human and Chimpanzee Microsatellites Using Genome Sequences. Mol. Biol. Evol. 2006, 23, 598–607. [Google Scholar] [CrossRef] [PubMed]
- Bagshaw, A.T.M. Functional Mechanisms of Microsatellite DNA in Eukaryotic Genomes. Genome Biol. Evol. 2017, 9, 2428–2443. [Google Scholar] [CrossRef]
- Liu, L.; Dybvig, K.; Panangala Victor, S.; van Santen Vicky, L.; French Christopher, T. GAA Trinucleotide Repeat Region Regulates M9/pMGA Gene Expression in Mycoplasma gallisepticum. Infect. Immun. 2000, 68, 871–876. [Google Scholar] [CrossRef]
- Miret, J.J.; Pessoa-Brandão, L.; Lahue, R.S. Orientation-dependent and sequence-specific expansions of CTG/CAG trinucleotide repeats in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1998, 95, 12438–12443. [Google Scholar] [CrossRef]
- Lantto, J.; Ohlin, M. Uneven Distribution of Repetitive Trinucleotide Motifs in Human Immunoglobulin Heavy Variable Genes. J. Mol. Evol. 2002, 54, 346–353. [Google Scholar] [CrossRef] [PubMed]
- Saeed, D.-A.; Wang, R.; Wang, S. Microsatellites in Pursuit of Microbial Genome Evolution. Front. Microbiol. 2016, 6, 1462. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, K.; Zhou, C.; Wang, L.; Zhang, C.; Yang, Z.; Hu, Z.; Yue, B.; Wu, Y. Comprehensive Comparative Analysis Sheds Light on the Patterns of Microsatellite Distribution across Birds Based on the Chromosome-Level Genomes. Animals 2023, 13, 655. https://doi.org/10.3390/ani13040655
Feng K, Zhou C, Wang L, Zhang C, Yang Z, Hu Z, Yue B, Wu Y. Comprehensive Comparative Analysis Sheds Light on the Patterns of Microsatellite Distribution across Birds Based on the Chromosome-Level Genomes. Animals. 2023; 13(4):655. https://doi.org/10.3390/ani13040655
Chicago/Turabian StyleFeng, Kaize, Chuang Zhou, Lei Wang, Chunhui Zhang, Zhixiong Yang, Zhengrui Hu, Bisong Yue, and Yongjie Wu. 2023. "Comprehensive Comparative Analysis Sheds Light on the Patterns of Microsatellite Distribution across Birds Based on the Chromosome-Level Genomes" Animals 13, no. 4: 655. https://doi.org/10.3390/ani13040655
APA StyleFeng, K., Zhou, C., Wang, L., Zhang, C., Yang, Z., Hu, Z., Yue, B., & Wu, Y. (2023). Comprehensive Comparative Analysis Sheds Light on the Patterns of Microsatellite Distribution across Birds Based on the Chromosome-Level Genomes. Animals, 13(4), 655. https://doi.org/10.3390/ani13040655









