Effects of the CDC10 (Septin 7) Gene on the Proliferation and Differentiation of Bovine Intramuscular Preadipocyte and 3T3-L1 Cells
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Standards
2.2. Cell Culture
2.3. Transfection of siRNA and Lentivirus Infection
2.4. RNA Isolation and Quantitative Real-Time PCR
2.5. Western Blot Analysis
2.6. EdU and Flow Cytometry Assay
2.7. Oil Red O Staining
2.8. Statistical Analysis
3. Results
3.1. Growth Curve of Primary Cultured BIMP and Identification of Adipogenic Marker Genes
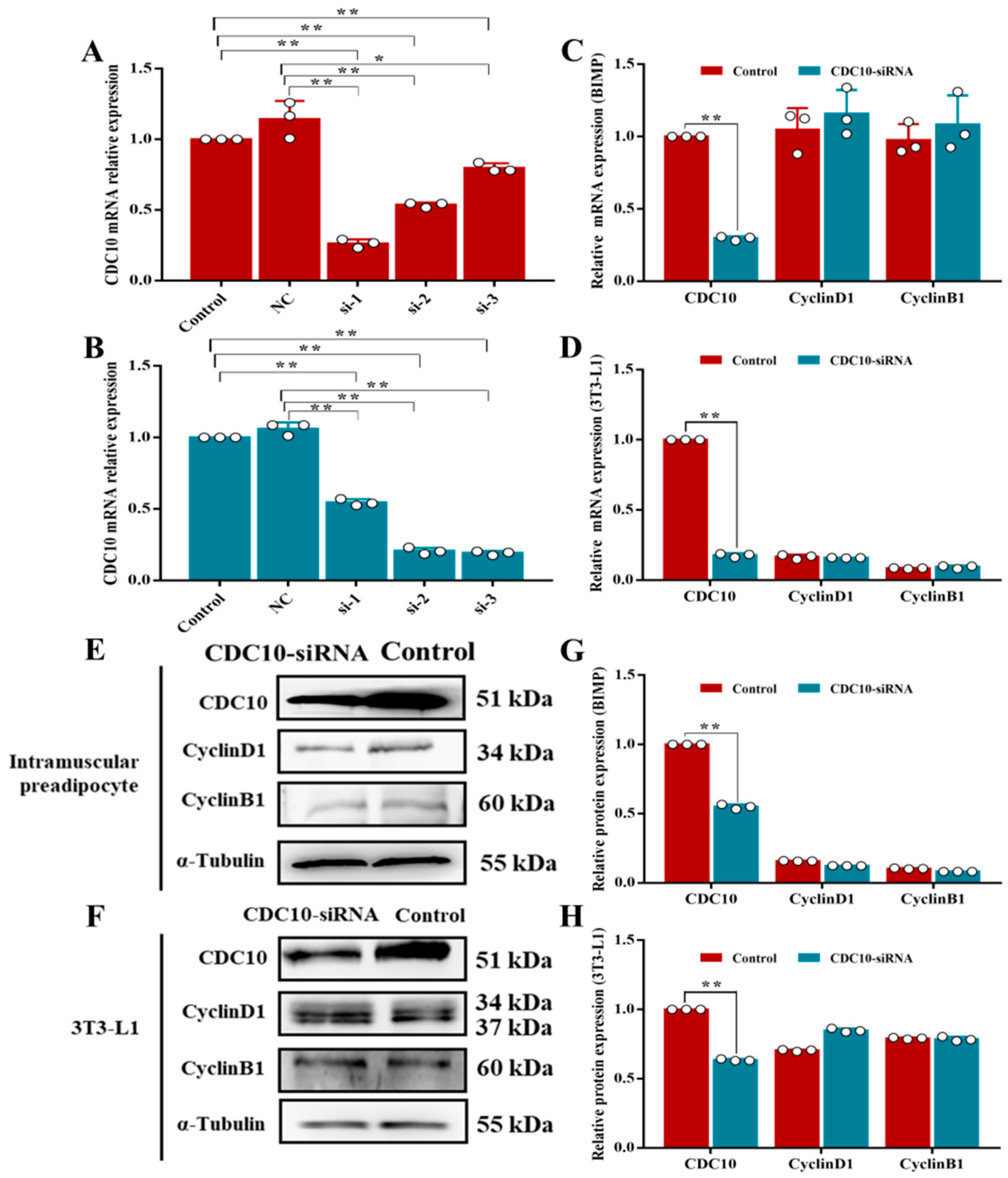
3.2. Expression of the CDC10 Gene in the Proliferation and Differentiation of BIMP and 3T3-L1 Cells
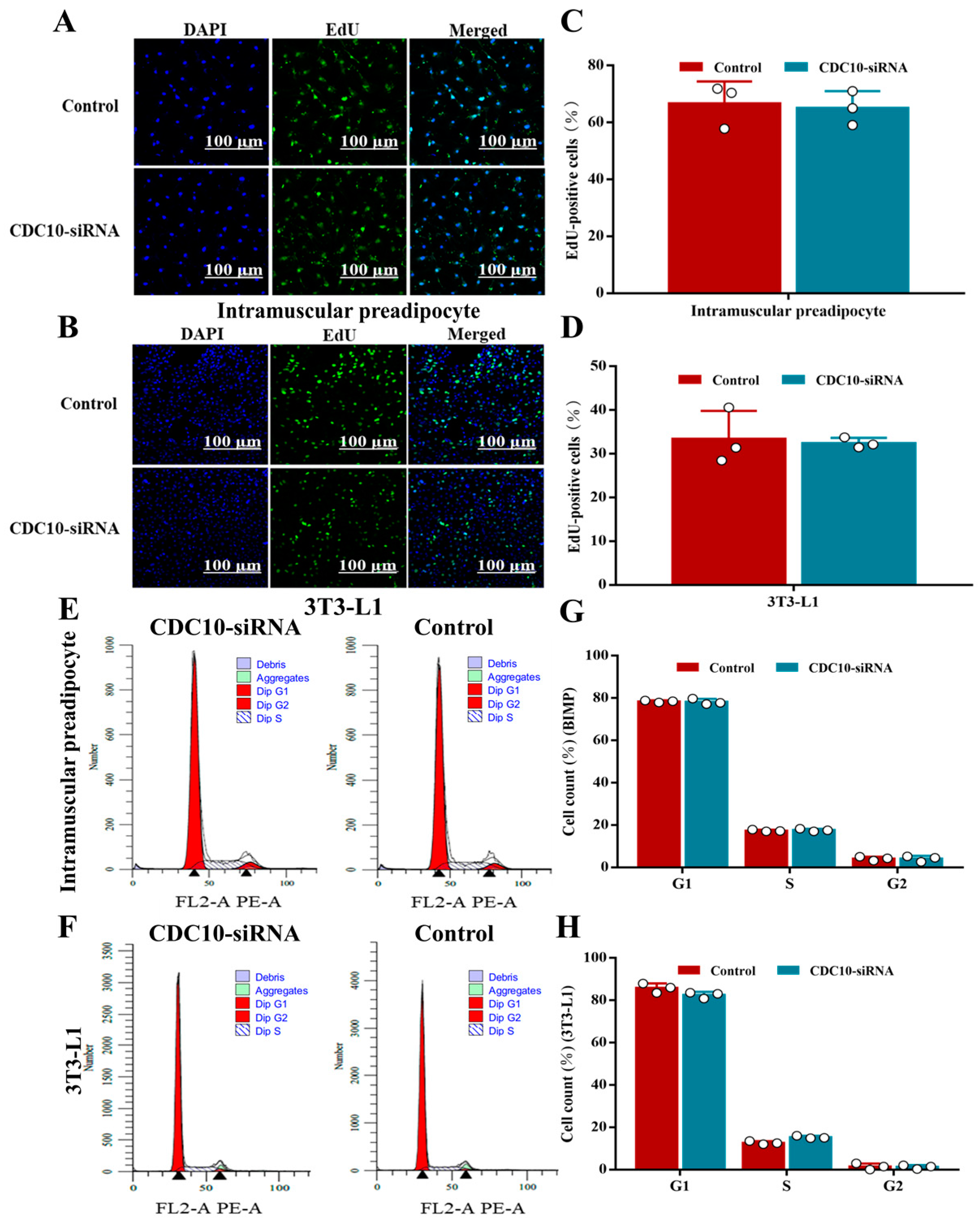
3.3. Knockdown of CDC10 Had No Significant Effect on the Proliferation of BIMP and 3T3-L1 Cells
3.4. Overexpression of CDC10 Had No Significant Effect on the Proliferation of BIMP and 3T3-L1 Cells
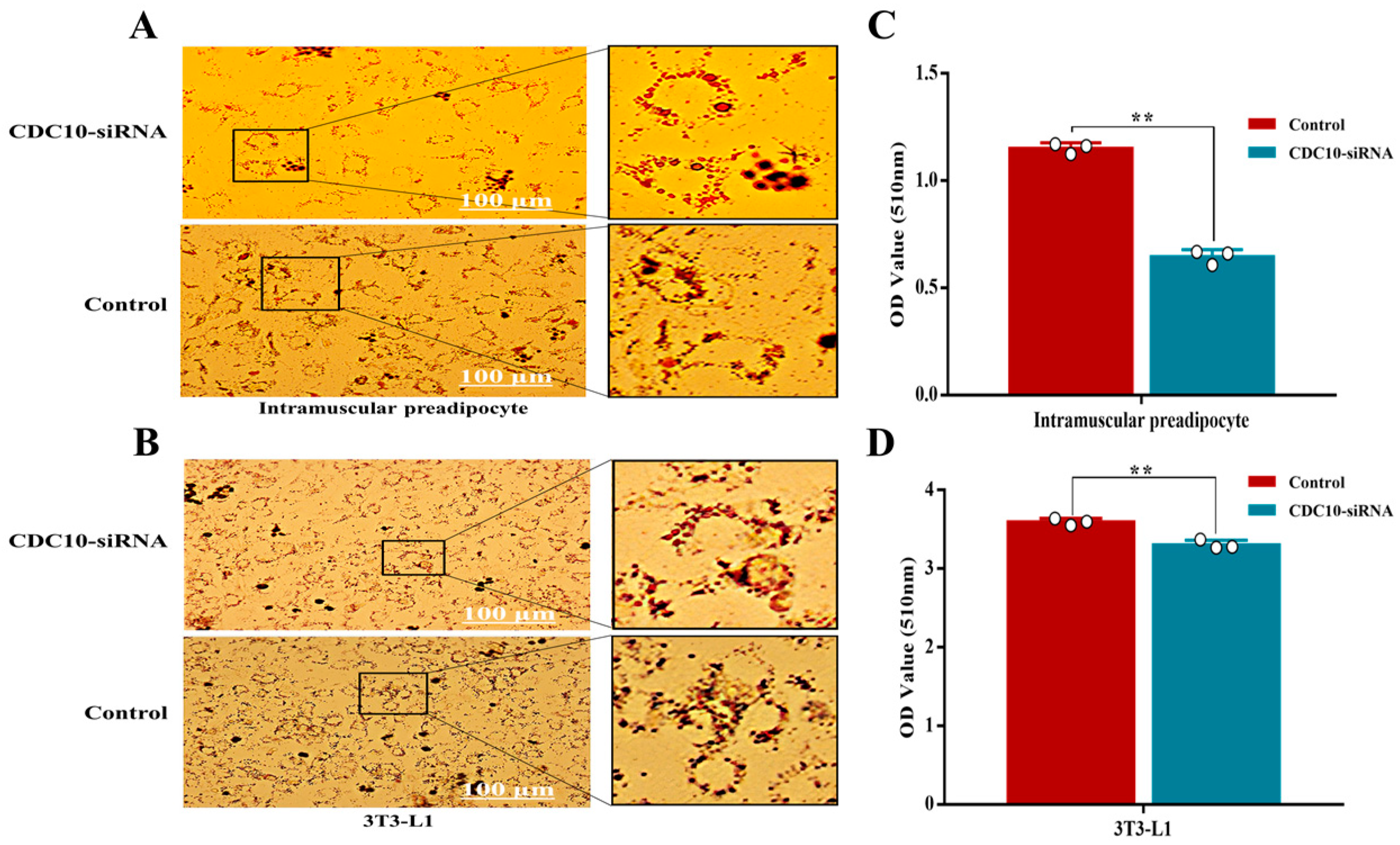
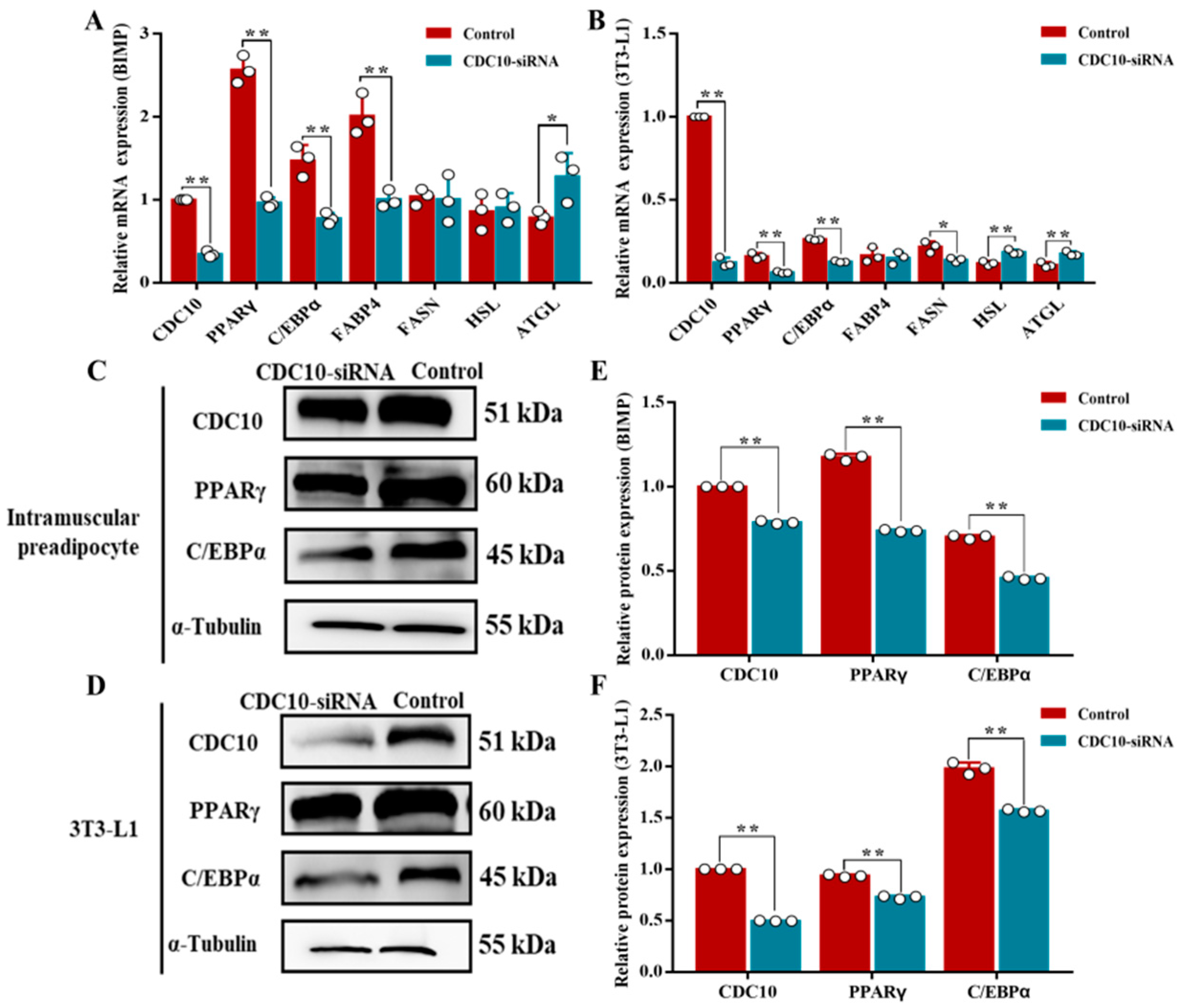
3.5. Knockdown of CDC10 Inhibits Differentiation of BIMP and 3T3-L1 Cells
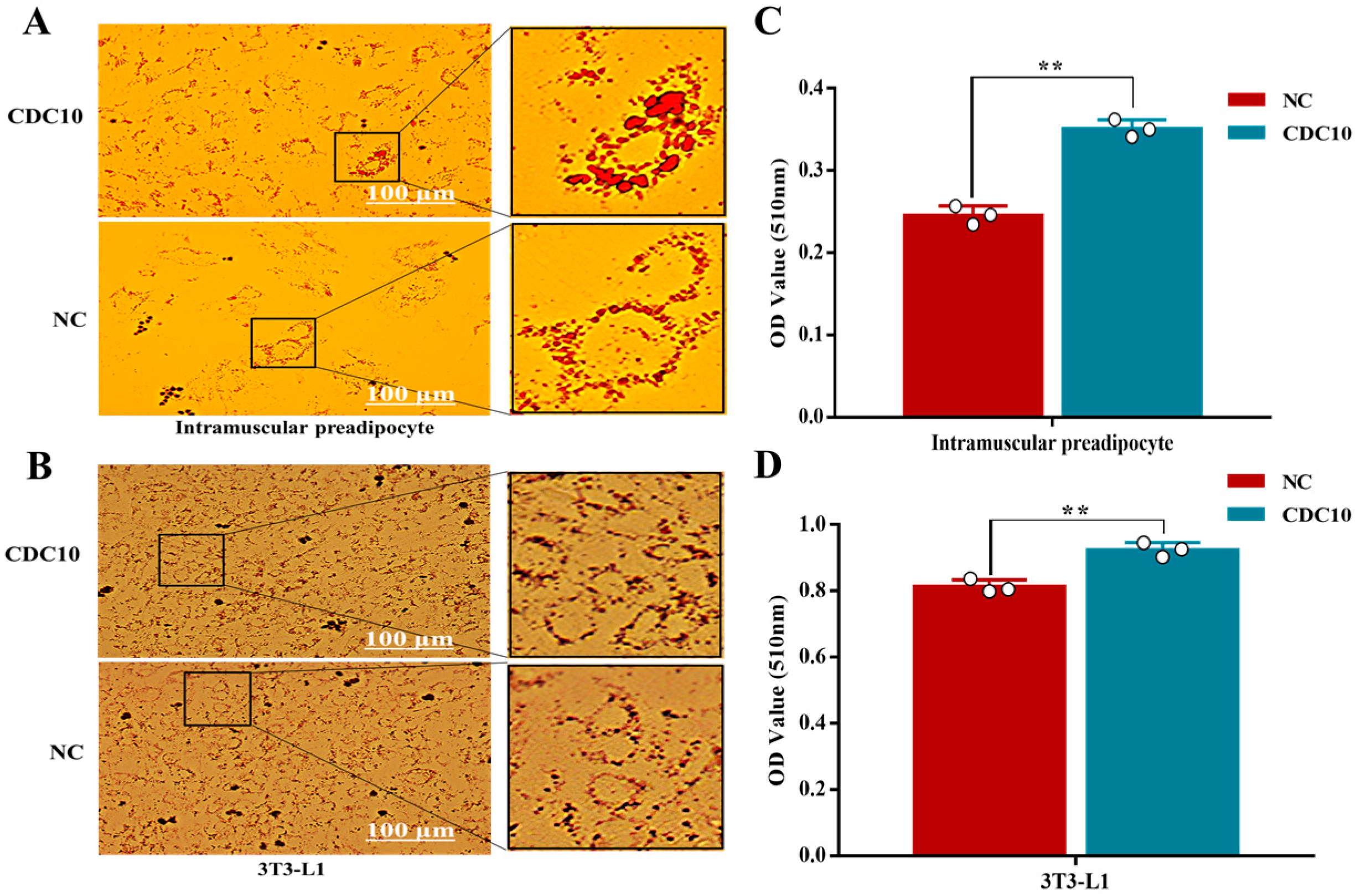
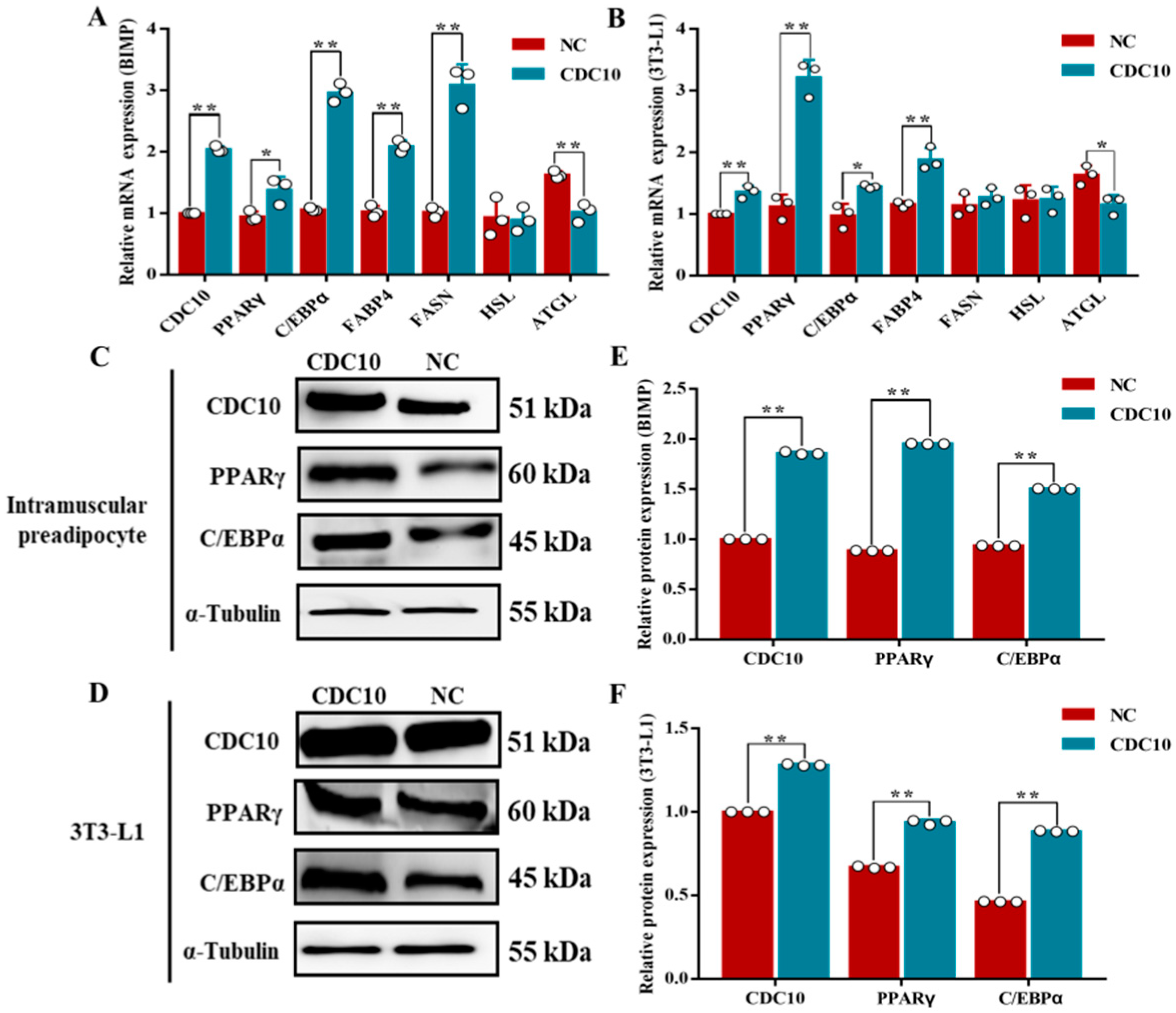
3.6. Overexpression of CDC10 Promotes Differentiation of BIMP and 3T3-L1 Cells
4. Discussion
4.1. Effect of the CDC10 Gene on Proliferation of BIMP and 3T3-L1 Cells
4.2. Effect of the CDC10 Gene on the Differentiation of BIMP and 3T3-L1 Cells
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, Y.; Zhou, J.; Wang, G.; Cai, S.; Zeng, X.; Qiao, S. Advances in low-protein diets for swine. J. Anim. Sci. Biotechnol. 2018, 9, 60. [Google Scholar] [CrossRef]
- Bahelka, I.; Oravcová, M.; Peškovičová, D.; Tomka, J.; Hanusová, E.; Lahučký, R.; Demo, P. Comparison of accuracy of intramuscular fat prediction in live pigs using five different ultrasound intensity levels. Animal 2009, 3, 1205–1211. [Google Scholar] [CrossRef] [PubMed]
- Cho, I.C.; Park, H.B.; Ahn, J.S.; Han, S.H.; Lee, J.B.; Lim, H.T.; Yoo, C.K.; Jung, E.J.; Kim, D.H.; Sun, W.S.; et al. A functional regulatory variant of MYH3 influences muscle fiber-type composition and intramuscular fat content in pigs. PLoS Genet. 2019, 15, e1008279. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.V.; Nguyen, O.C.; Malau-Aduli, A.E.O. Main regulatory factors of marbling level in beef cattle. Vet. Anim. Sci. 2021, 14, 100219. [Google Scholar] [CrossRef]
- Gui, L.; Jiang, B.; Zhang, Y.; Zan, L. Sequence variants in the bovine silent information regulator 6, their linkage and their associations with body measurements and carcass quality traits in Qinchuan cattle. Gene 2015, 559, 16–21. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, W.; Gui, L.; Wang, H.; Zan, L. Association and expression analyses of the Ucp2 and Ucp3 gene polymorphisms with body measurement and meat quality traits in Qinchuan cattle. J. Genet. 2016, 95, 939–946. [Google Scholar] [CrossRef]
- Gui, L.; Wu, H.; Raza, S.H.A.; Schreurs, N.M.; Shah, M.A. The effect of haplotypes in the promoter region of SIRT4 gene on the ultrasound traits in Qinchuan cattle. Trop. Anim. Health Prod. 2019, 51, 1877–1882. [Google Scholar] [CrossRef]
- Raza, S.H.A.; Khan, R.; Abdelnour, S.A.; Abd El-Hack, M.E.; Khafaga, A.F.; Taha, A.; Ohran, H.; Mei, C.; Schreurs, N.M.; Zan, L. Advances of Molecular Markers and Their Application for Body Variables and Carcass Traits in Qinchuan Cattle. Genes 2019, 10, 717. [Google Scholar] [CrossRef]
- Li, Y.; Cheng, G.; Yamada, T.; Liu, J.; Zan, L.; Tong, B. Effect of Expressions and SNPs of Candidate Genes on Intramuscular Fat Content in Qinchuan Cattle. Animals 2020, 10, 1370. [Google Scholar] [CrossRef]
- Gui, L.S.; Raza, S.H.A.; Memon, S.; Li, Z.; Abd El-Aziz, A.H.; Ullah, I.; Jahejo, A.R.; Shoorei, H.; Khan, R.; Quan, G.; et al. Association of hormone-sensitive lipase (HSL) gene polymorphisms with the intramuscular fat content in two Chinese beef cattle breeds. Genomics 2020, 112, 3883–3889. [Google Scholar] [CrossRef]
- Raza, S.H.A.; Khan, S.; Amjadi, M.; Abdelnour, S.A.; Ohran, H.; Alanazi, K.M.; Abd El-Hack, M.E.; Taha, A.E.; Khan, R.; Gong, C.; et al. Genome-wide association studies reveal novel loci associated with carcass and body measures in beef cattle. Arch. Biochem. Biophys. 2020, 694, 108543. [Google Scholar] [CrossRef] [PubMed]
- Weirich, C.S.; Erzberger, J.P.; Barral, Y. The septin family of GTPases: Architecture and dynamics. Nat. Rev. Mol. Cell Biol. 2008, 9, 478–489. [Google Scholar] [CrossRef] [PubMed]
- Katharina, N.; Barbara, Z. The Mammalian Septin Interactome. Front. Cell Dev. Biol. 2017, 5, 3. [Google Scholar]
- Macara, I.G.; Baldarelli, R.; Field, C.M.; Glotzer, M.; Hayashi, Y.; Hsu, S.C.; Kennedy, M.B.; Kinoshita, M.; Longtine, M.; Low, C.; et al. Mammalian septins nomenclature. Mol. Biol. Cell 2002, 13, 4111–4113. [Google Scholar] [CrossRef] [PubMed]
- Makoto, K. The septins. Genome. Biol. 2003, 4, 236. [Google Scholar]
- Beites, C.L.; Xie, H.; Bowser, R.; Trimble, W.S. The septin CDCrel-1 binds syntaxin and inhibits exocytosis. Nat. Neurosci. 1999, 2, 434–439. [Google Scholar] [CrossRef]
- Field, C.M.; Kellogg, D. Septins: Cytoskeletal polymers or signalling GTPases? Trends. Cell Biol. 1999, 9, 387–394. [Google Scholar] [CrossRef]
- Larisch, S.; Yi, Y.; Lotan, R.; Kerner, H.; Eimerl, S.; Tony Parks, W.; Gottfried, Y.; Birkey Reffey, S.; de Caestecker, M.P.; Danielpour, D.; et al. A novel mitochondrial septin-like protein, ARTS, mediates apoptosis dependent on its P-loop motif. Nat. Cell Biol. 2000, 2, 915–921. [Google Scholar] [CrossRef]
- Kartmann, B.; Roth, D. Novel roles for mammalian septins: From vesicle trafficking to oncogenesis. J. Cell Sci. 2001, 114, 839–844. [Google Scholar] [CrossRef]
- Jia, Z.F.; Huang, Q.; Kang, C.S.; Yang, W.D.; Wang, G.X.; Yu, S.Z.; Jiang, H.; Pu, P.Y. Overexpression of septin 7 suppresses glioma cell growth. J. Neurooncol. 2010, 98, 329–340. [Google Scholar] [CrossRef]
- Menon, M.B.; Sawada, A.; Chaturvedi, A.; Mishra, P.; Schuster-Gossler, K.; Galla, M.; Schambach, A.; Gossler, A.; Förster, R.; Heuser, M.; et al. Genetic deletion of SEPT7 reveals a cell type-specific role of septins in microtubule destabilization for the completion of cytokinesis. PLoS Genet. 2016, 10, e1004558. [Google Scholar] [CrossRef]
- Zhang, N.; Liu, L.; Fan, N.; Zhang, Q.; Wang, W.; Zheng, M.; Ma, L.; Li, Y.; Shi, L. The requirement of SEPT2 and SEPT7 for migration and invasion in human breast cancer via MEK/ERK activation. Oncotarget 2016, 7, 61587–61600. [Google Scholar] [CrossRef]
- Chen, T.Y.; Lin, T.C.; Kuo, P.L.; Chen, Z.R.; Cheng, H.L.; Chao, Y.Y.; Syu, J.S.; Lu, F.I.; Wang, C.Y. Septin 7 is a centrosomal protein that ensures S phase entry and microtubule nucleation by maintaining the abundance of p150(glued). J. Cell Physiol. 2021, 236, 2706–2724. [Google Scholar] [CrossRef]
- Prusty, D.; Park, B.H.; Davis, K.E.; Farmer, S.R. Activation of MEK/ERK signaling promotes adipogenesis by enhancing peroxisome proliferator-activated receptor gamma (PPARgamma) and C/EBPalpha gene expression during the differentiation of 3T3-L1 preadipocytes. J. Biol. Chem. 2002, 277, 46226–46232. [Google Scholar] [CrossRef]
- Chambard, J.C.; Lefloch, R.; Pouyssegur, J.; Lenormand, P. ERK implication in cell cycle regulation. Biochim. Biophys. Acta 2007, 1773, 1299–1310. [Google Scholar] [CrossRef]
- Kadereit, B.; Kumar, P.; Wang, W.J.; Miranda, D.; Snapp, E.L.; Severina, N.; Torregroza, I.; Evans, T.; Silver, D.L. Evolutionarily conserved gene family important for fat storage. Proc. Natl. Acad. Sci. USA 2008, 105, 94–99. [Google Scholar] [CrossRef]
- Chen, F.; Yan, B.; Ren, J.; Lyu, R.; Wu, Y.; Guo, Y.; Li, D.; Zhang, H.; Hu, J. FIT2 organizes lipid droplet biogenesis with ER tubule-forming proteins and septins. J. Cell Biol. 2021, 220, e201907183. [Google Scholar] [CrossRef]
- Wang, S.; Zhou, G.; Shu, G.; Wang, L.; Zhu, X.; Gao, P.; Xi, Q.; Zhang, Y.; Yuan, L.; Jiang, Q. Glucose Utilization, Lipid Metabolism and BMP-Smad Signaling Pathway of Porcine Intramuscular Preadipocytes Compared with Subcutaneous Preadipocytes. Cell Physiol. Biochem. 2013, 31, 981–996. [Google Scholar] [CrossRef]
- Poleti, M.D.; Regitano, L.C.A.; Souza, G.H.M.F.; Cesar, A.S.M.; Simas, R.C.; Silva-Vignato, B.; Oliveira, G.B.; Andrade, S.C.S.; Cameron, L.C.; Coutinho, L.L. Longissimus dorsi muscle label-free quantitative proteomic reveals biological mechanisms associated with intramuscular fat deposition. J. Proteom. 2018, 179, 30–41. [Google Scholar] [CrossRef]
- Li, J.; Wang, G.; Jiang, J.; Zhang, L.; Zhou, P.; Ren, H. MicroRNA-127-3p regulates myoblast proliferation by targeting Sept7. Biotechnol. Lett. 2020, 42, 1633–1644. [Google Scholar] [CrossRef]
- Auwerx, J. PPARgamma, the ultimate thrifty gene. Diabetologia 1999, 42, 1033–1049. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Rosen, E.D.; Brun, R.; Hauser, S.; Adelmant, G.; Troy, A.E.; McKeon, C.; Darlington, G.J.; Spiegelman, B.M. Cross-regulation of C/EBP alpha and PPAR gamma controls the transcriptional pathway of adipogenesis and insulin sensitivity. Mol. Cell 1999, 3, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Yeh, W.C.; Cao, Z.; Classon, M.; McKnight, S.L. Cascade regulation of terminal adipocyte differentiation by three members of the C/EBP family of leucine zipper proteins. Genes Dev. 1995, 9, 168–181. [Google Scholar] [CrossRef] [PubMed]
- Cerk, I.K.; Wechselberger, L.; Oberer, M. Adipose Triglyceride Lipase Regulation: An Overview. Curr. Protein Pept. Sci. 2018, 19, 221–233. [Google Scholar] [CrossRef]
- Zimmermann, R.; Strauss, J.G.; Haemmerle, G.; Schoiswohl, G.; Birner-Gruenberger, R.; Riederer, M.; Lass, A.; Neuberger, G.; Eisenhaber, F.; Hermetter, A.; et al. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science 2004, 306, 1383–1386. [Google Scholar] [CrossRef]
- Smirnova, E.; Goldberg, E.B.; Makarova, K.S.; Lin, L.; Brown, W.J.; Jackson, C.L. ATGL has a key role in lipid droplet/adiposome degradation in mammalian cells. EMBO Rep. 2006, 7, 106–113. [Google Scholar] [CrossRef]
- Wang, M.; Wang, J.J.; Li, J.; Park, K.; Qian, X.; Ma, J.X.; Zhang, S.X. Pigment epithelium-derived factor suppresses adipogenesis via inhibition of the MAPK/ERK pathway in 3T3-L1 preadipocytes. Am. J. Physiol. Endocrinol. Metab. 2009, 297, E1378–E1387. [Google Scholar] [CrossRef]
- Tong, B.; Gao, G.Q.; Muramatsu, Y.; Ohta, T.; Kose, H.; Li, G.P.; Fatchiyah, F.; Yamada, T. Association of the expression levels in the longissimus muscle and a SNP in the CDC10 gene with marbling in Japanese Black beef cattle. Meat Sci. 2015, 108, 28–31. [Google Scholar] [CrossRef]




| Name | Target Sequence (5′-3′) |
|---|---|
| si-B-CDC10-01 | CAGAGGAATGCCAACAGTT |
| si-B-CDC10-02 | CCTGATAACAGAGTGCAAT |
| si-B-CDC10-03 | GCTGTGGTAGGTAGTAATA |
| si-M-CDC10-01 | GCTTTGCCAACCTCCCAAA |
| si-M-CDC10-02 | CCAGAGTATCCAGGACCTT |
| si-M-CDC10-03 | GGACATGGACTTAAACCAT |
| Name | Primer Sequence-F (5′-3′) | Primer Sequence-R (5′-3′) |
|---|---|---|
| B-CDC10 | TCTGAAGCTGAGCTCCAA | AAATTGGTCAAACGGATCCA |
| B-CyclinD1 | CTGGTCCTGGTGAACAAACT | ACAGAGGGCAACGAAGGT |
| B-CyclinB1 | GGATACCTATGTGCCCAAGAAG | GACGGCGACCCAGACTAAA |
| B-PPARγ | TGGAGACCGCCCAGGTTTGC | AGCTGGGAGGACTCGGGGTG |
| B-C/EBPα | TGCGCAAGAGCCGGGACAAG | ACCAGGGAGCTCTCGGGCAG |
| B-FABP4 | TCCTTCAAATTGGGCCAGGAA | CCCTTGGCTTATGCTCTCTCA |
| B-HSL | GAGTTTGAGCGGATCATTCA | TGAGGCCATGTTTGCTAGAG |
| B-ATGL | TCTGCCTGCTGATTGCTATG | GGCCTGGATAAGCTCCTCTT |
| B-FASN | TAAGGTTCAAATTGCTGCGT | TCCAGAGCGAAGGAGAGATT |
| B-β-actin | ACCACACCTTCTACAACGAG | GAACATGATCTGGGTCATCTTC |
| M-CDC10 | CTTACACCAGAGGAATGCCAA | TTTTCAACTTCAGCCACACC |
| M-CyclinD1 | ATGTTCGTGGCCTCTAAGATGAAG | CAGAAGCAGTTCCATTTGCAGCAG |
| M-CyclinB1 | AAGGTGCCTGTGTGTGAACC | GTCAGCCCCATCATCTGCG |
| M-PPARγ | TTCGCTGATGCACTGCCTAT | GGAATGCGAGTGGTCTTCCA |
| M-CEBPα | GTGTGCACGTCTATGCTAAACCA | GCCGTTAGTGAAGAGTCTCAGTTTG |
| M-FABP4 | TGAAATCACCGCAGACGACA | ACACATTCCACCACCAGCTT |
| M-HSL | TGCCCAGGATTGGATGGTTT | GTGAGAACGCTGAGGCTTTG |
| M-ATGL | GGAGGAATGGCCTACTGAACC | ATCCTCTTCCTGGGGGACAA |
| M-FASN | TTGGCCTACACCCAGAGCTA | TTGTGGTAGAAGGACACGGC |
| M-β-actin | GGCTGTATTCCCCTCCATCG | CCAGTTGGTAACAATGCCATGT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, Z.; Li, X.; Bao, S.; Yamada, T.; Cao, G.; Liu, J.; Chen, A.; Tong, B. Effects of the CDC10 (Septin 7) Gene on the Proliferation and Differentiation of Bovine Intramuscular Preadipocyte and 3T3-L1 Cells. Animals 2023, 13, 609. https://doi.org/10.3390/ani13040609
Cheng Z, Li X, Bao S, Yamada T, Cao G, Liu J, Chen A, Tong B. Effects of the CDC10 (Septin 7) Gene on the Proliferation and Differentiation of Bovine Intramuscular Preadipocyte and 3T3-L1 Cells. Animals. 2023; 13(4):609. https://doi.org/10.3390/ani13040609
Chicago/Turabian StyleCheng, Zixuan, Xihe Li, Siqin Bao, Takahisa Yamada, Guifang Cao, Jianfeng Liu, Aorigele Chen, and Bin Tong. 2023. "Effects of the CDC10 (Septin 7) Gene on the Proliferation and Differentiation of Bovine Intramuscular Preadipocyte and 3T3-L1 Cells" Animals 13, no. 4: 609. https://doi.org/10.3390/ani13040609
APA StyleCheng, Z., Li, X., Bao, S., Yamada, T., Cao, G., Liu, J., Chen, A., & Tong, B. (2023). Effects of the CDC10 (Septin 7) Gene on the Proliferation and Differentiation of Bovine Intramuscular Preadipocyte and 3T3-L1 Cells. Animals, 13(4), 609. https://doi.org/10.3390/ani13040609





