Expression Profile of miRNA from High, Middle, and Low Stress-Responding Sheep during Bacterial Endotoxin Challenge
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Endotoxin Challenge and Sample Collection
2.2. miRNA Isolation and cDNA Synthesis
2.3. miRNA Expression Analysis
2.4. Target Gene Prediction and Pathway Analysis
2.5. Statistical Analysis
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Herman, J.P.; Figueiredo, H.; Mueller, N.K.; Ulrich-Lai, Y.; Ostrander, M.M.; Choi, D.C.; Cullinan, W.E. Central Mechanisms of Stress Integration: Hierarchical Circuitry Controlling Hypothalamo-Pituitary-Adrenocortical Responsiveness. Front. Neuroendocrinol. 2003, 24, 151–180. [Google Scholar] [CrossRef] [PubMed]
- Cockrem, J.F. Individual Variation in Glucocorticoid Stress Responses in Animals. Gen. Comp. Endocrinol. 2013, 181, 45–58. [Google Scholar] [CrossRef]
- Naylor, D.; Sharma, A.; Li, Z.; Monteith, G.; Sullivan, T.; Canovas, A.; Mallard, B.A.; Baes, C.; Karrow, N.A. Short Communication: Characterizing Ovine Serum Stress Biomarkers during Endotoxemia. J. Dairy Sci. 2020, 103, 5501–5508. [Google Scholar] [CrossRef] [PubMed]
- Pant, S.D.; You, Q.; Schenkel, L.C.; Voort, G.V.; Schenkel, F.S.; Wilton, J.; Cain, L.; Karrow, N.A. A Genome-Wide Association Study to Identify Chromosomal Regions Influencing Ovine Cortisol Response. Livest. Sci. 2016, 187, 40–47. [Google Scholar] [CrossRef]
- Larzul, C.; Terenina, E.; Foury, A.; Billon, Y.; Louveau, I.; Merlot, E.; Mormede, P. The Cortisol Response to ACTH in Pigs, Heritability and Influence of Corticosteroid-Binding Globulin. Animal 2015, 9, 1929–1934. [Google Scholar] [CrossRef]
- Overli, O.; Winberg, S.; Pottinger, T.G. Behavioral and Neuroendocrine Correlates of Selection for Stress Responsiveness in Rainbow Trout—A Review. Integr. Comp. Biol. 2005, 45, 463–474. [Google Scholar] [CrossRef]
- Herry, V.; Gitton, C.; Tabouret, G.; Répérant, M.; Forge, L.; Tasca, C.; Gilbert, F.B.; Guitton, E.; Barc, C.; Staub, C.; et al. Local Immunization Impacts the Response of Dairy Cows to Escherichia Coli Mastitis. Sci. Rep. 2017, 7, 3441. [Google Scholar] [CrossRef]
- Abaker, J.A.; Xu, T.L.; Jin, D.; Chang, G.J.; Zhang, K.; Shen, X.Z. Lipopolysaccharide Derived from the Digestive Tract Provokes Oxidative Stress in the Liver of Dairy Cows Fed a High-Grain Diet. J. Dairy Sci. 2017, 100, 666–678. [Google Scholar] [CrossRef]
- Cao, H.; Kabaroff, L.C.; You, Q.; Rodriguez, A.; Boermans, H.J.; Karrow, N.A. Characterization of Ovine Hepatic Gene Expression Profiles in Response to Escherichia coli Lipopolysaccharide Using a Bovine CDNA Microarray. BMC Vet. Res. 2006, 2, 34. [Google Scholar] [CrossRef]
- Chen, I.-Y.; Chang, S.C.; Wu, H.-Y.; Yu, T.-C.; Wei, W.-C.; Lin, S.; Chien, C.-L.; Chang, M.-F. Upregulation of the Chemokine (C-C Motif) Ligand 2 via a Severe Acute Respiratory Syndrome Coronavirus Spike-ACE2 Signaling Pathway. J. Virol. 2010, 84, 7703–7712. [Google Scholar] [CrossRef]
- Sharma, A.; Shandilya, U.K.; Sullivan, T.; Naylor, D.; Canovas, A.; Mallard, B.A.; Karrow, N.A. Identification of Ovine Serum MiRNAs Following Bacterial Lipopolysaccharide Challenge. Int. J. Mol. Sci. 2020, 21, 7920. [Google Scholar] [CrossRef] [PubMed]
- Naylor, D.; Sharma, A.; Li, Z.; Monteith, G.; Mallard, B.A.; Bergeron, R.; Baes, C.; Karrow, N.A. Endotoxin-Induced Cytokine, Chemokine and White Blood Cell Profiles of Variable Stress-Responding Sheep. Stress 2021, 24, 888–897. [Google Scholar] [CrossRef]
- Leung, A.K.L.; Sharp, P.A. MicroRNA Functions in Stress Responses. Mol. Cell 2010, 40, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Sengar, G.S.; Deb, R.; Singh, U.; Raja, T.V.; Kant, R.; Sajjanar, B.; Alex, R.; Alyethodi, R.R.; Kumar, A.; Kumar, S.; et al. Differential Expression of MicroRNAs Associated with Thermal Stress in Frieswal (Bos taurus x Bos indicus) Crossbred Dairy Cattle. Cell Stress Chaperones 2018, 23, 155–170. [Google Scholar] [CrossRef] [PubMed]
- Hashimi, S.T.; Fulcher, J.A.; Chang, M.H.; Gov, L.; Wang, S.; Lee, B. MicroRNA Profiling Identifies MiR-34a and MiR-21 and Their Target Genes JAG1 and WNT1 in the Coordinate Regulation of Dendritic Cell Differentiation. Blood 2009, 114, 404–414. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Ranade, K.; Talker, R.; Jallal, B.; Shen, N.; Yao, Y. MicroRNA-Mediated Regulation of Innate Immune Response in Rheumatic Diseases. Arthritis Res. 2013, 15, 210. [Google Scholar] [CrossRef] [PubMed]
- Javidi, M.A.; Ahmadi, A.H.; Bakhshinejad, B.; Nouraee, N.; Babashah, S.; Sadeghizadeh, M. Cell-Free MicroRNAs as Cancer Biomarkers: The Odyssey of MiRNAs through Body Fluids. Med. Oncol. 2014, 31, 295. [Google Scholar] [CrossRef]
- Arrese, M.; Eguchi, A.; Feldstein, A.E. Circulating MicroRNAs: Emerging Biomarkers of Liver Disease. Semin. Liver Dis. 2015, 35, 43–54. [Google Scholar] [CrossRef]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating MicroRNAs as Stable Blood-Based Markers for Cancer Detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef]
- Ortega, F.J.; Mercader, J.M.; Catalán, V.; Moreno-Navarrete, J.M.; Pueyo, N.; Sabater, M.; Gómez-Ambrosi, J.; Anglada, R.; Fernández-Formoso, J.A.; Ricart, W.; et al. Targeting the Circulating MicroRNA Signature of Obesity. Clin. Chem. 2013, 59, 781–792. [Google Scholar] [CrossRef]
- Yang, Z.; Chen, H.; Si, H.; Li, X.; Ding, X.; Sheng, Q.; Chen, P.; Zhang, H. Serum MiR-23a, a Potential Biomarker for Diagnosis of Pre-Diabetes and Type 2 Diabetes. Acta Diabetol. 2014, 51, 823–831. [Google Scholar] [CrossRef] [PubMed]
- Kalia, D.; Merey, G.; Nakayama, S.; Zheng, Y.; Zhou, J.; Luo, Y.; Guo, M.; Roembke, B.T.; Sintim, H.O. Nucleotide, c-Di-GMP, c-Di-AMP, CGMP, CAMP, (p)PpGpp Signaling in Bacteria and Implications in Pathogenesis. Chem. Soc. Rev. 2013, 42, 305–341. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- D’haene, B.; Mestdagh, P.; Hellemans, J.; Vandesompele, J. MiRNA Expression Profiling: From Reference Genes to Global Mean Normalization. Methods Mol. Biol. 2012, 822, 261–272. [Google Scholar] [CrossRef]
- Wu, J.; Cai, H.; Xiang, Y.-B.; Matthews, C.E.; Ye, F.; Zheng, W.; Cai, Q.; Shu, X.-O. Intra-Individual Variation of MiRNA Expression Levels in Human Plasma Samples. Biomarkers 2018, 23, 339–346. [Google Scholar] [CrossRef]
- Kucherenko, M.M.; Shcherbata, H.R. MiRNA Targeting and Alternative Splicing in the Stress Response—Events Hosted by Membrane-Less Compartments. J. Cell Sci. 2018, 131, jcs202002. [Google Scholar] [CrossRef]
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. MiRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells 2020, 9, 276. [Google Scholar] [CrossRef]
- Solé, C.; Domingo, S.; Ferrer, B.; Moliné, T.; Ordi-Ros, J.; Cortés-Hernández, J. MicroRNA Expression Profiling Identifies MiR-31 and MiR-485-3p as Regulators in the Pathogenesis of Discoid Cutaneous Lupus. J. Investig. Dermatol. 2019, 139, 51–61. [Google Scholar] [CrossRef]
- Han, J.M.; Patterson, S.J.; Levings, M.K. The Role of the PI3K Signaling Pathway in CD4+ T Cell Differentiation and Function. Front. Immunol. 2012, 3, 245. [Google Scholar] [CrossRef] [PubMed]
- Shibata, N.; Glass, C.K. Regulation of Macrophage Function in Inflammation and Atherosclerosis. J. Lipid Res. 2009, 50, S277–S281. [Google Scholar] [CrossRef]
- Sun, F.; Li, S.-G.; Zhang, H.-W.; Hua, F.-W.; Sun, G.-Z.; Huang, Z. MiRNA-411 Attenuates Inflammatory Damage and Apoptosis Following Spinal Cord Injury. Eur. Rev. Med. Pharm. Sci. 2020, 24, 491–498. [Google Scholar] [CrossRef]
- Vasilescu, C.; Dragomir, M.; Tanase, M.; Giza, D.; Purnichescu-Purtan, R.; Chen, M.; Yeung, S.-C.J.; Calin, G.A. Circulating MiRNAs in Sepsis-A Network under Attack: An in-Silico Prediction of the Potential Existence of MiRNA Sponges in Sepsis. PLoS ONE 2017, 12, e0183334. [Google Scholar] [CrossRef]
- Dumache, R.; Rogobete, A.F.; Bedreag, O.H.; Sarandan, M.; Cradigati, A.C.; Papurica, M.; Dumbuleu, C.M.; Nartita, R.; Sandesc, D. Use of MiRNAs as Biomarkers in Sepsis. Anal. Cell. Pathol. 2015, 2015, 186716. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Zhang, C.-L.; Liao, X.-X.; Chen, D.; Wang, W.-Q.; Zhu, Y.-H.; Geng, X.-H.; Ji, D.-J.; Mao, Y.-J.; Gong, Y.-C.; et al. Transcriptome MicroRNA Profiling of Bovine Mammary Glands Infected with Staphylococcus Aureus. Int. J. Mol. Sci. 2015, 16, 4997–5013. [Google Scholar] [CrossRef] [PubMed]
- Luoreng, Z.-M.; Wang, X.-P.; Mei, C.-G.; Zan, L.-S. Comparison of MicroRNA Profiles between Bovine Mammary Glands Infected with Staphylococcus aureus and Escherichia coli. Int. J. Biol. Sci. 2018, 14, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Reid, G.; Kirschner, M.B.; van Zandwijk, N. Circulating MicroRNAs: Association with Disease and Potential Use as Biomarkers. Crit. Rev. Oncol. Hematol. 2011, 80, 193–208. [Google Scholar] [CrossRef]
- Hu, Y.; Yang, C.; Yang, S.; Cheng, F.; Rao, J.; Wang, X. MiR-665 Promotes Hepatocellular Carcinoma Cell Migration, Invasion, and Proliferation by Decreasing Hippo Signaling through Targeting PTPRB. Cell Death Dis. 2018, 9, 954. [Google Scholar] [CrossRef]
- Rottiers, V.; Näär, A.M. MicroRNAs in Metabolism and Metabolic Disorders. Nat. Rev. Mol. Cell Biol. 2012, 13, 239–250. [Google Scholar] [CrossRef]
- Croston, T.L.; Nayak, A.P.; Lemons, A.R.; Goldsmith, W.T.; Gu, J.K.; Germolec, D.R.; Beezhold, D.H.; Green, B.J. Influence of Aspergillus Fumigatus Conidia Viability on Murine Pulmonary MicroRNA and MRNA Expression Following Subchronic Inhalation Exposure. Clin. Exp. Allergy 2016, 46, 1315–1327. [Google Scholar] [CrossRef]
- Melnik, B.C. Milk: An Epigenetic Amplifier of FTO-Mediated Transcription? Implications for Western Diseases. J. Transl. Med. 2015, 13, 385. [Google Scholar] [CrossRef]
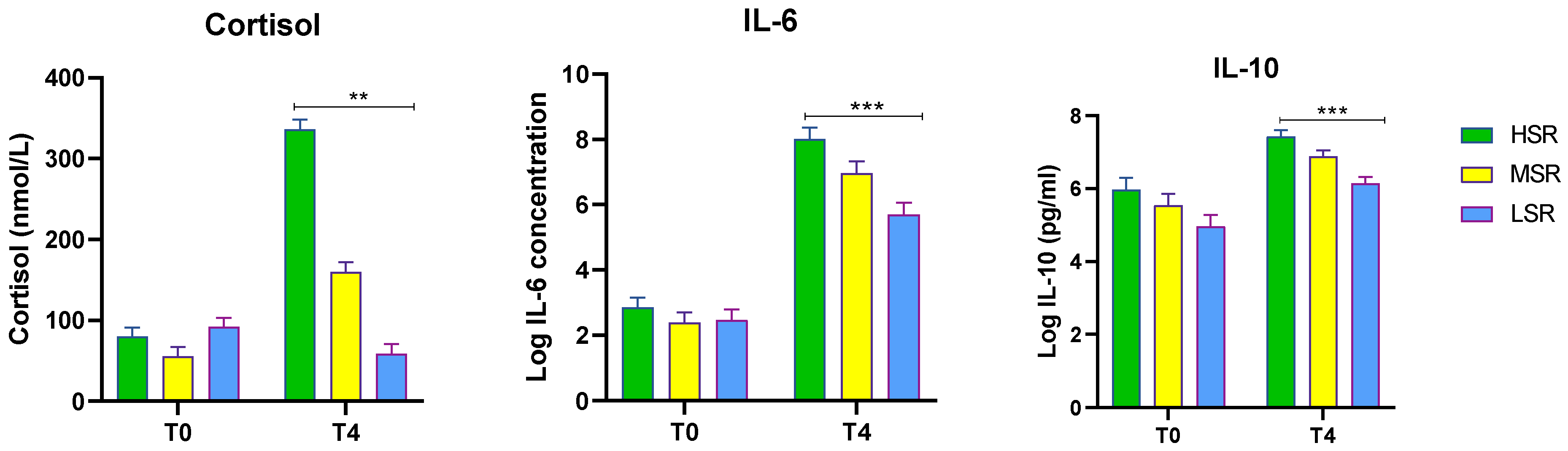
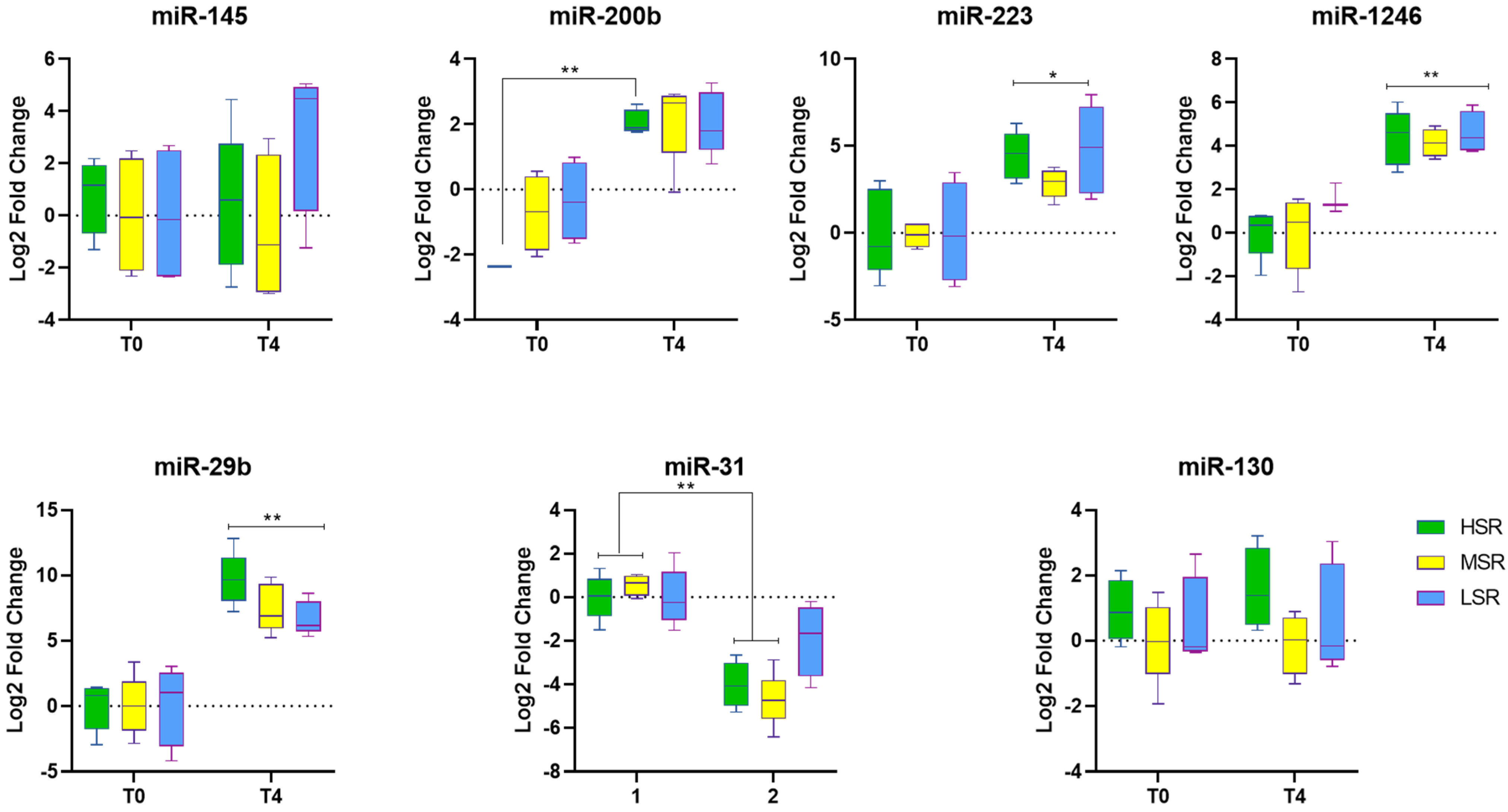
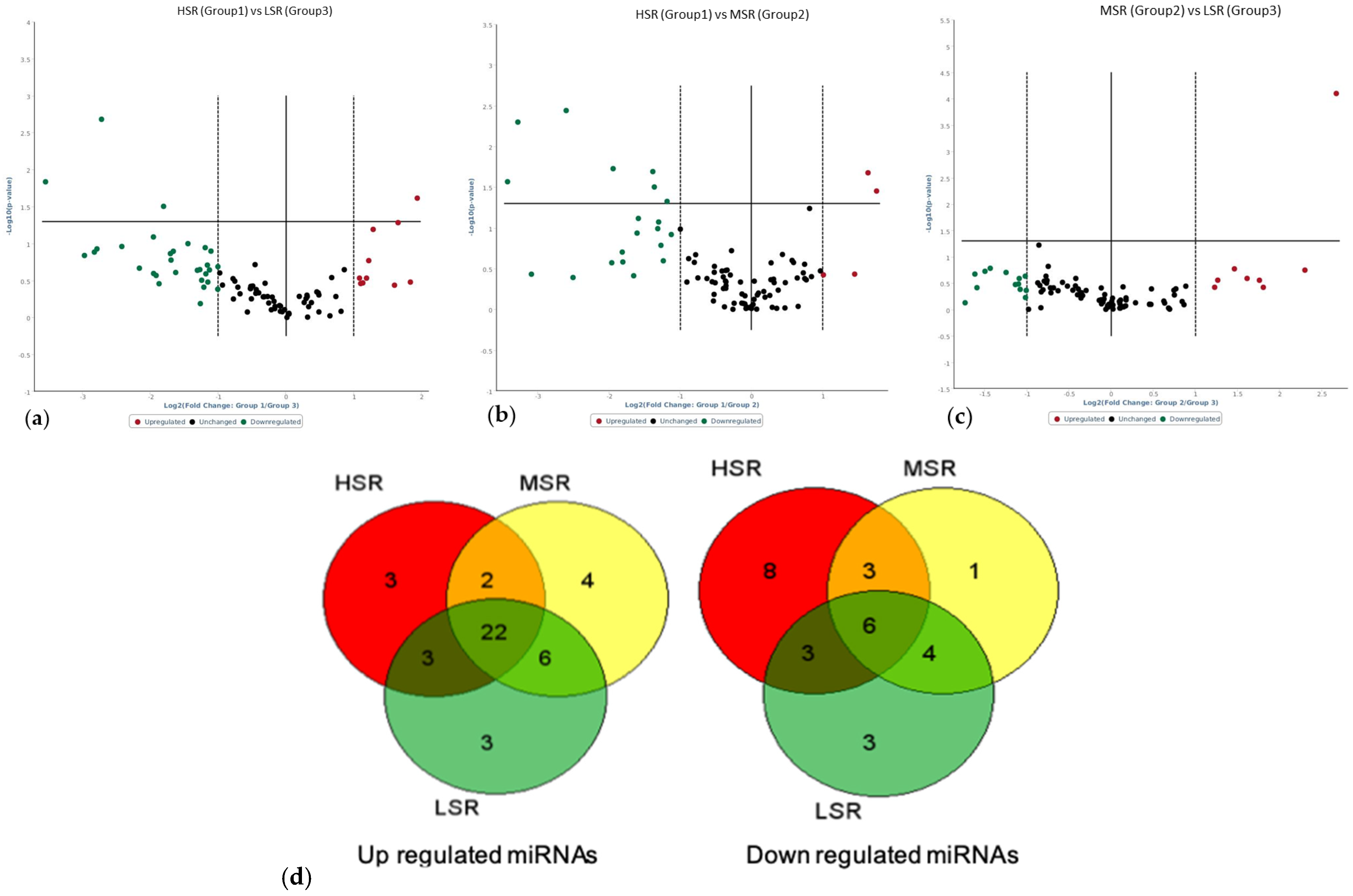
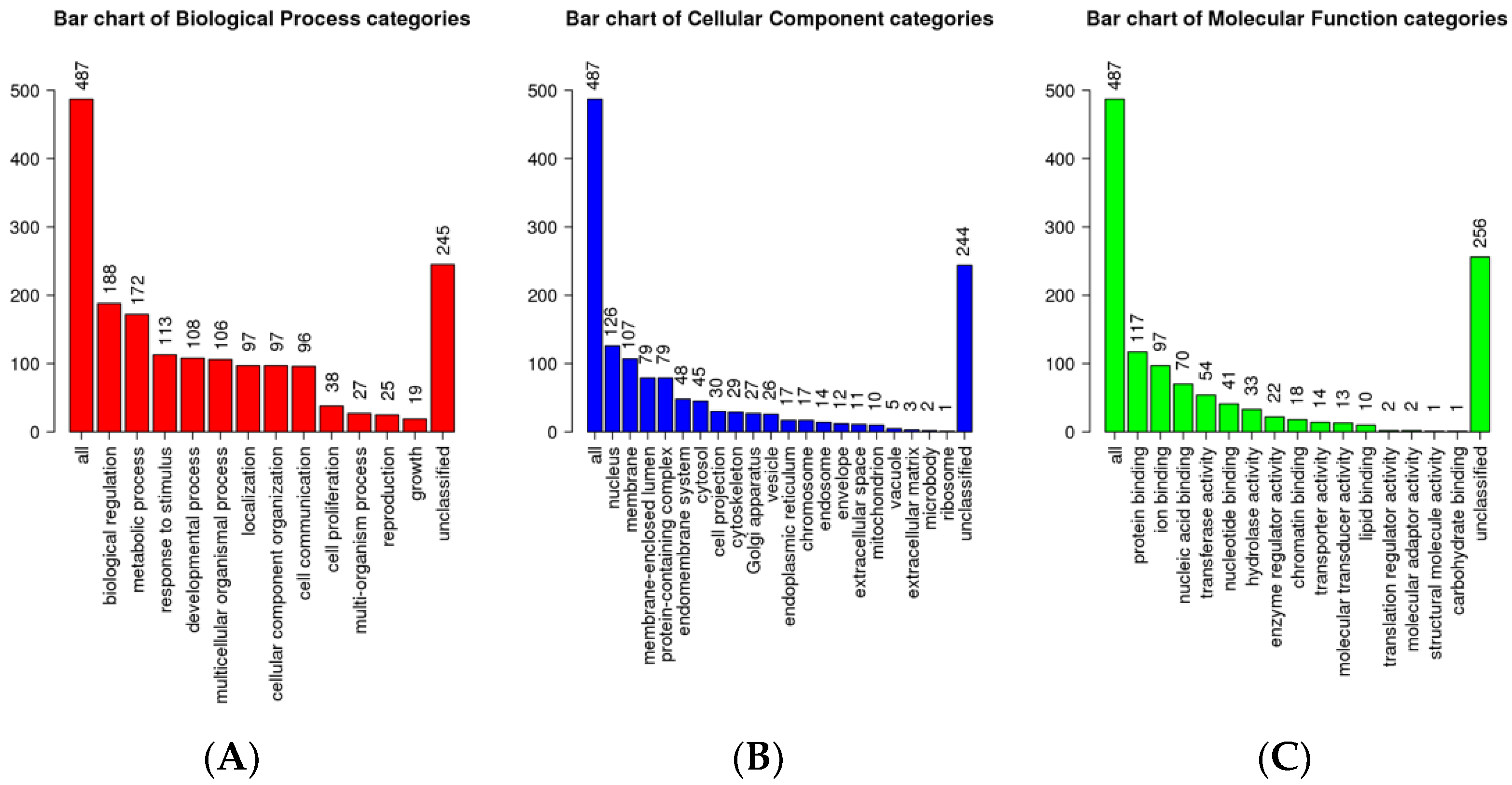
| S.No | HSR Log FC | MSR Log FC | LSR Log FC | |
|---|---|---|---|---|
| Upregulated miRNAs | ||||
| 1 | oar-miR-485-3p | 24.894 (0.009) | 7.367 (0.034) | 6.518 (0.0070) |
| 2 | oar-miR-543-3p | 22.743 (0.028) | 23.775 (0.007) | 22.089 (0.022) |
| 3 | oar-miR-655-3p | 6.601 (0.033) | 8.480 (0.0009) | 14.236 (0.020) |
| 4 | oar-miR-3957-5p | 4.631 (0.015) | 5.156 (0.359) | 1.4751 (0.344) |
| 5 | oar-miR-329b-3p | 2.402 (0.051) | 1.777 (0.169) | 5.456 (0.172) |
| 6 | oar-miR-369-3p | 13.139 (0.393) | 32.476 (0.047) | 35.838 (0.05) |
| 7 | oar-miR-411a-5p | 4.871 (0.087) | 9.739 (0.026) | 7.021 (0.158) |
| 8 | oar-miR-411a-3p | 4.504 (0.110) | 6.420 (0.023) | 10.881 (0.132) |
| 9 | oar-miR-487b-3p | 5.588 (0.098) | 7.206 (0.023) | 9.443 (0.148) |
| 10 | oar-miR-758-3p | 1.107 (0.630) | 3.332 (0.001) | −1.912 (0.092) |
| 11 | oar-miR-668-3p | 8.781 (0.08) | 5.237 (0.05) | 4.847 (0.05) |
| 12 | oar-miR-376c-3p | 1.329 (0.90) | 3.475 (0.090 | 4.650 (0.05) |
| 13 | oar-miR-381-3p | −1.668 (0.466) | 2.308 (0.05) | −1.041 (0.810) |
| Downregulated miRNAs | ||||
| 14 | oar-miR-665-5p | −2.015 (0.014) | −3.547 (0.003 | −2.074 (0.05) |
| 15 | oar-miR-379-5p | −2.034 (0.039) | −1.603 (0.121) | −1.480 (0.147) |
| 16 | oar-miR-154b-5p | −1.966 (0.057) | −1.964 (0.050 | −1.837 (0.097) |
| 17 | oar-miR-3958-5p | 1.045 (0.524) | −2.612 (0.0660 | −2.896 (0.068) |
| 18 | oar-miR-496-5p | −1.636 (0.23) | −2.122 (0.05) | −2.030 (0.0310 |
| 19 | oar-miR-329b-5p | −1.180 (0.993) | 1.003 (0.808) | −2.745 (0.050) |
| HSR vs. LSR | HSR vs. MSR | MSR vs. LSR | |
|---|---|---|---|
| Upregulated | oar-miR-485-3p (3.82, 0.0243) oar-miR-1193-5p (2.43, 0.064) oar-miR-3957-5p (3.14, 0.052) | oar-miR-485-3p (3.38, 0.0352) oar-miR-1193-5p (3.11, 0.021) | oar-miR-758-3p (6.37, 0.0000794) |
| Downregulated | oar-miR-376b-3p (−6.6, 0.0020) oar-miR-376c-3p (−3.5, 0.0313) oar-miR-411b-5p (−11.69, 0.014) | oar-miR-376a-3p (−2.28, 0.047) oar-miR-376b-3p (−6.08, 0.003) oar-miR-381-3p (−3.85, 0.018) oar-miR-758-3p (−3.01, 0.076) |
| S. No | Term | Corrected p-Value (FDR) | Database |
|---|---|---|---|
| Pathways enriched in all treated groups compared to control | |||
| 1 | MAPK signaling pathway | 0.00000000775 | KEGG PATHWAY |
| 2 | Signaling pathways regulating pluripotency of stem cells | 0.0000364 | KEGG PATHWAY |
| 3 | EGFR tyrosine kinase inhibitor resistance | 0.0000542 | KEGG PATHWAY |
| 4 | PI3K-Akt signaling pathway | 0.0000674 | KEGG PATHWAY |
| 5 | Longevity regulating pathway | 0.00012941 | KEGG PATHWAY |
| 6 | Ras signaling pathway | 0.00014182 | KEGG PATHWAY |
| 7 | Longevity regulating pathway—multiple species | 0.0003686 | KEGG PATHWAY |
| 8 | Autophagy—animal | 0.00051481 | KEGG PATHWAY |
| 9 | AMPK signaling pathway | 0.00120604 | KEGG PATHWAY |
| 10 | mTOR signaling pathway | 0.00154106 | KEGG PATHWAY |
| Enriched pathways specific to HSR group | |||
| 1 | Gene expression (transcription) | 0.00256736 | Reactome |
| 2 | Immune system | 0.00740769 | Reactome |
| 3 | Ras signaling pathway | 0.0105625 | KEGG PATHWAY |
| 4 | Signal transduction | 0.02158305 | Reactome |
| 5 | MAPK signaling pathway | 0.02705895 | KEGG PATHWAY |
| 6 | Adaptive immune system | 0.03477747 | Reactome |
| 7 | Cytokine signaling in immune system | 0.00232352 | Reactome |
| Enriched pathways specific to LSR group | |||
| 8 | Metabolic pathways | 0.01151519 | KEGG PATHWAY |
| 9 | Glucagon signaling in metabolic regulation | 0.04716364 | Reactome |
| 10 | Transport of inorganic cations/anions and amino acids/oligopeptides | 0.06204974 | Reactome |
| 11 | Integration of energy metabolism | 0.06204974 | Reactome |
| 12 | Post-translational protein modification | 0.07675048 | Reactome |
| 13 | Longevity regulating pathway | 0.07740826 | Reactome |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shandilya, U.K.; Sharma, A.; Naylor, D.; Canovas, A.; Mallard, B.; Karrow, N.A. Expression Profile of miRNA from High, Middle, and Low Stress-Responding Sheep during Bacterial Endotoxin Challenge. Animals 2023, 13, 508. https://doi.org/10.3390/ani13030508
Shandilya UK, Sharma A, Naylor D, Canovas A, Mallard B, Karrow NA. Expression Profile of miRNA from High, Middle, and Low Stress-Responding Sheep during Bacterial Endotoxin Challenge. Animals. 2023; 13(3):508. https://doi.org/10.3390/ani13030508
Chicago/Turabian StyleShandilya, Umesh K., Ankita Sharma, Danielle Naylor, Angela Canovas, Bonnie Mallard, and Niel A. Karrow. 2023. "Expression Profile of miRNA from High, Middle, and Low Stress-Responding Sheep during Bacterial Endotoxin Challenge" Animals 13, no. 3: 508. https://doi.org/10.3390/ani13030508
APA StyleShandilya, U. K., Sharma, A., Naylor, D., Canovas, A., Mallard, B., & Karrow, N. A. (2023). Expression Profile of miRNA from High, Middle, and Low Stress-Responding Sheep during Bacterial Endotoxin Challenge. Animals, 13(3), 508. https://doi.org/10.3390/ani13030508







