Inclusion of Camelina sativa Seeds in Ewes’ Diet Modifies Rumen Microbiota
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Diets and Experimental Design
2.2. Sample Collection
2.3. Fatty Acid Determination
2.4. DNA Extraction
2.5. Metagenomic NGS Analysis
2.6. RT-qPCR Platform for Selected Rumen Microorganisms
2.7. Statistical Analysis
3. Results
3.1. Rumen Fatty Acid Profile
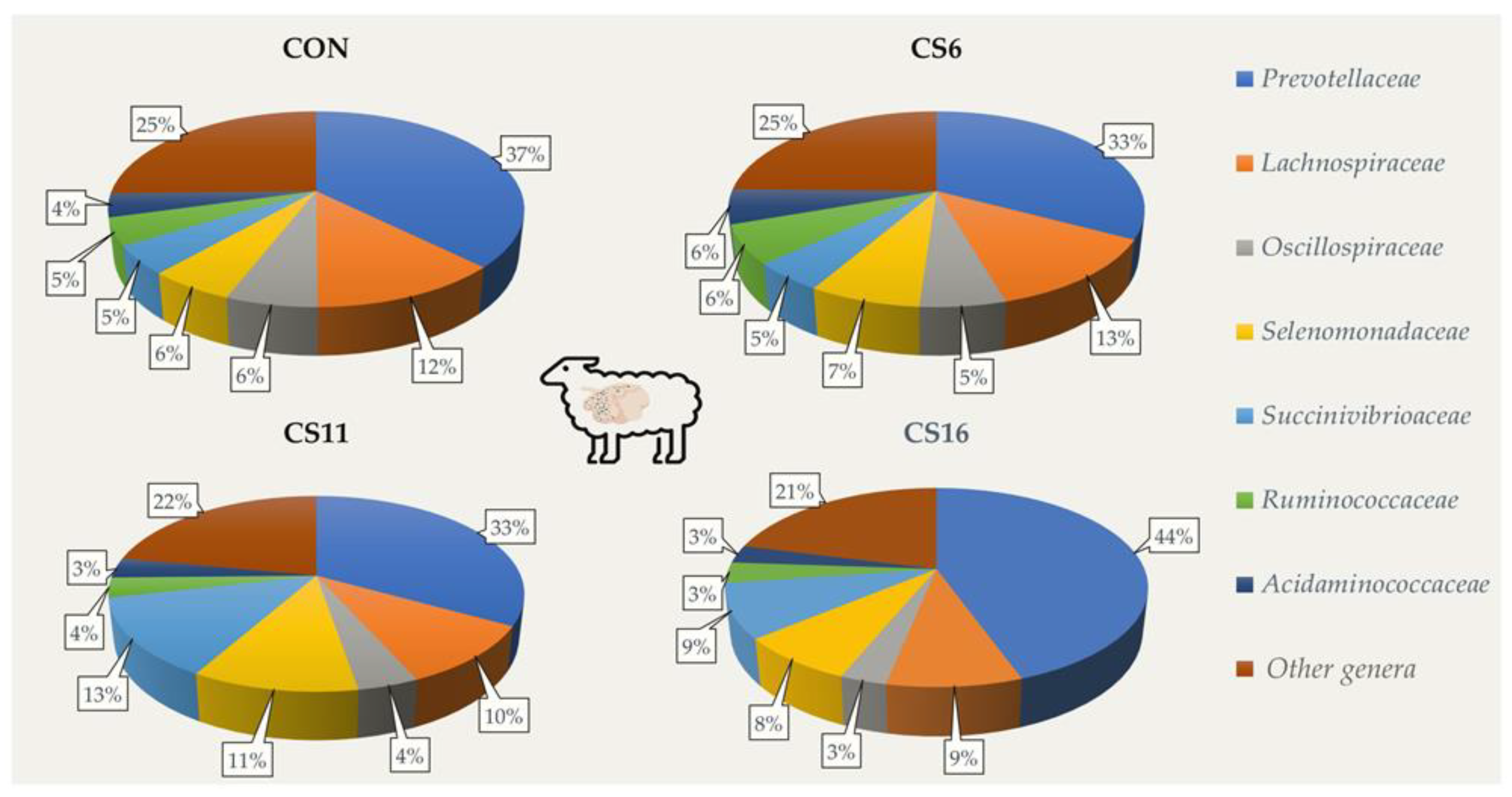
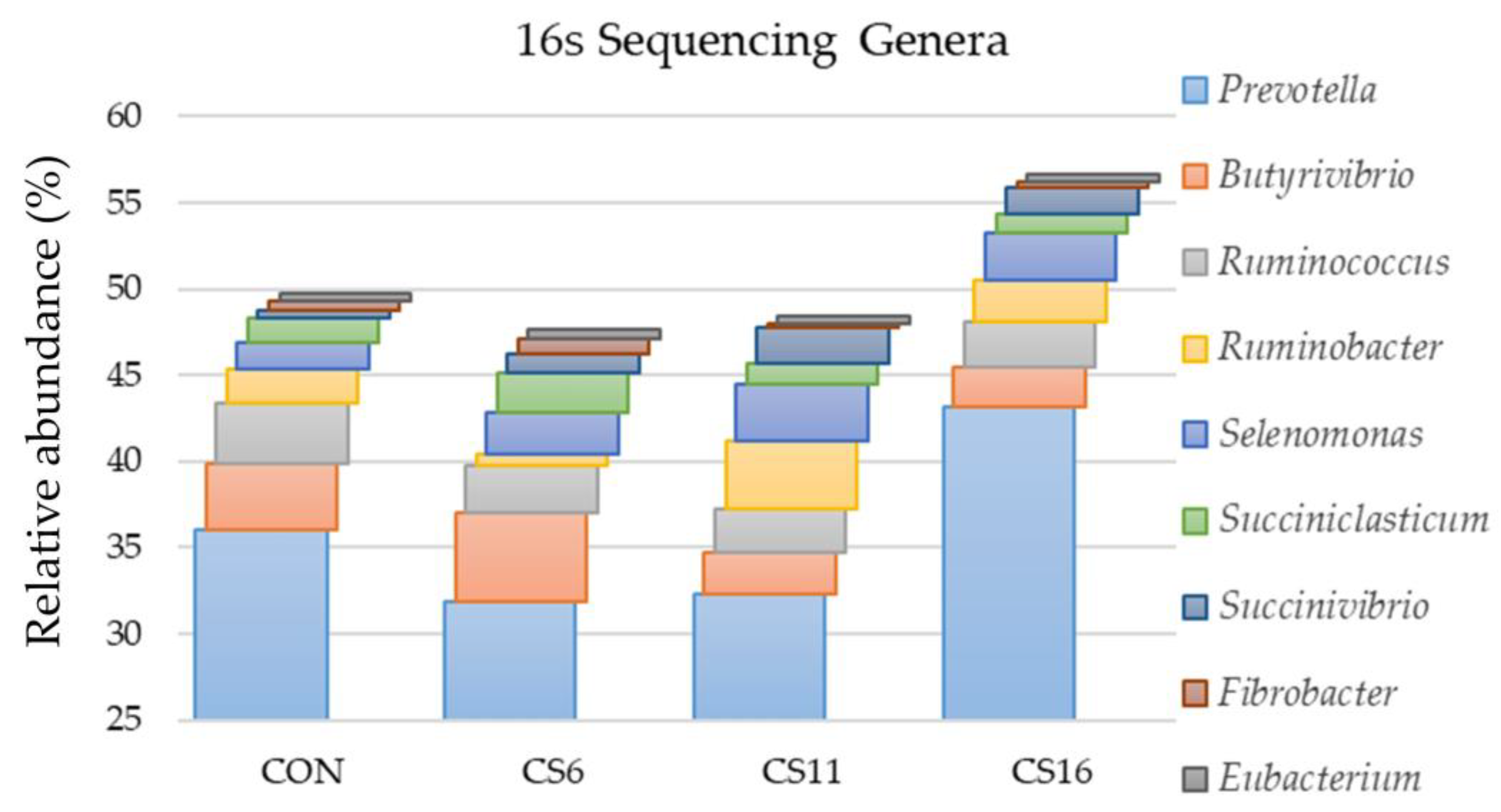
3.2. 16S Amplicon Sequencing
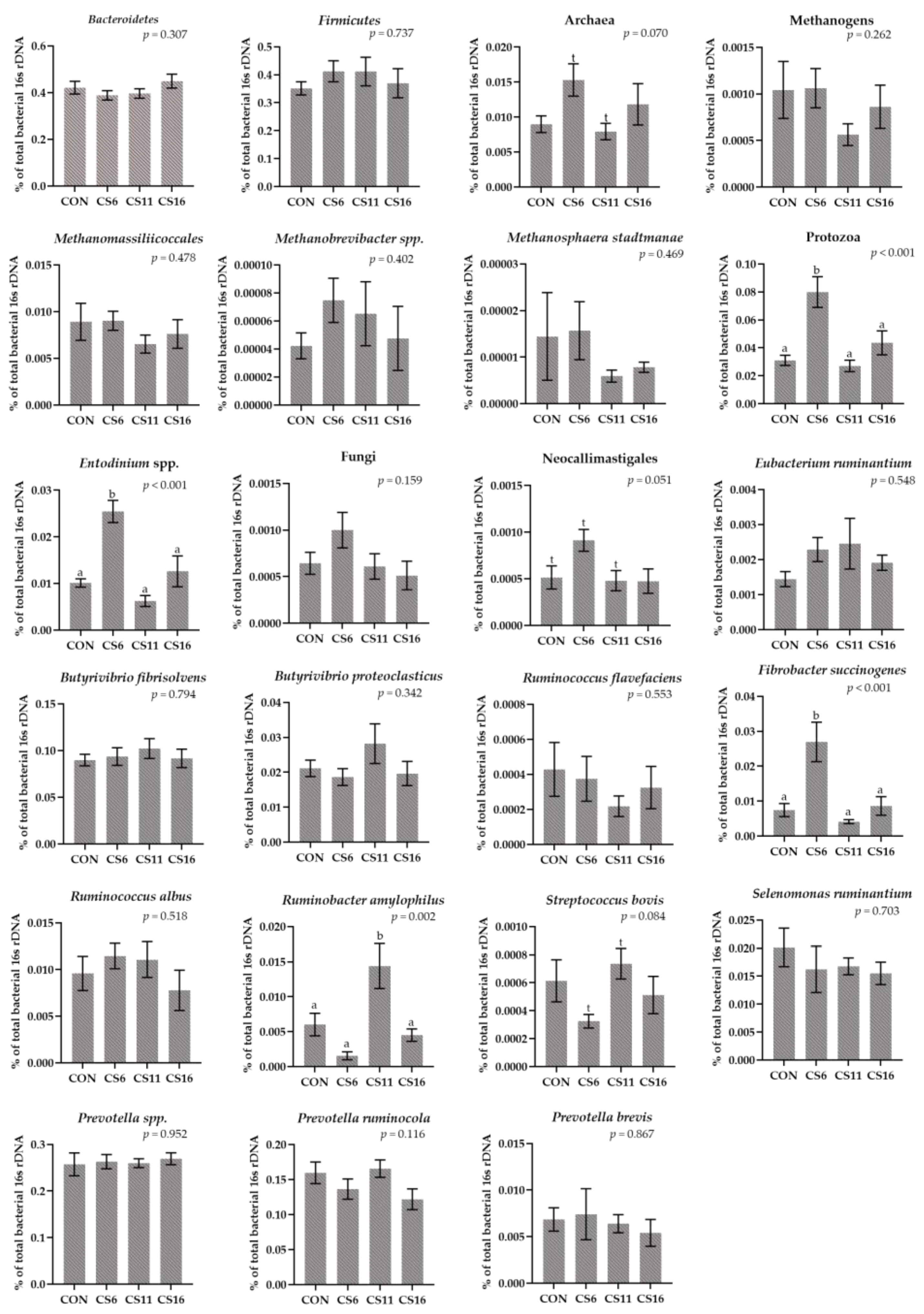
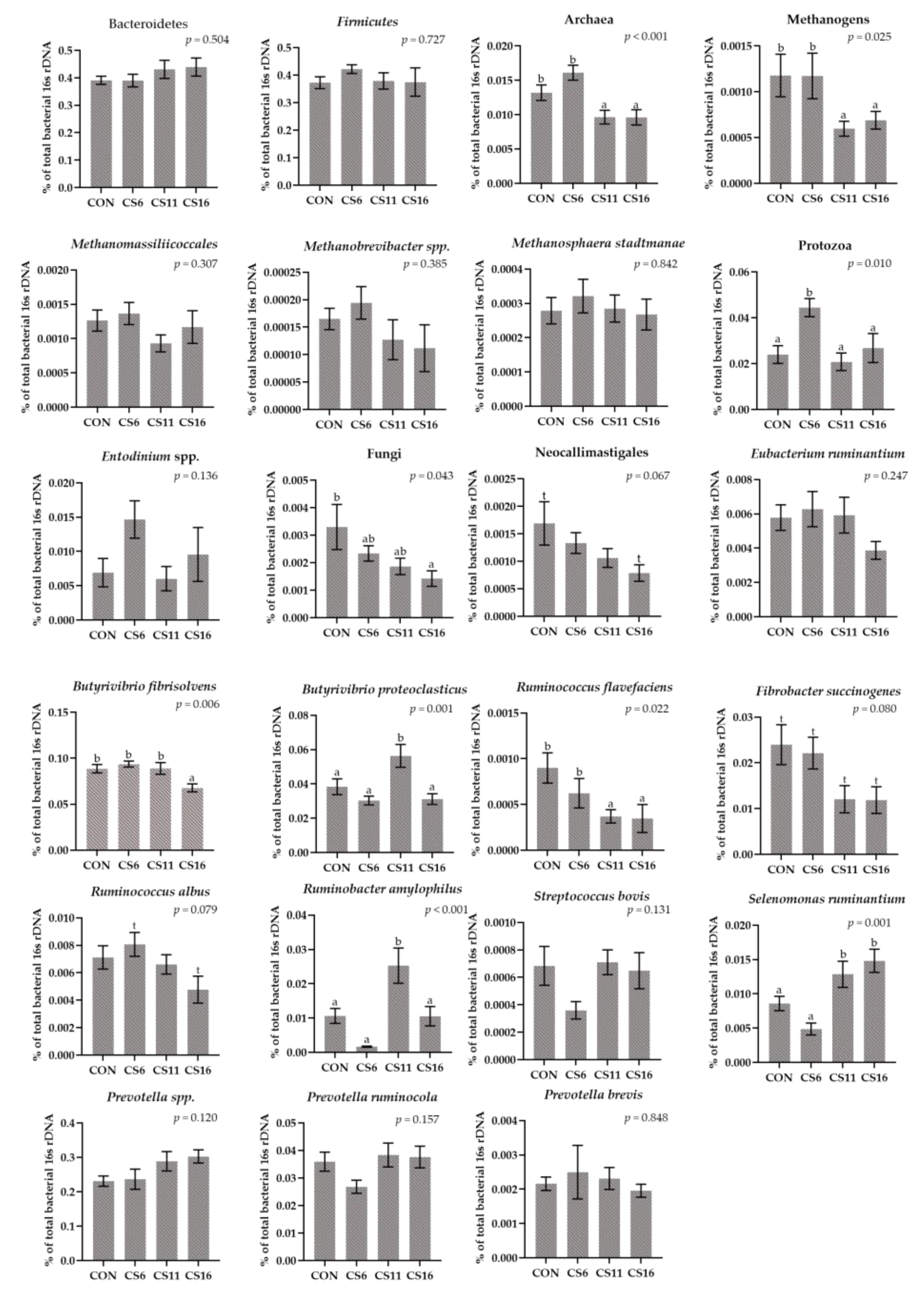
3.3. Relative Abundance of Selected Microorganisms in the Rumen Fluid Samples Using a RT-qPCR Platform
3.4. Relative Abundance of Selected Microorganisms in the Rumen Solid Particle Using RT-qPCR Platform
4. Discussion
4.1. The Effect of Camelina Sativa Seeds Supplementation in Rumen Fatty Acids Profile
4.2. The Camelina Sativa Seeds Supplementation Demonstrated Important Findings in the Modification of the Rumen Microorganisms
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Huws, S.A.; Creevey, C.J.; Oyama, L.B.; Mizrahi, I.; Denman, S.E.; Popova, M.; Muñoz-Tamayo, R.; Forano, E.; Waters, S.M.; Hess, M.; et al. Addressing global ruminant agricultural challenges through understanding the rumen microbiome: Past, present, and future. Front. Microbiol. 2018, 9, 2161. [Google Scholar] [CrossRef] [PubMed]
- Johnson, K.A.; Johnson, D.E. Methane emissions from cattle. J. Anim. Sci. 1995, 73, 2483–2492. [Google Scholar] [CrossRef] [PubMed]
- Food and Agriculture Organization of the United Nations (FAO). Global Perspective Studies—Food and Agriculture Projections to 2050. 2018. Available online: http://www.fao.org/global-perspectives-studies/food-agriculture-projections-to-2050/en/ (accessed on 6 December 2022).
- Nguyen, Q.V.; Malau-Aduli, B.S.; Cavalieri, J.; Malau-Aduli, A.E.O.; Nichols, P.D. Enhancing Omega-3 Long-Chain Polyunsaturated Fatty Acid Content of Dairy-Derived Foods for Human Consumption. Nutrients 2019, 11, 743. [Google Scholar] [CrossRef] [PubMed]
- Höglund-Isaksson, L.; Gómez-Sanabria, A.; Klimont, Z.; Rafaj, P.; Schöpp, W. Technical potentials and costs for reducing global anthropogenic methane emissions in the 2050 timeframe—Results from the gains model. Environ. Res. Commun. 2020, 2. [Google Scholar] [CrossRef]
- Bodas, R.; Prieto, N.; García-González, R.; Andrés, S.; Giráldez, F.J.; López, S. Manipulation of rumen fermentation and methane production with plant secondary metabolites. Anim. Feed Sci. Technol. 2012, 176, 78–93. [Google Scholar] [CrossRef]
- Ebrahimi, M. Production of Omega-3 Polyunsaturated Fatty Acids Enriched Choven Using Treated Oil Palm (Elaeis guineensis jacq) Fronds. Ph.D. Thesis, Universiti Putra Malaysia, Serdang, Malaysia, 2012. [Google Scholar]
- Abuelfatah, K.; Zuki, A.B.; Goh, Y.M.; Sazili, A.Q.; Abubakr, A. Effects of feeding whole linseed on ruminal fatty acid composition and microbial population in goats. Anim. Nutr. 2016, 2, 323–328. [Google Scholar] [CrossRef]
- Zhang, C.M.; Guo, Y.Q.; Yuan, Z.P.; Wu, Y.M.; Wang, J.K.; Liu, J.X.; Zhu, W.Y. Effect of octadeca carbon fatty acids on microbial fermentation, methanogenesis and microbial flora in vitro. Anim. Feed Sci. Technol. 2008, 146, 259–269. [Google Scholar] [CrossRef]
- Ivan, M.; Petit, H.V.; Chiquette, J.; Wright, A.D.G. Rumen fermentation and microbial population in lactating dairy cows receiving diets containing oilseeds rich in C-18 fatty acids. Br. J. Nutr. 2013, 109, 1211–1218. [Google Scholar] [CrossRef]
- Zhou, X.; Meile, L.; Kreuzer, M.; Zeitz, J.O. The effect of saturated fatty acids on methanogenesis and cell viability of methanobrevibacter ruminantium. Archaea 2013, 2013, 106916. [Google Scholar] [CrossRef]
- Liu, S.J.; Bu, D.P.; Wang, J.Q.; Liu, L.; Liang, S.; Wei, H.Y.; Zhou, L.Y.; Li, D.; Loor, J.J. Effect of incremental levels of fish oil supplementation on specific bacterial populations in bovine ruminal fluid. J. Anim. Physiol. Anim. Nutr. 2012, 96, 9–16. [Google Scholar] [CrossRef]
- Ebrahimi, M.; Rajion, M.A.; Adeyemi, K.D.; Jafari, S.; Jahromi, M.F.; Oskoueian, E. Dietary n-6:n-3 fatty acid ratios alter rumen fermentation parameters and microbial populations in goats. J. Agric. Food Chem. 2017, 65, 737–744. [Google Scholar] [CrossRef] [PubMed]
- Hurtaud, C.; Peyraud, J.L. Effects of feeding camelina (seeds or meal) on milk fatty acid composition and butter spreadability. J. Dairy Sci. 2007, 90, 5134–5145. [Google Scholar] [CrossRef] [PubMed]
- Zubr, J.; Matthaus, B. Effects of growth conditions on fatty acids and tocopherols in Camelina sativa oil. Ind. Crops Prod. 2002, 690, 155–162. [Google Scholar] [CrossRef]
- Berhow, M.; Vaughn, S.; Moser, B.; Belenli, D.; Polat, U. Evaluating the Phytochemical Potential of Camelina: An Emerging New Crop of Old World Origin. In Phytochemicals—Biosynthesis, Function and Application; Recent Advances in Phytochemistry 44; Jetter, R., Ed.; Springer: New York, NY, USA, 2014; pp. 129–148. ISBN 978-3-319-04045-5. [Google Scholar] [CrossRef]
- Bayat, A.R.; Kairenius, P.; Stefański, T.; Leskinen, H.; Comtet-Marre, S.; Forano, E.; Chaucheyras-Durand, F.; Shingfield, K.J. Effect of camelina oil or live yeasts (Saccharomyces cerevisiae) on ruminal methane production, Rumen fermentation, And milk fatty acid composition in lactating cows fed grass silage diets. J. Dairy Sci. 2015, 98, 3166–3181. [Google Scholar] [CrossRef] [PubMed]
- Ebeid, H.M.; Hassan, F.U.; Li, M.; Peng, L.; Peng, K.; Liang, X.; Yang, C. Camelina sativa L. Oil Mitigates Enteric in vitro Methane Production, Modulates Ruminal Fermentation, and Ruminal Bacterial Diversity in Buffaloes. Front. Vet. Sci. 2020, 7, 550. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Weimer, P.J.; Dill-McFarland, K.A.; Brandao, V.L.N.; Suen, G.; Faciola, A.P. Camelina seed supplementation at two dietary fat levels change ruminal bacterial community composition in a dual-flow continuous culture system. Front. Microbiol. 2017, 8, 2147. [Google Scholar] [CrossRef] [PubMed]
- Mould, F.L.; Kliem, K.E.; Morgan, R.; Mauricio, R.M. In vitro microbial inoculum: A review of its function and properties. Anim. Feed Sci. Technol. 2005, 123–124 Pt 1, 31–50. [Google Scholar] [CrossRef]
- Ramos-Morales, E.; Arco-Pérez, A.; Martín-García, A.I.; Yáñez-Ruiz, D.R.; Frutos, P.; Hervás, G. Use of stomach tubing as an alternative to rumen cannulation to study ruminal fermentation and microbiota in sheep and goats. Anim. Feed Sci. Technol. 2014, 198, 57–66. [Google Scholar] [CrossRef]
- O’Fallon, J.V.; Busboom, J.R.; Nelson, M.L.; Gaskins, C.T. A direct method for fatty acid methyl ester synthesis: Application to wet meat tissues, oils, and feedstuffs. J. Anim. Sci. 2007, 85, 1511–1521. [Google Scholar] [CrossRef]
- Christodoulou, C.; Mavrommatis, A.; Mitsiopoulou, C.; Symeon, G.; Dotas, V.; Sotirakoglou, K.; Kotsampasi, B.; Tsiplakou, E. Assessing the optimum level of supplementation with camelina seeds in ewes’ diets to improve milk quality. Foods 2021, 10, 2076. [Google Scholar] [CrossRef]
- Mavrommatis, A.; Skliros, D.; Sotirakoglou, K.; Flemetakis, E.; Tsiplakou, E. The effect of forage-to-concentrate ratio on schizochytrium spp.-supplemented goats: Modifying rumen microbiota. Animals 2021, 11, 2746. [Google Scholar] [CrossRef] [PubMed]
- Carberry, C.A.; Kenny, D.A.; Han, S.; McCabe, M.S.; Waters, S.M. Effect of phenotypic residual feed intake and dietary forage content on the rumen microbial community of beef cattle. Appl. Environ. Microbiol. 2012, 78, 4949–4958. [Google Scholar] [CrossRef] [PubMed]
- Denman, S.E.; McSweeney, C.S. Development of a real-time PCR assay for monitoring anaerobic fungal and cellulolytic bacterial populations within the rumen. FEMS Microbiol. Ecol. 2006, 58, 572–582. [Google Scholar] [CrossRef] [PubMed]
- Sereika, M.; Kirkegaard, R.H.; Karst, S.M.; Michaelsen, T.Y.; Sørensen, E.A.; Wollenberg, R.D.; Albertsen, M. Oxford Nanopore R10.4 long-read sequencing enables the generation of near-finished bacterial genomes from pure cultures and metagenomes without short-read or reference polishing. Nat. Methods 2022, 19, 823–826. [Google Scholar] [CrossRef]
- Loor, J.J.; Bandara, A.B.P.A.; Herbein, J.H. Characterization of 18:1 and 18:2 isomers produced during microbial biohydrogenation of unsaturated fatty acids from canola and soya bean oil in the rumen of lactating cows. J. Anim. Physiol. Anim. Nutr. 2002, 86, 422–432. [Google Scholar] [CrossRef]
- Jami, E.; Mizrahi, I. Composition and similarity of bovine rumen microbiota across individual animals. PLoS ONE 2012, 7, e33306. [Google Scholar] [CrossRef]
- Jami, E.; White, B.A.; Mizrahi, I. Potential role of the bovine rumen microbiome in modulating milk composition and feed efficiency. PLoS ONE 2014, 9, e85423. [Google Scholar] [CrossRef]
- Luton, P.E.; Wayne, J.M.; Sharp, R.J.; Riley, P.W. The mcrA gene as an alternative to 16S rRNA in the phylogenetic analysis of methanogen populations in landfill. Microbiology 2002, 148, 3521–3530. [Google Scholar] [CrossRef]
- Patra, A.; Park, T.; Kim, M.; Yu, Z. Rumen methanogens and mitigation of methane emission by anti-methanogenic compounds and substances. J. Anim. Sci. Biotechnol. 2017, 8, 13. [Google Scholar] [CrossRef]
- Soliva, C.R.; Meile, L.; Hindrichsen, I.K.; Kreuzer, M.; Machmüller, A. Myristic acid supports the immediate inhibitory effect of lauric acid on ruminal methanogens and methane release. Anaerobe 2004, 10, 269–276. [Google Scholar] [CrossRef]
- Moate, P.J.; Williams, S.R.O.; Hannah, M.C.; Eckard, R.J.; Auldist, M.J.; Ribaux, B.E.; Jacobs, J.L.; Wales, W.J. Effects of feeding algal meal high in docosahexaenoic acid on feed intake, milk production, and methane emissions in dairy cows. J. Dairy Sci. 2013, 96, 3177–3188. [Google Scholar] [CrossRef] [PubMed]
- Patra, A.K.; Yu, Z. Effects of coconut and fish oils on ruminal methanogenesis, Fermentation, And abundance and diversity of microbial populations in vitro. J. Dairy Sci. 2013, 96, 1782–1792. [Google Scholar] [CrossRef] [PubMed]
- Lillis, L.; Boots, B.; Kenny, D.A.; Petrie, K.; Boland, T.M.; Clipson, N.; Doyle, E.M. The effect of dietary concentrate and soya oil inclusion on microbial diversity in the rumen of cattle. J. Appl. Microbiol. 2011, 111, 1426–1435. [Google Scholar] [CrossRef]
- Yuyama, K.T.; Rohde, M.; Molinari, G.; Stadler, M.; Abraham, W.R. Unsaturated fatty acids control biofilm formation of staphylococcus aureus and other gram-positive bacteria. Antibiotics 2020, 9, 788. [Google Scholar] [CrossRef] [PubMed]
- Abbott, D.W.; Aasen, I.M.; Beauchemin, K.A.; Grondahl, F.; Gruninger, R.; Hayes, M.; Huws, S.; Kenny, D.A.; Krizsan, S.J.; Kirwan, S.F.; et al. Seaweed and seaweed bioactives for mitigation of enteric methane: Challenges and opportunities. Animals 2020, 10, 2432. [Google Scholar] [CrossRef]
- Tymensen, L.D.; Beauchemin, K.A.; McAllister, T.A. Structures of free-living and protozoa-associated methanogen communities in the bovine rumen differ according to comparative analysis of 16S rRNA and mcrA genes. Microbiology 2012, 158, 1808–1817. [Google Scholar] [CrossRef] [PubMed]
- Jin, W.; Cheng, Y.F.; Mao, S.Y.; Zhu, W.Y. Discovery of a novel rumen methanogen in the anaerobic fungal culture and its distribution in the rumen as revealed by real-time PCR. BMC Microbiol. 2014, 14, 104. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.M. Dietary fatty acids and human health. Anim. Res. 2000, 49, 165–180. [Google Scholar] [CrossRef]
- Guyader, J.; Eugène, M.; Nozière, P.; Morgavi, D.P.; Doreau, M.; Martin, C. Influence of rumen protozoa on methane emission in ruminants: A meta-analysis approach. Animal 2014, 8, 1816–1825. [Google Scholar] [CrossRef] [PubMed]
- Newbold, C.J.; De la Fuente, G.; Belanche, A.; Ramos-Morales, E.; McEwan, N.R. The role of ciliate protozoa in the rumen. Front. Microbiol. 2015, 6, 1313. [Google Scholar] [CrossRef] [PubMed]
- Newbold, C.J.; Lassalas, B.; Jouany, J.P. The importance of methanogens associated with ciliate protozoa in ruminal methane production in vitro. Lett. Appl. Microbiol. 1995, 21, 230–234. [Google Scholar] [CrossRef] [PubMed]
- Machmüller, A.; Soliva, C.R.; Kreuzer, M. Effect of coconut oil and defaunation treatment on methanogenesis in sheep. Reprod. Nutr. Dev. 2003, 43, 41–55. [Google Scholar] [CrossRef] [PubMed]
- Beauchemin, K.A.; McGinn, S.M.; Benchaar, C.; Holtshausen, L. Crushed sunflower, flax, or canola seeds in lactating dairy cow diets: Effects on methane production, rumen fermentation, and milk production. J. Dairy Sci. 2009, 92, 2118–2127. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.F.; Edwards, J.E.; Allison, G.G.; Zhu, W.Y.; Theodorou, M.K. Diversity and activity of enriched ruminal cultures of anaerobic fungi and methanogens grown together on lignocellulose in consecutive batch culture. Bioresour. Technol. 2009, 100, 4821–4828. [Google Scholar] [CrossRef] [PubMed]
- Matthews, C.; Crispie, F.; Lewis, E.; Reid, M.; O’Toole, P.W.; Cotter, P.D. The rumen microbiome: A crucial consideration when optimising milk and meat production and nitrogen utilisation efficiency. Gut Microbes 2019, 10, 115–132. [Google Scholar] [CrossRef] [PubMed]
- Maia, M.R.G.; Chaudhary, L.C.; Figueres, L.; Wallace, R.J. Metabolism of polyunsaturated fatty acids and their toxicity to the microflora of the rumen. Antonie van Leeuwenhoek 2007, 91, 303–314. [Google Scholar] [CrossRef] [PubMed]
- Forsberg, C.W.; Cheng, K.-J.; White, B.A. Polysaccharide Degradation in the Rumen and Large Intestine. In Gastrointestinal Microbiology; Mackie, R.O., White, B.A., Eds.; Springer: Berlin/Heidelberg, Germany, 1997; pp. 319–379. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Shinkai, T.; Koike, S. Ecological and physiological characterization shows that Fibrobacter succinogenes is important in rumen fiber digestion—Review. Folia Microbiol. 2008, 53, 195–200. [Google Scholar] [CrossRef]
- Maia, M.R.; Chaudhary, L.C.; Bestwick, C.S.; Richardson, A.J.; McKain, N.; Larson, T.R.; Graham, I.A.; Wallace, R.J. Toxicity of unsaturated fatty acids to the biohydrogenating ruminal bacterium, Butyrivibrio fibrisolvens. BMC Microbiol. 2010, 10, 52. [Google Scholar] [CrossRef]
- Maia, M.R.G.; Fonseca, A.J.M.; Oliveira, H.M.; Mendonça, C.; Cabrita, A.R.J. The potential role of seaweeds in the natural manipulation of rumen fermentation and methane production. Sci. Rep. 2016, 6, 32321. [Google Scholar] [CrossRef]
- Christodoulou, C.; Mavrommatis, A.; Simoni, M.; Righi, F.; Prandi, B.; Tedeschi, T.; Sforza, S.; Tsiplakou, E. The amino acid profile of Camelina sativa seeds correlates with the strongest immune response in dairy ewes. Animal 2022, 16, 100621. [Google Scholar] [CrossRef]
- Kudo, H.; Cheng, K.J.; Costerton, J.W. Interactions between Treponema bryantii and cellulolytic bacteria in the in vitro degradation of straw cellulose. Can. J. Microbiol. 1987, 33, 244–248. [Google Scholar] [CrossRef] [PubMed]
- Fondevila, M.; Dehority, B.A. Interactions between Fibrobacter succinogenes, Prevotella ruminicola, and Ruminococcus flavefaciens in the Digestion of Cellulose from Forages. J. Anim. Sci. 1996, 74, 678–684. [Google Scholar] [CrossRef] [PubMed]
- Wolin, M.J.; Miller, T.L.; Stewart, C.S. Microbe–Microbe Interactions. In The Rumen Microbial Ecosystem, 2nd ed.; Hobson, P.N., Stewart, C.S., Eds.; Blackie Academic and Professional: London, UK, 1997; pp. 467–491. ISBN 978-94-010-7149-9. [Google Scholar]
- Sawanon, S.; Kobayashi, Y. Synergistic fibrolysis in the rumen by cellulolytic Ruminococcus flavefaciens and non-cellulolytic Selenomonas ruminantium: Evidence in defined cultures. Anim. Sci. J. 2006, 77, 208–214. [Google Scholar] [CrossRef]
- Sawanon, S.; Koike, S.; Kobayashi, Y. Evidence for the possible involvement of Selenomonas ruminantium in rumen fiber digestion. FEMS Microbiol. Lett. 2011, 325, 170–179. [Google Scholar] [CrossRef]
- Scheifinger, C.C.; Wolin, M.J. Propionate Formation from Cellulose and Soluble Sugars by Combined Cultures of Bacteroides succinogenes and Selenomonas ruminantium. Appl. Microbiol. 1973, 26, 789–795. [Google Scholar] [CrossRef] [PubMed]
- Nagaraja, T.G. Microbiology of the Rumen. In Rumenology; Millen, D., De Beni Arrigoni, M., Lauritano Pacheco, R., Eds.; Springer: Cham, Switzerland, 2016; pp. 39–61. [Google Scholar] [CrossRef]
| Daily Feed Intake (g/Ewe/Day as Fed) | Dietary Treatments (D) | |||
| CON | CS6 | CS11 | CS16 | |
| Wheat Straw | 200 | 200 | 200 | 200 |
| Alfalfa Hay | 1500 | 1500 | 1500 | 1500 |
| Concentrate | 1500 | 1500 | 1500 | 1500 |
| Nutrients intake (g/ewe/day) | Dietary treatments (D) | |||
| CON | CS6 | CS11 | CS16 | |
| Dry Matter | 2880 | 2874 | 2882 | 2888 |
| Crude Protein | 601 | 600 | 602 | 620 |
| Ether Extract | 40 | 80 | 107 | 133 |
| Neutral Detergent Fiber | 1284 | 1285 | 1287 | 1301 |
| Acid Detergent Fiber | 665 | 711 | 712 | 719 |
| Dietary Treatments (D) | ||||
|---|---|---|---|---|
| Main Fatty Acids Intake (g/Ewe/Day) of the Total Diet | CON | CS6 | CS11 | CS16 |
| C14:0 | 0.18 | 0.24 | 0.27 | 0.25 |
| C16:0 | 7.50 | 9.57 | 10.74 | 11.78 |
| C16:1 n-7 | 0.20 | 0.23 | 0.25 | 0.27 |
| C18:0 | 1.34 | 2.14 | 2.62 | 3.08 |
| cis-9 C18:1 | 7.47 | 13.57 | 18.35 | 22.41 |
| cis C18:2 n-6 | 19.94 | 29.08 | 31.74 | 37.67 |
| C20:0 | 0.11 | 0.33 | 0.51 | 0.67 |
| C18:3 n-3 | 2.02 | 14.40 | 23.01 | 30.88 |
| C20:1 n-9 | 0.21 | 6.71 | 11.41 | 15.63 |
| C20:2 n-6 | 0.16 | 0.85 | 1.38 | 1.80 |
| C20:3 n-3 | 0.14 | 0.24 | 0.34 | 0.41 |
| C22:0 | 0.07 | 0.13 | 0.18 | 0.16 |
| C24:0 | 0.22 | 0.26 | 0.31 | 0.37 |
| Dietary Treatment | ||||||
|---|---|---|---|---|---|---|
| Fatty Acid | CON | CS6 | CS11 | CS16 | SEM a | p |
| C14:0 | 1.22 | 0.91 | 0.72 | 0.57 | 0.11 | 0.235 |
| C14:1 | 1.51 | 1.26 | 0.88 | 1.00 | 0.11 | 0.180 |
| C15:0 | 0.86 | 0.87 | 1.02 | 0.94 | 0.08 | 0.887 |
| C16:0 | 24.33 ab | 29.32 b | 21.09 a | 21.84 a | 0.10 | 0.007 |
| C16:1 n-7 | 0.26 | 0.04 | 0.09 | 0.18 | 0.04 | 0.310 |
| C17:0 | 0.22 | 0.00 | 0.04 | 0.10 | 0.03 | 0.138 |
| C18:0 | 46.37 | 42.78 | 45.07 | 42.18 | 0.87 | 0.308 |
| trans C18:1 | 0.82 | 1.16 | 1.68 | 1.39 | 0.24 | 0.644 |
| trans-11 C18:1 | 5.93 | 5.32 | 8.01 | 7.78 | 0.50 | 0.137 |
| cis-9 C18:1 | 8.03 a | 9.02 a | 11.84 b | 12.40 b | 0.49 | 0.001 |
| trans C18:2 n-6 | 0.00 | 0.00 | 0.06 | 0.07 | 0.02 | 0.622 |
| cis C18:2 n-6 | 5.91 | 4.71 | 4.58 | 4.73 | 0.26 | 0.265 |
| C20:0 | 0.39 | 0.50 | 0.23 | 0.80 | 0.14 | 0.275 |
| C18:3 n-3 | 0.48 a | 0.59 a | 1.53 b | 1.30 b | 0.13 | 0.003 |
| cis-9, trans-11 C18:2 | 3.54 | 3.32 | 2.55 | 3.71 | 0.26 | 0.395 |
| C20:2 n-6 | 0.11 | 0.10 | 0.05 | 0.22 | 0.04 | 0.413 |
| C20:4 n-6 | 0.38 | 0.16 | 0.56 | 0.81 | 0.14 | 0.445 |
| Dietary Treatments | Phyla (Relative Abundance,%) | |||
|---|---|---|---|---|
| Bacteroidetes | Firmicutes | Proteobacteria | Firmicutes: Bacteroidetes | |
| CON | 39.3 | 51.0 | 7.7 | 1.30 |
| CS6 | 35.0 | 56.4 | 6.5 | 1.61 |
| CS11 | 34.2 | 45.7 | 18.6 | 1.34 |
| CS16 | 46.7 | 37.6 | 14.2 | 0.81 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Christodoulou, C.; Mavrommatis, A.; Loukovitis, D.; Symeon, G.; Dotas, V.; Kotsampasi, B.; Tsiplakou, E. Inclusion of Camelina sativa Seeds in Ewes’ Diet Modifies Rumen Microbiota. Animals 2023, 13, 377. https://doi.org/10.3390/ani13030377
Christodoulou C, Mavrommatis A, Loukovitis D, Symeon G, Dotas V, Kotsampasi B, Tsiplakou E. Inclusion of Camelina sativa Seeds in Ewes’ Diet Modifies Rumen Microbiota. Animals. 2023; 13(3):377. https://doi.org/10.3390/ani13030377
Chicago/Turabian StyleChristodoulou, Christos, Alexandros Mavrommatis, Dimitris Loukovitis, George Symeon, Vassilios Dotas, Basiliki Kotsampasi, and Eleni Tsiplakou. 2023. "Inclusion of Camelina sativa Seeds in Ewes’ Diet Modifies Rumen Microbiota" Animals 13, no. 3: 377. https://doi.org/10.3390/ani13030377
APA StyleChristodoulou, C., Mavrommatis, A., Loukovitis, D., Symeon, G., Dotas, V., Kotsampasi, B., & Tsiplakou, E. (2023). Inclusion of Camelina sativa Seeds in Ewes’ Diet Modifies Rumen Microbiota. Animals, 13(3), 377. https://doi.org/10.3390/ani13030377









