Analysis on Changes and Influencing Factors of the Intestinal Microbiota of Alpine Musk Deer between the Place of Origin and Migration
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Environment and Experimental Animals
2.2. Sampling and Experimentation
2.2.1. Fresh Feces Collection
2.2.2. DNA Extraction, Polymerase Chain Reaction (PCR) Amplification, and 16S-rRNA Gene Sequencing
2.2.3. Bioinformatics and Statistical Analyses
3. Results
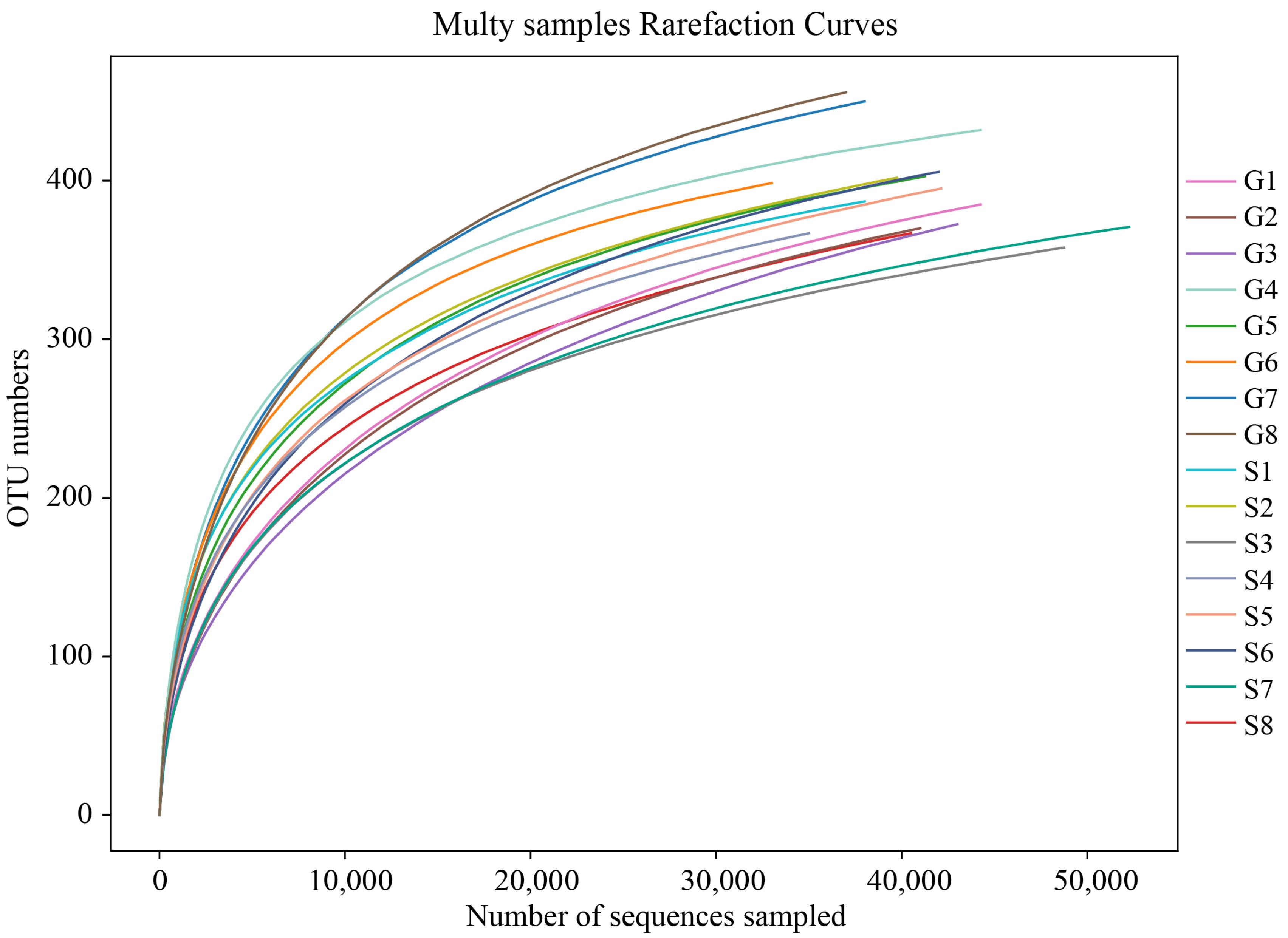
3.1. 16srRNA Gene Sequencing Results
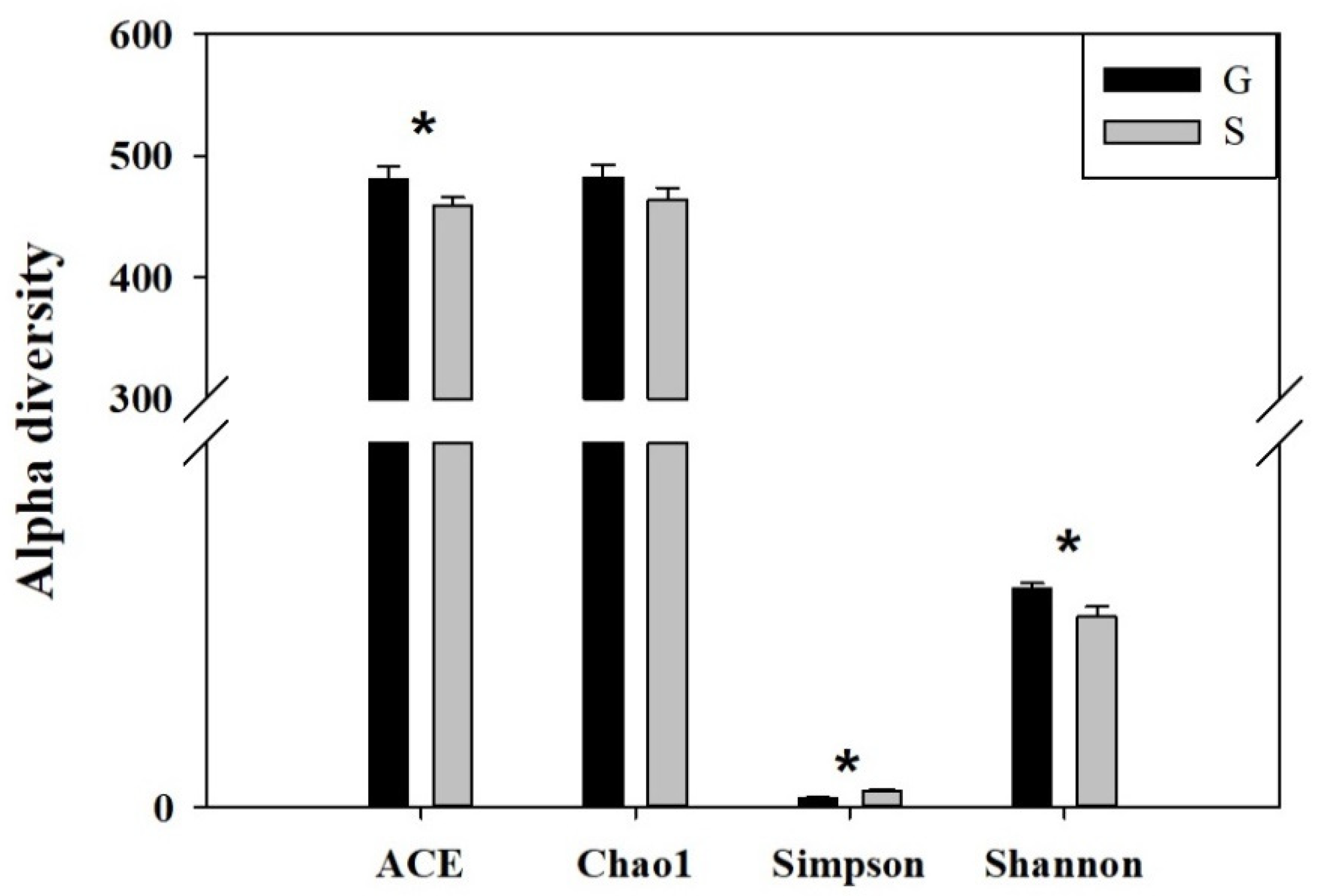
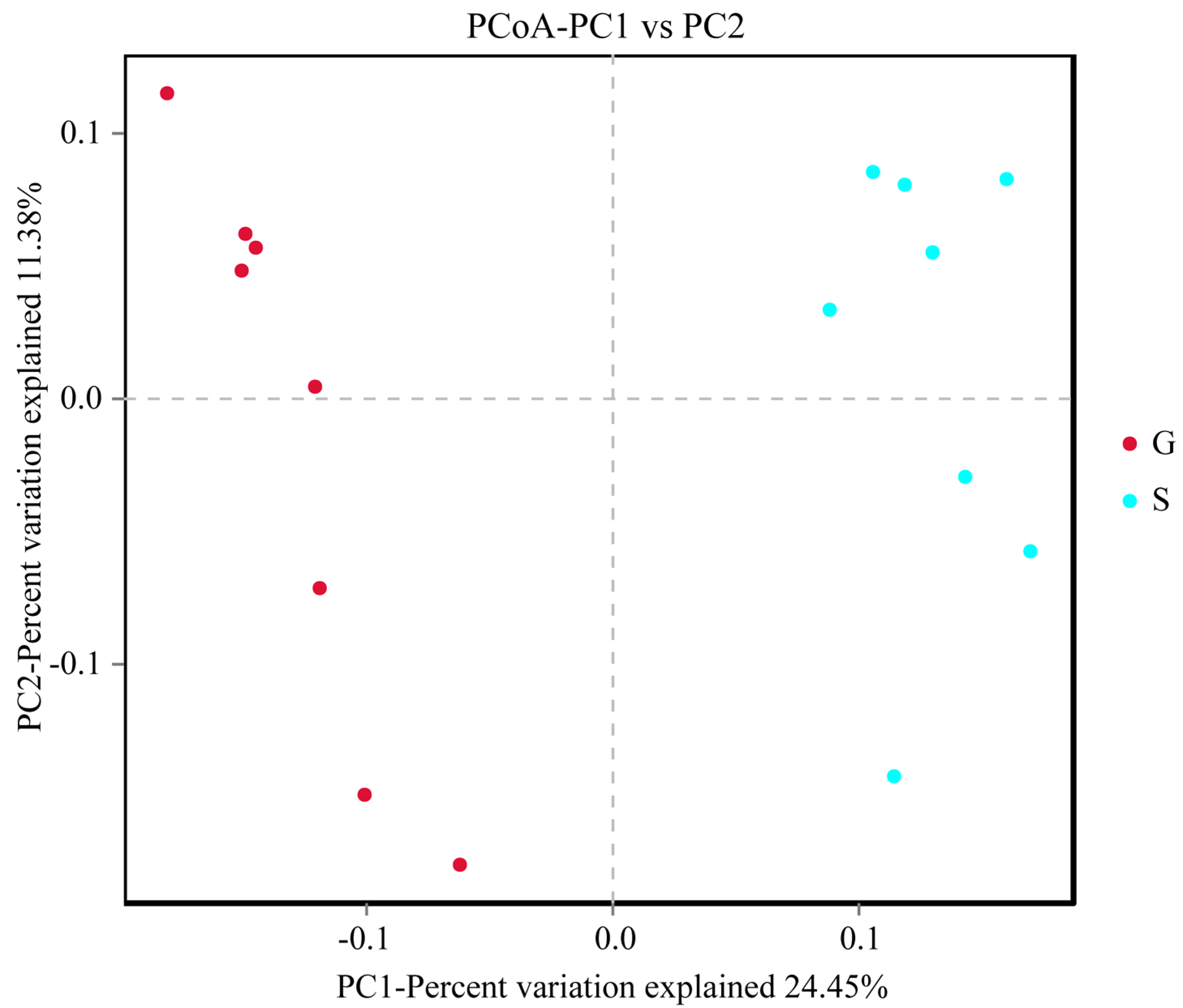
3.2. Analysis of Intestinal Microbiome Diversity
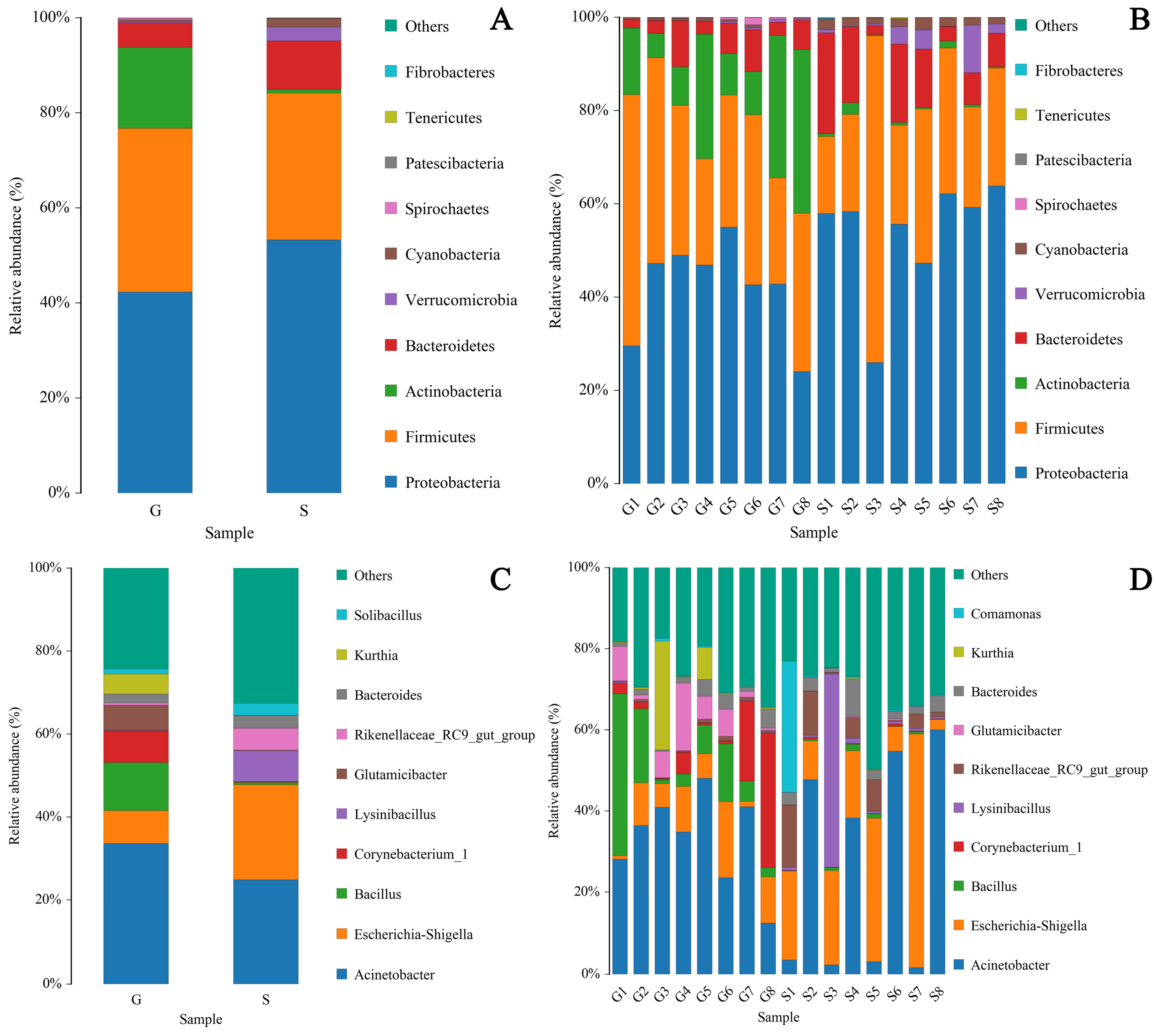
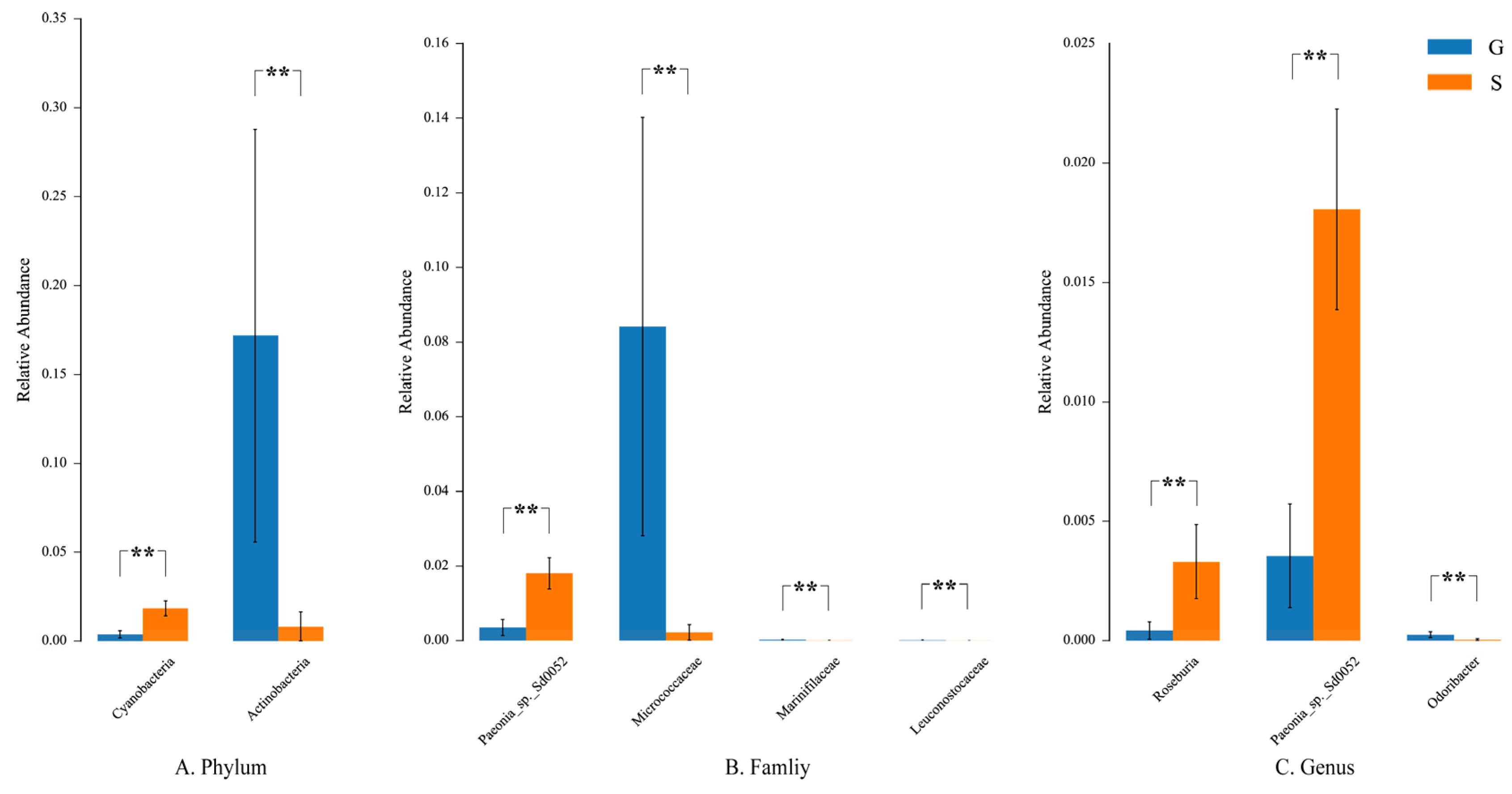
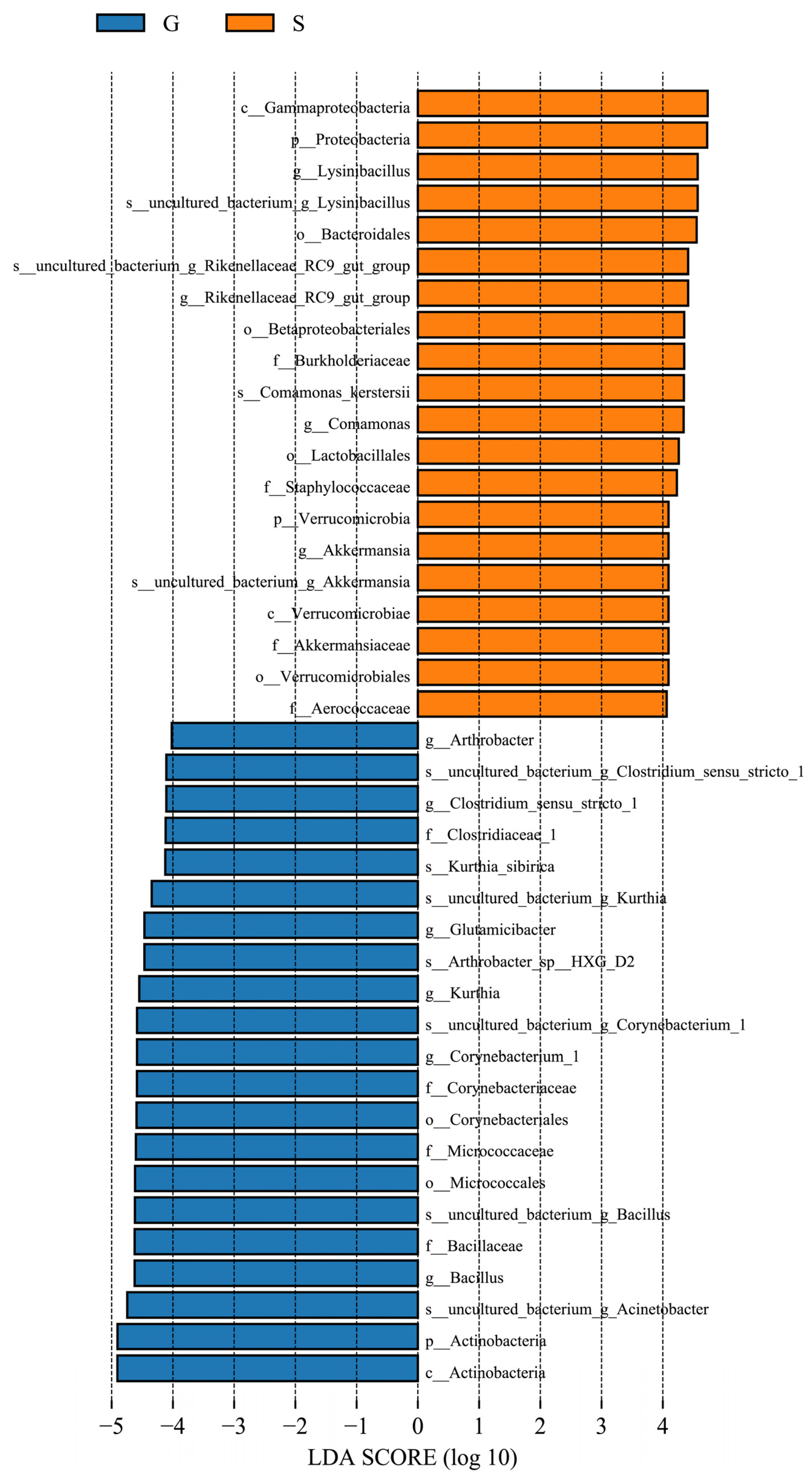
3.3. Differences in Microbiome Composition
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wu, J.; Wang, W. The Musk Deer of China, 1st ed.; China Forestry Publishing House: Beijing, China, 2006; pp. 62–78. (In Chinese) [Google Scholar]
- Sheng, H.L.; Liu, Z.X. The Musk Deer in China, 1st ed.; Shanghai Science and Technology Publishing House: Shanghai, China, 2007; pp. 156–197. (In Chinese) [Google Scholar]
- Wang, J.; Sun, J.P.; Xu, T.; Qi, J.; Zhang, Y.L.; Zhang, X.Y.; Meng, X.X. Population distribution, quantitative characteristics and influencing factors of the wild alpine musk deer in Xinglongshan National Nature Reserve, Gansu Province. Acta Ecol. Sin. 2009, 40, 7997–8004. [Google Scholar]
- Gao, H.X.; Shen, L.Q.; Liu, R.; Wang, G.; Zhang, A.P.; Chen, L.; Zhang, Y.Z.; Zhang, X.Y.; Qi, J.; Wang, C.L.; et al. Population Dynamics of Alpine Musk Deer in Xinglongshan National Nature Reserve and the Relationships to the Summer Habitat Suitability. Chin. J. Wildl. 2023, 44, 284–289. (In Chinese) [Google Scholar]
- Yang, Y.T.; Yang, Y.Y.; Na, H.Y.; Deng, E.; Yuan, L.L. Study on the survival status of alpine musk deer on the western slope of Helan Mountain. Agr. Tech. 2017, 37, 31. (In Chinese) [Google Scholar]
- Ding, Y.K.; Yao, Z.C.; Zhao, C.; Zhang, Z.R.; Chen, J.D.; Teng, L.W.; Liu, Z.S. Habitat suitability assessment of Moschus chrysogaster in Helan Moutain. Acta Ecol. Sin. 2023, 43, 3150–3156. (In Chinese) [Google Scholar]
- Liu, Z.X.; Li, Q.; Kang, F.G.; Sheng, H.L. Some ecological characteristics of the isolated population of alpine musk deer (Moschus chrysogaster) in the Xinglong forest, Gansu Province. Acta Ecol. Sin. 2001, 21, 964–968. (In Chinese) [Google Scholar]
- Hooper, L.V.; Littman, D.R.; Macpherson, A.J. Interactions between the microbiota and the immune system. Science 2012, 336, 1268–1273. [Google Scholar] [CrossRef] [PubMed]
- Warne, R.W.; Kirschman, L.; Zeglin, L. Manipulation of gut microbiota during critical developmental windows affects host physiological performance and disease susceptibility across ontogeny. J. Anim. Ecol. 2019, 88, 845–856. [Google Scholar] [CrossRef]
- Zhu, L.; Wu, Q.; Dai, J.; Zhang, S.; Wie, F. Evidence of cellulose metabolism by the giant panda gut microbiome. Proc. Natl. Acad. Sci. USA 2011, 108, 17714–17719. [Google Scholar] [CrossRef]
- Li, Y.M.; Hu, X.L.; Yang, S.; Zhou, J.T.; Qi, L.; Sun, X.N.; Fan, M.Y.; Xu, S.H.; Cha, M.H.; Zhang, M.S.; et al. Comparison Between the Fecal Bacterial Microbiota of Healthy and Diarrheic Captive Musk Deer. Front. Microbiol. 2018, 9, 300. [Google Scholar] [CrossRef]
- Zhao, W.; Ren, Z.W.; Luo, Y.; Cheng, J.G.; Wang, J.; Wang, Y.; Yang, Z.X.; Yao, X.P.; Zhong, Z.J.; Yang, W.; et al. Metagenomics analysis of the gut microbiome in healthy and bacterial pneumonia forest musk deer. Genes Genom. 2021, 43, 43–53. [Google Scholar] [CrossRef]
- Hicks, A.L.; Lee, K.J.; Couto-Rodriguez, M.; Patel, J.; Sinha, R.; Guo, C.; Olson, S.H.; Seimon, A.; Seimon, T.A.; Ondzie, A.U.; et al. Gut microbiomes of wild great apes fluctuate seasonally in response to diet. Nat. Commun. 2018, 9, 1786. [Google Scholar] [CrossRef] [PubMed]
- Muegge, B.D.; Kuczynski, J.; Knights, D.; Clemente, J.C.; Gonzalez, A.; Fontana, L.; Henrissat, B.; Knight, R.; Gordon, J.I. Diet Drives Convergence in Gut Microbiome Functions Across Mammalian Phylogeny and Within Humans. Science 2011, 332, 970. [Google Scholar] [CrossRef]
- Dennis, K.L.; Wang, Y.; Blatner, N.R.; Wang, S.; Saadalla, A.; Trudeau, E.; Roers, A.; Weaver, C.T.; Lee, J.J.; Gilbert, J.A.; et al. Adenomatous polyps are driven by microbe-instigated focal inflammation and are controlled by il-10–producing T cells. Cancer Res. 2013, 73, 5905–5913. [Google Scholar] [CrossRef] [PubMed]
- Magoc, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and webbased tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing Mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. AEM 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Williams, C.L.; Caraballo-Rodríguez, A.M.; Allaband, C.; Zarrinpar, A.; Knight, R.; Gauglitz, J.M. Wild-life-microbiome interactions and disease: Exploring opportunities for disease mitigation across ecological scales. Drug Discov. Today 2018, 28, 105–115. [Google Scholar]
- Gibson, K.M.; Nguyen, B.N.; Neumann, L.M.; Miller, M.; Buss, P.; Daniels, S.; Ahn, M.J.; Crandall, K.A.; Pukazhenthi, B. Gut microbiome differences between wild and captive black rhinoceros–implications for rhino health. Sci. Rep. 2019, 9, 7570. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Holmes, E.; Kinross, J.; Burcelin, R.; Gibson, G.; Jia, W.; Pettersson, S. Host-gut microbiota metabolic interactions. Science 2012, 336, 1262–1267. [Google Scholar] [CrossRef] [PubMed]
- Ottman, N.; Smidt, H.; De Vos, W.M.; Belzer, C. The function of our microbiota: Who is out there and what do they do? Front. Cell Infect Microbiol. 2012, 2, 104. [Google Scholar] [CrossRef] [PubMed]
- Houttu, V.; Boulund, U.; Grefhorst, A.; Soeters, M.R.; Pinto-Sietsma, S.; Nieuwdorp, M.; Holleboom, A.G. The role of the gut microbiome and exercise in non-alcoholic fatty liver disease. Therap. Adv. Gastroenterol. 2020, 13, 1756284820941745. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wang, K.; Wang, X.; Pang, Y.; Jiang, C. The role of the gut microbiome and its metabolites in metabolic diseases. Protein Cell. 2021, 12, 360–373. [Google Scholar] [CrossRef] [PubMed]
- Jayachandran, M.; Chung, S.S.M.; Xu, B. A critical review of the relationship between dietary components, the gut microbe Akkermansia muciniphila, and human health. Crit. Rev. Food Sci. Nutr. 2020, 60, 2265–2276. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.A.; Stombaugh, J.I.; Gordon, J.I.; Jansson, J.K.; Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 2012, 489, 220–230. [Google Scholar] [CrossRef] [PubMed]
- Mosca, A.; Leclerc, M.; Hugot, J.P. Gut microbiota diversity and human diseases: Should we reintroduce key predators in our ecosystem? Front. Microbiol. 2016, 7, 455. [Google Scholar] [CrossRef]
- Jiang, F.; Gao, H.M.; Qin, W.; Song, P.F.; Wang, H.J.; Zhang, J.J.; Liu, D.X.; Wang, D.; Zhang, T.Z. Marked Seasonal Variation in Structure and Function of Gut Microbiota in Forest and Alpine Musk Deer. Front. Microbiol. 2021, 12, 699797. [Google Scholar] [CrossRef]
- Berlemont, R.; Martiny, A.C. Phylogenetic distribution of potential cellulases in bacteria. AEM 2013, 79, 1545–1554. [Google Scholar] [CrossRef]
- Sun, Y.W.; Sun, Y.J.; Shi, Z.H.; Liu, Z.S.; Zhao, C.; Lu, T.F.; Gao, H.; Zhu, F.; Chen, R.; Zhang, J.; et al. Gut microbiota of wild and captive Alpine musk deer (Moschus chrysogaster). Front. Microbiol. 2020, 10, 3156. [Google Scholar] [CrossRef]
- Singh, A.; Tiwari, U.; Berrocoso, J.; Dersjant-Li, Y.; Awati, A.; Jha, R. Effects of a combination of xylanase, amylase and protease, and probiotics on major nutrients including amino acids and non-starch polysaccharides utilization in broilers fed different level of fibers. Poult. Sci. 2019, 98, 5571–5581. [Google Scholar] [CrossRef] [PubMed]
- Kong, C.; Gao, R.Y.; Yan, X.B.; Huang, L.S.; Qin, H.L. Probiotics improve gut microbiota dysbiosis in obese mice fed a high-fat or high-sucrose diet. Nutrition 2019, 60, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.X.; Taylor, L.; Shommu, N.; Ghosh, S.; Reimer, R.; Panaccione, R.; Kaur, S.; Hyun, J.E.; Cai, C.; Deehan, E.C.; et al. A diversified dietary pattern is associated with a balanced gut microbial composition of Faecalibacterium and Escherichia/Shigella in patients with Crohn’s disease in remission. J. Crohns Colitis. 2020, 14, 1547–1557. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.L.; Wang, C.Y.; Li, F.C. Impact of dietary fiber/starch ratio in shaping caecal microbiota in rabbits. Can. J. Microbiol. 2015, 61, 771–784. [Google Scholar] [CrossRef] [PubMed]
- Méndez-Salazar, E.O.; Ortiz-López, M.G.; Granados-Silvestre, M.Á.; Palacios-González, B.; Menjivar, M. Altered gut microbiota and compositional changes in Firmicutes and Proteobacteria in Mexican undernourished and obese children. Front. Microbiol. 2018, 9, 2494. [Google Scholar]
- Fei, N.; Zhao, L.P. An opportunistic pathogen isolated from the gut of an obese human causes obesity in germfree mice. ISME J. 2013, 7, 880–884. [Google Scholar] [CrossRef]
- Shin, N.R.; Whon, T.W.; Bae, J.W. Proteobacteria: Microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015, 33, 496–503. [Google Scholar] [CrossRef]
- Jeong, M.Y.; Jang, H.M.; Kim, D.H. High-fat diet causes psychiatric disorders in mice by increasing Proteobacteria population. Neurosci. Lett. 2019, 698, 51–57. [Google Scholar] [CrossRef]
- Carvalheira, A.; Silva, J.; Teixeira, P. Acinetobacter spp. in food and drinking water—A review. Food Microbiol. 2020, 95, 103675. [Google Scholar] [CrossRef]
- Khba, B.; Jdaia, C.; Fm, D. Heterogeneity in enterotoxigenic Escherichia coli and shigella infections in children under 5 years of age from 11 African countries: A subnational approach quantifying risk, mortality, morbidity, and stunting. Lancet Glob. Health 2020, 8, e101–e112. [Google Scholar]
- Althani, A.A.; Marei, H.E.; Hamdi, W.S.; Nasrallah, G.K.; El Zowalaty, M.E.; Al Khodor, S.; Al-Asmakh, M.; Abdel-Aziz, H.; Cenciarelli, C. Human microbiome and its association with health and diseases. J. Cell Physiol. 2016, 231, 1688–1694. [Google Scholar] [CrossRef] [PubMed]
- Delgado, M.L.; Singh, P.; Funk, J.A.; Moore, J.A.; Cannell, E.M.; Kanesfsky, J.; Manning, S.D.; Scribner, K.T. Intestinal microbial community dynamics of white-tailed deer (Odocoileus virginianus) in an agroecosystem. Microb. Ecol. 2017, 74, 496–506. [Google Scholar] [CrossRef] [PubMed]
- Weese, J.S.; Shury, T.; Jelinski, M.D. The fecal microbiota of semi-free-ranging wood bison (Bison bison athabascae). BMC Vet. Res. 2014, 10, 120. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hu, X.L.; Liu, G.; Shafer, A.B.A.; Wei, Y.T.; Zhou, J.T.; Lin, S.B.; Wu, H.B.; Mi, Z.; Hu, D.F.; Liu, S.Q. Comparative analysis of the gut microbial communities in forest and alpine musk deer using high-throughput sequencing. Front. Microbiol. 2017, 8, 572. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.M.; Hu, X.L.; Yang, S.; Zhou, J.T.; Zhang, T.X.; Qi, L.; Sun, X.N.; Fan, M.Y.; Xu, S.H.; Cha, M.H.; et al. Comparative Analysis of the Gut Microbiota Composition between Captive and Wild Forest Musk Deer. Front. Microbiol. 2017, 8, 1705. [Google Scholar] [CrossRef] [PubMed]
- Sawaswong, V.; Praianantathavorn, K.; Chanchaem, P.; Khamwut, A.; Kemthong, T.; Hamada, Y.; Malaivijitnond, S.; Payungporn, S. Comparative analysis of oral-gut microbiota between captive and wild long-tailed macaque in Thailand. Sci. Rep. 2021, 11, 14280. [Google Scholar] [CrossRef] [PubMed]
- Binda, C.; Lopetuso, L.R.; Rizzatti, G.; Gibiino, G.; Cennamo, V.; Gasbarrini, A. Actinobacteria: A relevant minority for the maintenance of gut homeostasis. Dig. Liver Dis. 2018, 50, 421–428. [Google Scholar] [CrossRef]
- Kim, J.H.; Kim, Y.; Kim, Y.J.; Park, Y. Conjugated linoleic acid: Potential health benefits as a functional food ingredient. Annu. Rev. Food Sci. Technol. 2016, 7, 221–244. [Google Scholar] [CrossRef]
- Jeong, Y.; Kim, J.W.; You, H.J.; Park, S.J.; Lee, J.; Ju, J.H.; Park, M.S.; Jin, H.; Cho, M.L.; Kwon, B.; et al. Gut microbial composition and function are altered in patients with early rheumatoid arthritis. J. Clin. Med. 2019, 8, 693. [Google Scholar] [CrossRef]
- Sun, R.L.; Xu, K.; Ji, S.B.; Pu, Y.Q.; Man, Z.D.; Ji, J.H.; Chen, M.J.; Yin, L.H.; Zhang, J.; Pu, Y.P. Benzene exposure induces gut microbiota dysbiosis and metabolic disorder in mice. Sci. Total Environ. 2020, 705, 135879. [Google Scholar] [CrossRef]
- Weber, T.; Charusanti, P.; Musiol-Kroll, E.M.; Jiang, X.; Tong, Y.; Kim, H.U.; Lee, S.Y. Metabolic engineering of antibiotic factories: New tools for antibiotic production in actinomycetes. Trends Biotechnol. 2015, 33, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Elshaghabee, F.M.; Rokana, N.; Gulhane, R.D.; Sharma, C.; Panwar, H. Bacillus as potential probiotics: Status, concerns, and future perspectives. Front. Microbiol. 2017, 8, 1490. [Google Scholar] [CrossRef] [PubMed]
- Bernardeau, M.; Lehtinen, M.J.; Forssten, S.D.; Nurminen, P. Importance of the gastrointestinal life cycle of Bacillus for probiotic functionality. J. Food Sci. Technol. 2017, 54, 2570–2584. [Google Scholar] [CrossRef] [PubMed]
- West, A.G.; Waite, D.W.; Deines, P.; Bourne, D.G.; Digby, A.; McKenzie, V.J.; Taylor, M.W. The microbiome in threatened species conservation. Biol. Conserv. 2019, 229, 85–98. [Google Scholar] [CrossRef]
- Tsukayama, P.; Boolchandani, M.; Patel, S.; Pehrsson, E.C.; Gibson, M.K.; Chiou, K.L.; Jolly, C.J.; Rogers, J.; Phillips-Conroy, J.E.; Dantas, G. Characterization of wild and captive baboon gut microbiota and their antibiotic resistomes. Msystems 2018, 3, e00016-18. [Google Scholar] [CrossRef]
- Guo, W.; Mishra, S.; Wang, C.D.; Zhang, H.M.; Ning, R.H.; Kong, F.L.; Zeng, B.; Zhao, J.C.; Li, Y. Comparative study of gut microbiota in wild and captive giant pandas (Ailuropoda melanoleuca). Genes 2019, 10, 827. [Google Scholar] [CrossRef]


| Group | Chao1 | ACE | Shannon | Simpson |
|---|---|---|---|---|
| G | 482.33 ± 10.58 | 481.16 ± 10.17 | 2.99 ± 0.08 | 0.13 ± 0.01 |
| S | 463.53 ± 10.39 | 458.69 ± 6.98 | 2.61 ± 0.14 | 0.22 ± 0.03 |
| P | >0.05 | <0.05 | <0.05 | <0.05 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, B.; Shi, M.; Xu, S.; Zhang, H.; Li, Y.; Hu, D. Analysis on Changes and Influencing Factors of the Intestinal Microbiota of Alpine Musk Deer between the Place of Origin and Migration. Animals 2023, 13, 3791. https://doi.org/10.3390/ani13243791
Zhang B, Shi M, Xu S, Zhang H, Li Y, Hu D. Analysis on Changes and Influencing Factors of the Intestinal Microbiota of Alpine Musk Deer between the Place of Origin and Migration. Animals. 2023; 13(24):3791. https://doi.org/10.3390/ani13243791
Chicago/Turabian StyleZhang, Baofeng, Minghui Shi, Shanghua Xu, Haonan Zhang, Yimeng Li, and Defu Hu. 2023. "Analysis on Changes and Influencing Factors of the Intestinal Microbiota of Alpine Musk Deer between the Place of Origin and Migration" Animals 13, no. 24: 3791. https://doi.org/10.3390/ani13243791
APA StyleZhang, B., Shi, M., Xu, S., Zhang, H., Li, Y., & Hu, D. (2023). Analysis on Changes and Influencing Factors of the Intestinal Microbiota of Alpine Musk Deer between the Place of Origin and Migration. Animals, 13(24), 3791. https://doi.org/10.3390/ani13243791





