In-Line Detection of Clinical Mastitis by Identifying Clots in Milk Using Images and a Neural Network Approach
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Data
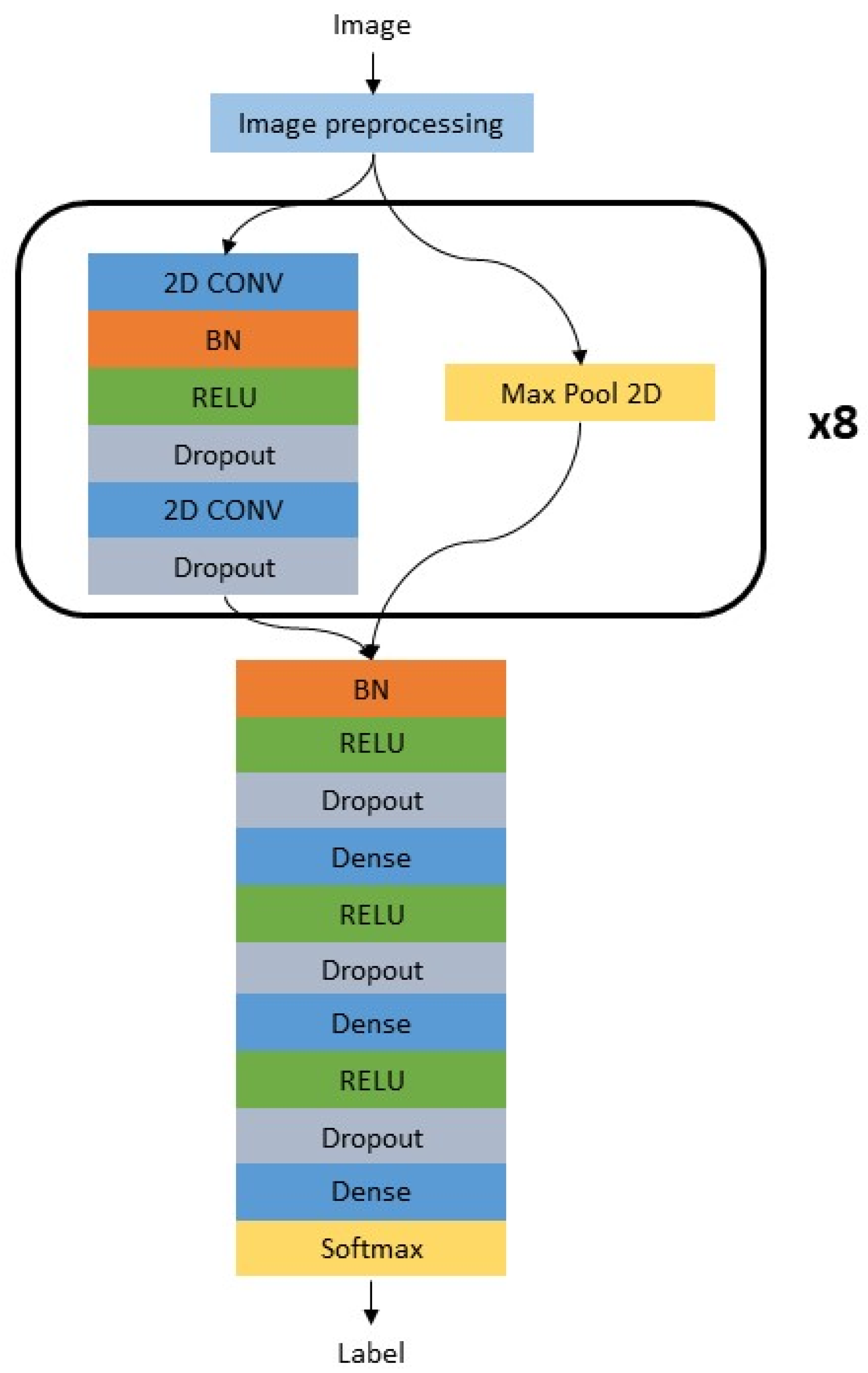
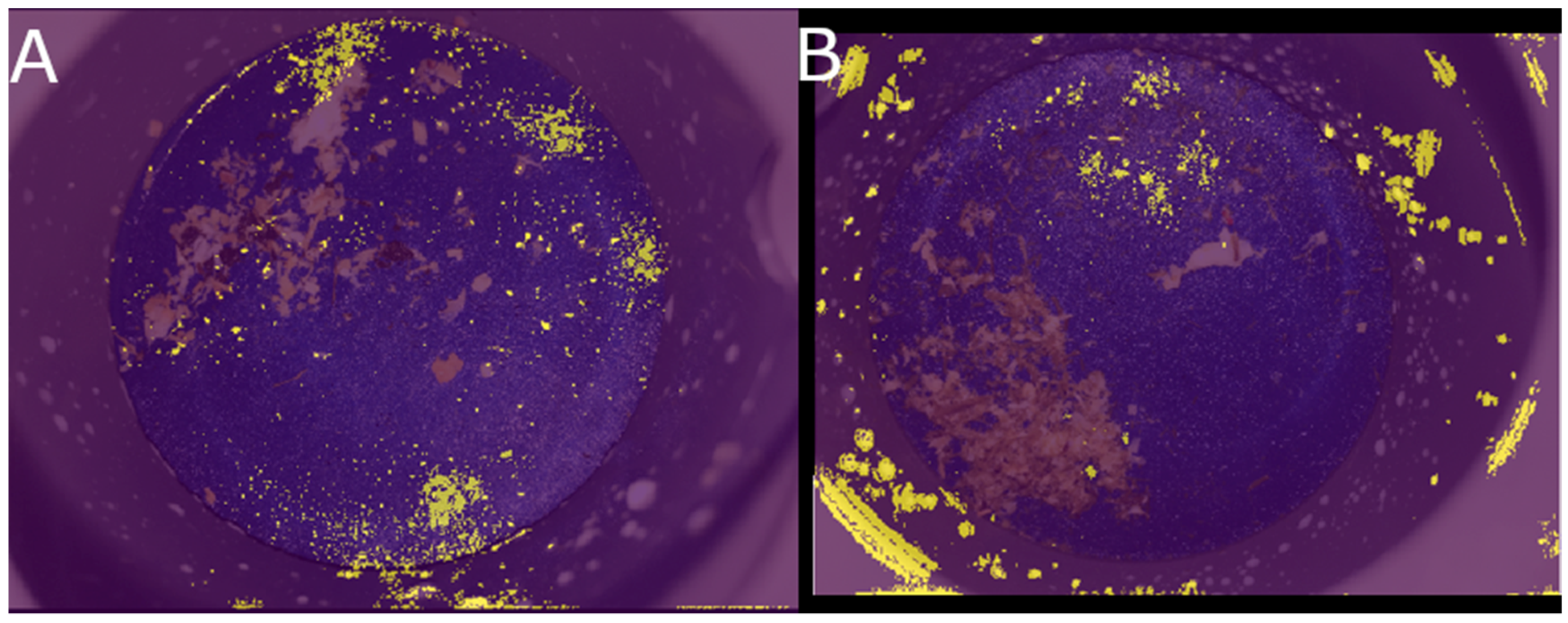
2.2. Image Analysis
2.3. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Halasa, T.; Huijps, K.; Østerås, O.; Hogeveen, H. Economic Effects of Bovine Mastitis and Mastitis Management: A Review. Vet. Q. 2007, 29, 18–31. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, N. Review on Mastitis and Its Economic Effect. Can. J. Res. 2017, 6, 13–22. [Google Scholar] [CrossRef]
- Getaneh, A.M.; Mekonnen, S.A.; Hogeveen, H. Stochastic Bio—Economic Modeling of Mastitis in Ethiopian Dairy Farms. Prev. Vet. Med. 2017, 138, 94–103. [Google Scholar] [CrossRef] [PubMed]
- Azooz, M.F.; El-Wakeel, S.A.; Yousef, H.M. Financial and Economic Analyses of the Impact of Cattle Mastitis on the Profitability of Egyptian Dairy Farms. Vet. World 2020, 13, 1750. [Google Scholar] [CrossRef] [PubMed]
- Hogeveen, H.; Van Der Voort, M. Assessing the Economic Impact of an Endemic Disease: The Case of Mastitis. Rev. Sci. Et Tech. Off. Int. Des Epizoot. 2017, 36, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Heikkilä, A.M.; Nousiainen, J.I.; Pyörälä, S. Costs of Clinical Mastitis with Special Reference to Premature Culling. J. Dairy Sci. 2012, 95, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Willits, S.; Infrared, R. Infrared Thermography for Screening and Early Detection of Mastitis Infections in Working Dairy Herds. InfraMation Proc. ITC 2005, 42, 1–5. [Google Scholar]
- Sandgren, C.H.; Emanuelson, U. Is There an Ideal Automatic Milking System Cow and How Is She Different from an Ideal Parlor Milked Cow? In Proceedings of the National Mastitis Council 56th Annual Meeting, St. Pete Beach, FL, USA, 28 January 2017; pp. 61–68. [Google Scholar]
- Penry, J.F. Mastitis Control in Automatic Milking Systems. Vet. Clin. North Am. Food Anim. Pract. 2018, 34, 439–456. [Google Scholar] [CrossRef]
- Khatun, M.; Thomson, P.C.; Kerrisk, K.L.; Lyons, N.A.; Clark, C.E.F.; Molfino, J.; García, S.C. Development of a New Clinical Mastitis Detection Method for Automatic Milking Systems. J. Dairy Sci. 2018, 101, 9385–9395. [Google Scholar] [CrossRef]
- ISO 20966:2007; Automatic Milking Installations—Requirements and Testing. ISO: Geneva, Switzerland, 2007.
- Fogsgaard, K.K.; Bennedsgaard, T.W.; Herskin, M.S. Behavioral Changes in Freestall-Housed Dairy Cows with Naturally Occurring Clinical Mastitis. J. Dairy Sci. 2015, 98, 1730–1738. [Google Scholar] [CrossRef]
- de Mol, R.M.; Ouweltjes, W. Detection Model for Mastitis in Cows Milked in an Automatic Milking System. Prev. Vet. Med. 2001, 49, 71–82. [Google Scholar] [CrossRef] [PubMed]
- Naqvi, S.A.; King, M.T.M.; Matson, R.D.; DeVries, T.J.; Deardon, R.; Barkema, H.W. Mastitis Detection with Recurrent Neural Networks in Farms Using Automated Milking Systems. Comput. Electron. Agric. 2022, 192, 106618. [Google Scholar] [CrossRef]
- Adkins, P.R.F.; Middleton, J.R. Methods for Diagnosing Mastitis. Vet. Clin. North Am. Food Anim. Pract. 2018, 34, 479–491. [Google Scholar] [CrossRef] [PubMed]
- Mein, G.A.; Rasmussen, M.D. Performance Evaluation of Systems for Automated Monitoring of Udder Health: Would the Real Gold Standard Please Stand Up? Brill Wageningen Academic: Paderborn, Germany, 2008; ISBN 9789086860852. [Google Scholar]
- Claycomb, R.W.; Johnstone, P.T.; Mein, G.A.; Sherlock, R.A. An Automated In-Line Clinical Mastitis Detection System Using Measurement of Conductivity from Foremilk of Individual Udder Quarters. New Zealand Vet. J. 2009, 57, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Wiethoff, M.; Suhr, O. Method and Device for Determining the Quality of Milk Produced by Machine Milking. US Patent, US8261597B2 2007. [Google Scholar]
- Hallén Sandgren, C.; Anglart, D.; Klaas, I.C.; Rönnegård, L.; Emanuelson, U. Homogeneity Density Scores of Quarter Milk in Automatic Milking Systems. J. Dairy Sci. 2021, 104, 10121–10130. [Google Scholar] [CrossRef] [PubMed]
- Deb, K.; Pratap, A.; Agarwal, S.; Meyarivan, T. A Fast and Elitist Multiobjective Genetic Algorithm: NSGA-II. IEEE Trans. Evol. Comput. 2002, 6, 182–197. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. In Proceedings of the ICLR 2015, San Diego, CA, USA, 7–9 May 2015; pp. 1–15. [Google Scholar]
- Chollet, F. Keras. Available online: https://keras.io (accessed on 22 September 2019).
- Hogeveen, H.; Klaas, I.C.; Dalen, G.; Honig, H.; Zecconi, A.; Kelton, D.F.; Sánchez Mainar, M. Novel Ways to Use Sensor Data to Improve Mastitis Management. J. Dairy Sci. 2021, 104, 11317–11332. [Google Scholar] [CrossRef]
- Mollenhorst, H.; Rijkaart, L.J.; Hogeveen, H. Mastitis Alert Preferences of Farmers Milking with Automatic Milking Systems. J. Dairy Sci. 2012, 95, 2523–2530. [Google Scholar] [CrossRef]
- Deng, Z.; Koop, G.; Lam, T.J.G.M.; van der Lans, I.A.; Vernooij, J.C.M.; Hogeveen, H. Farm-Level Risk Factors for Bovine Mastitis in Dutch Automatic Milking Dairy Herds. J. Dairy Sci. 2019, 102, 4522–4535. [Google Scholar] [CrossRef]
- Fawcett, T. An Introduction to ROC Analysis. Pattern Recognit. Lett. 2006, 27, 861–874. [Google Scholar] [CrossRef]
- van Steenkiste, G.; van Loon, G.; Crevecoeur, G. Transfer Learning in ECG Classification from Human to Horse Using a Novel Parallel Neural Network Architecture. Sci. Rep. 2020, 10, 186. [Google Scholar] [CrossRef] [PubMed]
- Sundararajan, M.; Taly, A.; Yan, Q. Axiomatic Attribution for Deep Networks. In Proceedings of the 34th International Conference on Machine Learning, ICML 2017, Sydney, NSW, Australia, 6–11 August 2017; Volume 7, pp. 5109–5118. [Google Scholar]
- Razzak, M.I.; Naz, S.; Zaib, A. Deep Learning for Medical Image Processing: Overview, Challenges and the Future; Springer: Berlin/Heidelberg, Germany, 2018; Volume 26, pp. 323–350. [Google Scholar]

| Augmentation Type | Value |
|---|---|
| Random rotation | 60° |
| Random shift in width | 10% |
| Random shift in height | 10% |
| Random zoom | 100–130% |
| Horizontal flip probability | 50% |
| Vertical flip probability | 50% |
| Shear range | 20° |
| Fill mode | nearest |
| Layer | Hyper Parameter | Value |
|---|---|---|
| All | Dropout | 0.2 |
| Conv 1.1 | Filters | 8 |
| Kernel size | 4 | |
| Subsampling | 2 | |
| Conv 1.2 | Filters | 8 |
| Kernel size | 1 | |
| Subsampling | 1 | |
| Conv 2.1 | Filters | 16 |
| Kernel size | 4 | |
| Subsampling | 1 | |
| Conv 2.2 | Filters | 16 |
| Kernel size | 1 | |
| Subsampling | 1 | |
| Conv 3.1 | Filters | 24 |
| Kernel size | 4 | |
| Subsampling | 1 | |
| Conv 3.2 | Filters | 24 |
| Kernel size | 1 | |
| Subsampling | 1 | |
| Conv 4.1 | Filters | 32 |
| Kernel size | 4 | |
| Subsampling | 1 | |
| Conv 4.2 | Filters | 32 |
| Kernel size | 1 | |
| Subsampling | 1 | |
| Conv 5.1 | Filters | 40 |
| Kernel size | 4 | |
| Subsampling | 1 | |
| Conv 5.2 | Filters | 40 |
| Kernel size | 1 | |
| Subsampling | 1 | |
| Conv 6.1 | Filters | 48 |
| Kernel size | 4 | |
| Subsampling | 1 | |
| Conv 6.2 | Filters | 48 |
| Kernel size | 1 | |
| Subsampling | 1 | |
| Conv 7.1 | Filters | 48 |
| Kernel size | 4 | |
| Subsampling | 2 | |
| Conv 7.2 | Filters | 48 |
| Kernel size | 1 | |
| Subsampling | 1 | |
| Conv 8.1 | Filters | 48 |
| Kernel size | 4 | |
| Subsampling | 1 | |
| Conv 8.2 | Filters | 48 |
| Kernel size | 1 | |
| Subsampling | 1 | |
| Dense 1 | Number of neurons | 64 |
| L2 regularization | 0.1 | |
| Dense 2 | Number of neurons | 32 |
| L2 regularization | 0.01 |
| Predicted Negative | Predicted Positive | |
|---|---|---|
| True negative | 182 | 0 |
| True positive | 0 | 153 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Van Steenkiste, G.; Van Den Brulle, I.; Piepers, S.; De Vliegher, S. In-Line Detection of Clinical Mastitis by Identifying Clots in Milk Using Images and a Neural Network Approach. Animals 2023, 13, 3783. https://doi.org/10.3390/ani13243783
Van Steenkiste G, Van Den Brulle I, Piepers S, De Vliegher S. In-Line Detection of Clinical Mastitis by Identifying Clots in Milk Using Images and a Neural Network Approach. Animals. 2023; 13(24):3783. https://doi.org/10.3390/ani13243783
Chicago/Turabian StyleVan Steenkiste, Glenn, Igor Van Den Brulle, Sofie Piepers, and Sarne De Vliegher. 2023. "In-Line Detection of Clinical Mastitis by Identifying Clots in Milk Using Images and a Neural Network Approach" Animals 13, no. 24: 3783. https://doi.org/10.3390/ani13243783
APA StyleVan Steenkiste, G., Van Den Brulle, I., Piepers, S., & De Vliegher, S. (2023). In-Line Detection of Clinical Mastitis by Identifying Clots in Milk Using Images and a Neural Network Approach. Animals, 13(24), 3783. https://doi.org/10.3390/ani13243783











