Early Colonization of the Intestinal Microbiome of Neonatal Piglets Is Influenced by the Maternal Microbiome
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
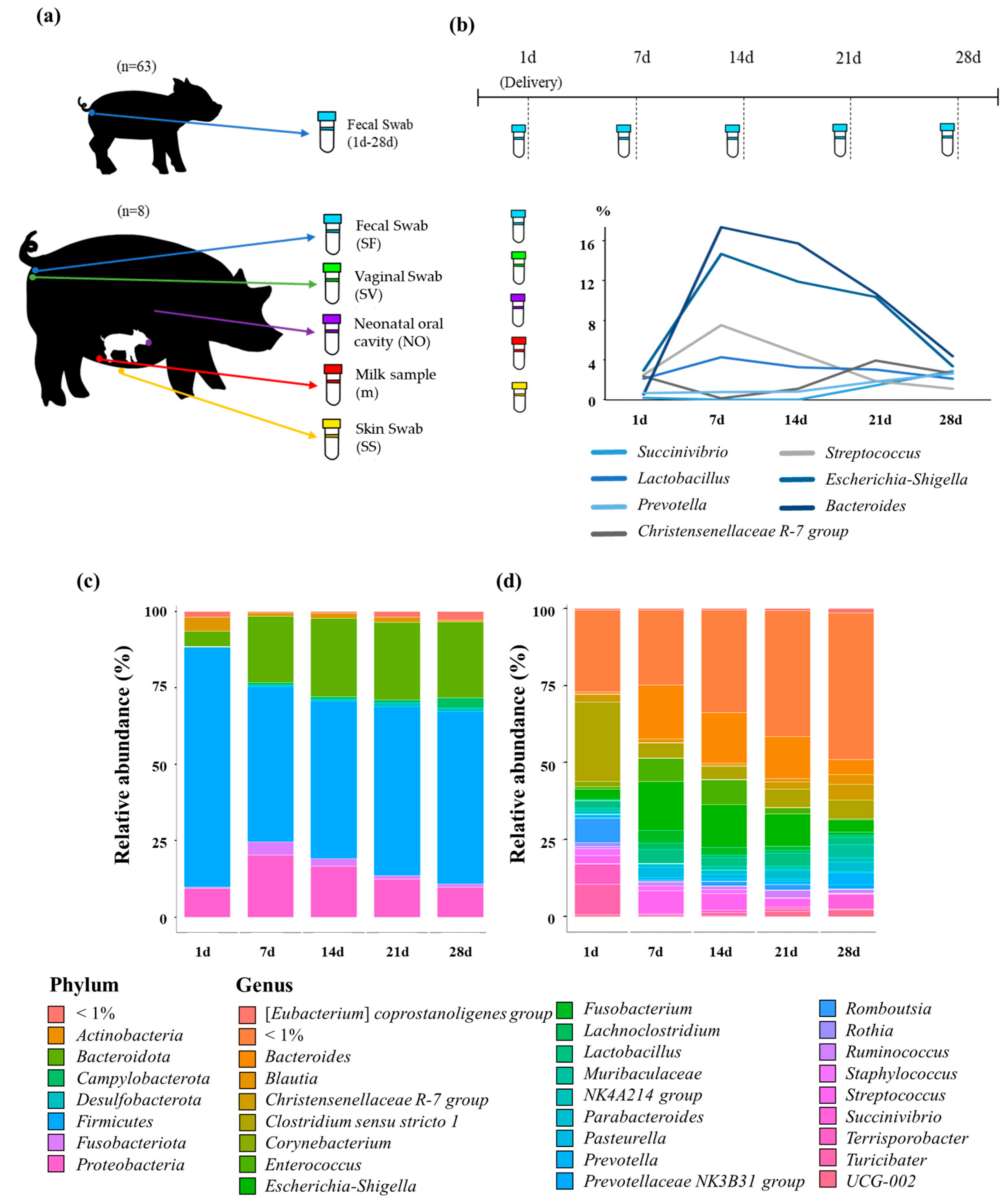
2.1. Animal Management and Experimental Design
2.2. Microbial DNA Extraction and 16S rRNA Gene Sequencing
2.3. Microbial Data Analysis
2.4. Statistical Analysis
3. Results
3.1. 16S rRNA Gene Sequencing for Species and Strain-Level Microbiome Analysis
3.2. Composition of the Gut Microbiome in Piglets
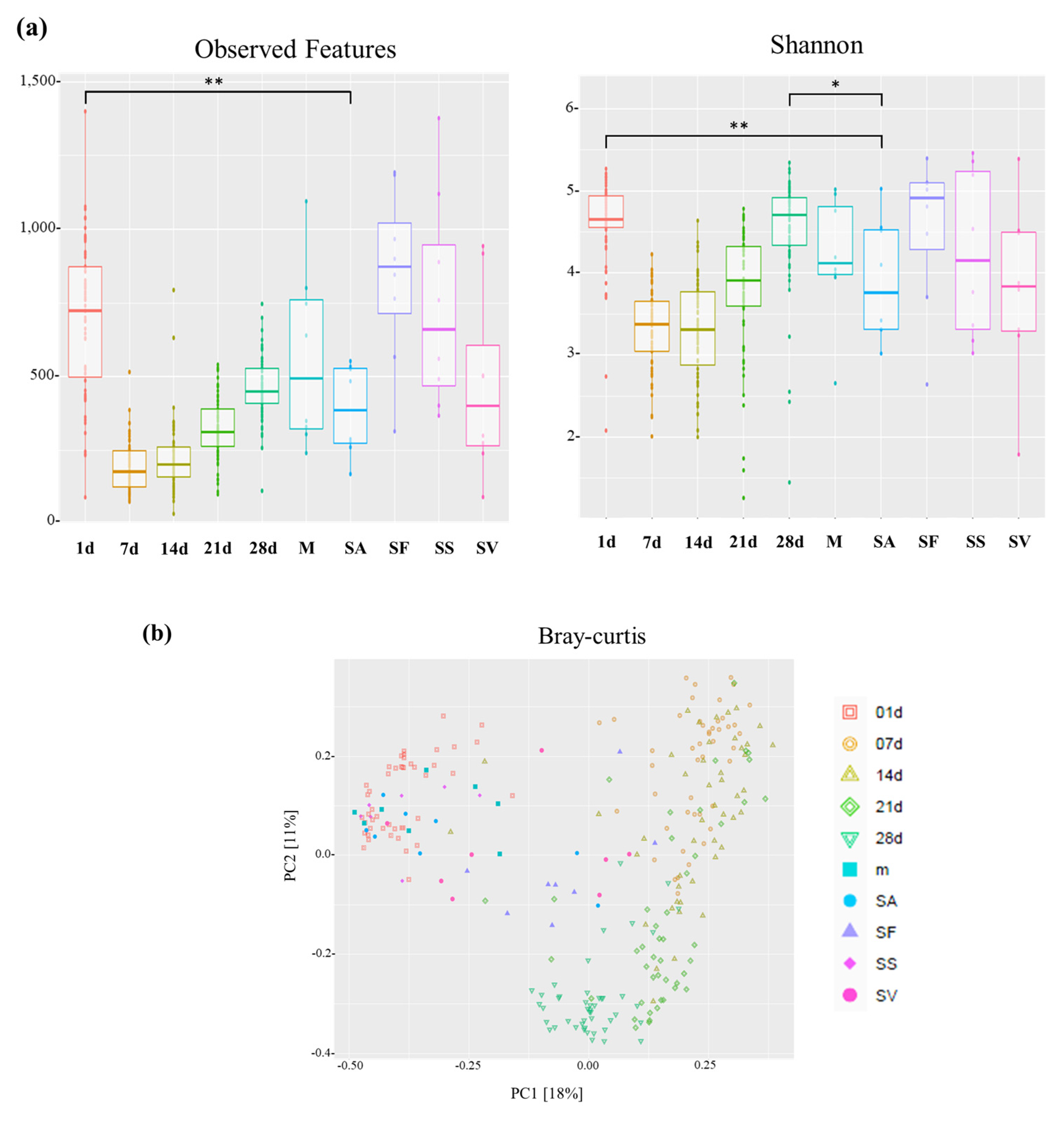
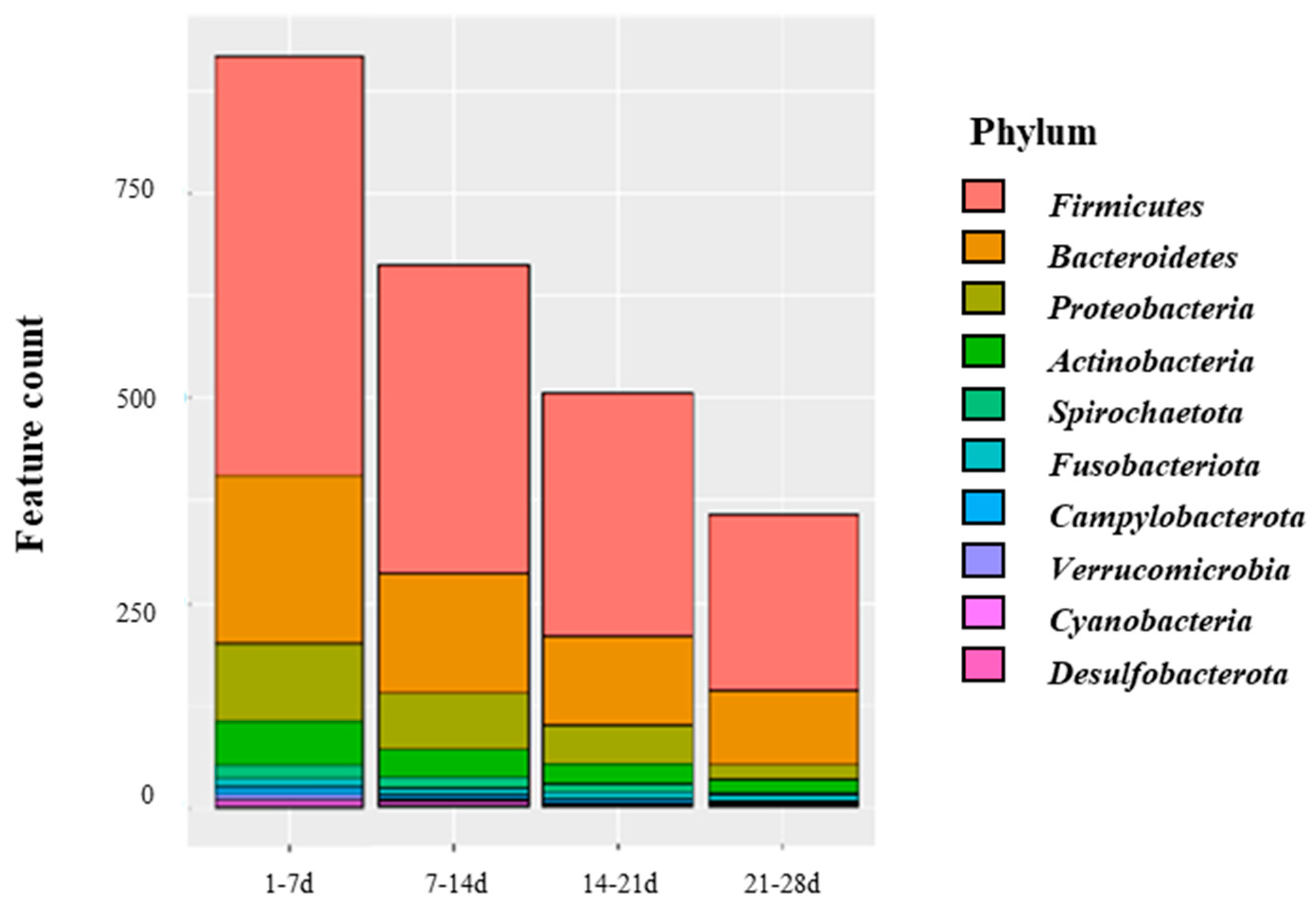
3.3. Microbial Diversity in Piglets from Birth to Weaning
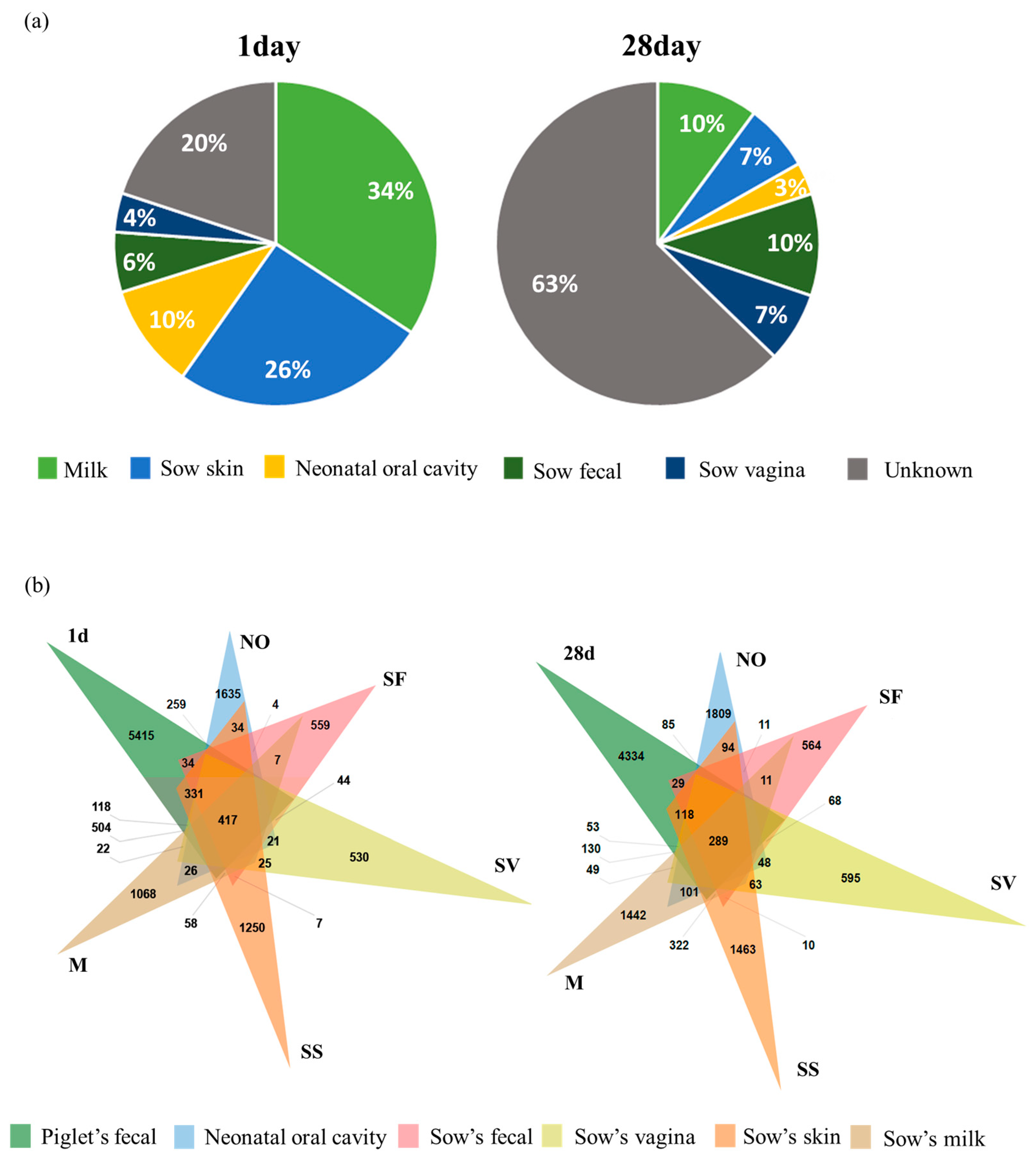
3.4. Transmission of the Maternal Microbiome from Sows to Piglets
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bassols, A.; Costa, C.; Eckersall, P.D.; Osada, J.; Sabria, J.; Tibau, J. The pig as an animal model for human pathologies: A proteomics perspective. Proteom.-Clin. Appl. 2014, 8, 715–731. [Google Scholar]
- Whitton, C.; Bogueva, D.; Marinova, D.; Phillips, C.J. Are we approaching peak meat consumption? Analysis of meat consumption from 2000 to 2019 in 35 countries and its relationship to gross domestic product. Animals 2021, 11, 3466. [Google Scholar]
- Yang, A.-Q.; Chen, B.; Ran, M.-L.; Yang, G.-M.; Zeng, C. The application of genomic selection in pig cross breeding. Yi Chuan=Hered. 2020, 42, 145–152. [Google Scholar]
- Lebret, B. Effects of feeding and rearing systems on growth, carcass composition and meat quality in pigs. Animal 2008, 2, 1548–1558. [Google Scholar]
- Jiang, H.; Fang, S.; Yang, H.; Chen, C. Identification of the relationship between the gut microbiome and feed efficiency in a commercial pig cohort. J. Anim. Sci. 2021, 99, skab045. [Google Scholar]
- Vasquez, R.; Oh, J.K.; Song, J.H.; Kang, D.-K. Gut microbiome-produced metabolites in pigs: A review on their biological functions and the influence of probiotics. J. Anim. Sci. Technol. 2022, 64, 671. [Google Scholar]
- Turnbaugh, P.J.; Ley, R.E.; Hamady, M.; Fraser-Liggett, C.M.; Knight, R.; Gordon, J.I. The human microbiome project. Nature 2007, 449, 804–810. [Google Scholar]
- Pickard, J.M.; Zeng, M.Y.; Caruso, R.; Núñez, G. Gut microbiota: Role in pathogen colonization, immune responses, and inflammatory disease. Immunol. Rev. 2017, 279, 70–89. [Google Scholar]
- Jandhyala, S.M.; Talukdar, R.; Subramanyam, C.; Vuyyuru, H.; Sasikala, M.; Reddy, D.N. Role of the normal gut microbiota. World J. Gastroenterol. 2015, 21, 8787. [Google Scholar]
- Collado, M.C.; Rautava, S.; Aakko, J.; Isolauri, E.; Salminen, S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci. Rep. 2016, 6, 23129. [Google Scholar]
- Aagaard, K.; Ma, J.; Antony, K.M.; Ganu, R.; Petrosino, J.; Versalovic, J. The placenta harbors a unique microbiome. Sci. Transl. Med. 2014, 6, 237ra265. [Google Scholar]
- The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar]
- Walker, W.A. The importance of appropriate initial bacterial colonization of the intestine in newborn, child, and adult health. Pediatr. Res. 2017, 82, 387–395. [Google Scholar]
- Hansen, C.H.F.; Nielsen, D.S.; Kverka, M.; Zakostelska, Z.; Klimesova, K.; Hudcovic, T.; Tlaskalova-Hogenova, H.; Hansen, A.K. Patterns of early gut colonization shape future immune responses of the host. PloS ONE 2012, 7, e34043. [Google Scholar]
- Guevarra, R.B.; Lee, J.H.; Lee, S.H.; Seok, M.-J.; Kim, D.W.; Kang, B.N.; Johnson, T.J.; Isaacson, R.E.; Kim, H.B. Piglet gut microbial shifts early in life: Causes and effects. J. Anim. Sci. Technol. 2019, 10, 1–10. [Google Scholar]
- de Goffau, M.C.; Lager, S.; Sovio, U.; Gaccioli, F.; Cook, E.; Peacock, S.J.; Parkhill, J.; Charnock-Jones, D.S.; Smith, G. Human placenta has no microbiome but can contain potential pathogens. Nature 2019, 572, 329–334. [Google Scholar]
- Mitchell, C.M.; Mazzoni, C.; Hogstrom, L.; Bryant, A.; Bergerat, A.; Cher, A.; Pochan, S.; Herman, P.; Carrigan, M.; Sharp, K. Delivery mode affects stability of early infant gut microbiota. Cell Rep. Med. 2020, 1, 100156. [Google Scholar]
- Laursen, M.F.; Pekmez, C.T.; Larsson, M.W.; Lind, M.V.; Yonemitsu, C.; Larnkjær, A.; Mølgaard, C.; Bode, L.; Dragsted, L.O.; Michaelsen, K.F. Maternal milk microbiota and oligosaccharides contribute to the infant gut microbiota assembly. ISME Commun. 2021, 1, 21. [Google Scholar]
- Cioffi, C.C.; Tavalire, H.F.; Neiderhiser, J.M.; Bohannan, B.; Leve, L.D. History of breastfeeding but not mode of delivery shapes the gut microbiome in childhood. PLoS ONE 2020, 15, e0235223. [Google Scholar]
- Sutera, A.M.; Arfuso, F.; Tardiolo, G.; Riggio, V.; Fazio, F.; Aiese Cigliano, R.; Paytuví, A.; Piccione, G.; Zumbo, A. Effect of a Co-Feed Liquid Whey-Integrated Diet on Crossbred Pigs’ Fecal Microbiota. Animals 2023, 13, 1750. [Google Scholar]
- Tardiolo, G.; Romeo, O.; Zumbo, A.; Di Marsico, M.; Sutera, A.M.; Cigliano, R.A.; Paytuví, A.; D’Alessandro, E. Characterization of the Nero Siciliano Pig Fecal Microbiota after a Liquid Whey-Supplemented Diet. Animals 2023, 13, 642. [Google Scholar]
- Jost, T.; Lacroix, C.; Braegger, C.P.; Rochat, F.; Chassard, C. Vertical mother–neonate transfer of maternal gut bacteria via breastfeeding. Environ. Microbiol. 2014, 16, 2891–2904. [Google Scholar]
- Mackie, R.I.; Sghir, A.; Gaskins, H.R. Developmental microbial ecology of the neonatal gastrointestinal tract. Am. J. Clin. Nutr. 1999, 69, 1035s–1045s. [Google Scholar]
- Lima, J.; Manning, T.; Rutherford, K.; Baima, E.; Dewhurst, R.; Walsh, P.; Roehe, R. Taxonomic annotation of 16S rRNA sequences of pig intestinal samples using MG-RAST and QIIME2 generated different microbiota compositions. J. Microbiol. Methods 2021, 186, 106235. [Google Scholar]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2′s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar]
- Bardou, P.; Mariette, J.; Escudié, F.; Djemiel, C.; Klopp, C. jvenn: An interactive Venn diagram viewer. BMC Bioinform. 2014, 15, 293. [Google Scholar]
- Bray, J.R.; Curtis, J.T. An ordination of the upland forest communities of southern Wisconsin. Ecol. Monogr. 1957, 27, 326–349. [Google Scholar]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar]
- Wilcoxon, F. Probability tables for individual comparisons by ranking methods. Biometrics 1947, 3, 119–122. [Google Scholar]
- Knights, D.; Kuczynski, J.; Charlson, E.S.; Zaneveld, J.; Mozer, M.C.; Collman, R.G.; Bushman, F.D.; Knight, R.; Kelley, S.T. Bayesian community-wide culture-independent microbial source tracking. Nat. Methods 2011, 8, 761–763. [Google Scholar]
- Varga, G.A.; Kolver, E.S. Microbial and animal limitations to fiber digestion and utilization. J. Nutr. 1997, 127, 819S–823S. [Google Scholar]
- Neis, E.P.; Dejong, C.H.; Rensen, S.S. The role of microbial amino acid metabolism in host metabolism. Nutrients 2015, 7, 2930–2946. [Google Scholar]
- Thakur, K.; Tomar, S.K.; De, S. Lactic acid bacteria as a cell factory for riboflavin production. Microb. Biotechnol. 2016, 9, 441–451. [Google Scholar]
- Hill, M. Intestinal flora and endogenous vitamin synthesis. Eur. J. Cancer Prev. 1997, 6, S43–S45. [Google Scholar]
- Rowland, I.; Gibson, G.; Heinken, A.; Scott, K.; Swann, J.; Thiele, I.; Tuohy, K. Gut microbiota functions: Metabolism of nutrients and other food components. Eur. J. Nutr. 2018, 57, 1–24. [Google Scholar]
- Van den Elsen, L.W.; Garssen, J.; Burcelin, R.; Verhasselt, V. Shaping the gut microbiota by breastfeeding: The gateway to allergy prevention? Front. Pediatr. 2019, 7, 47. [Google Scholar]
- Tamburini, S.; Shen, N.; Wu, H.C.; Clemente, J.C. The microbiome in early life: Implications for health outcomes. Nat. Med. 2016, 22, 713–722. [Google Scholar]
- Nowland, T.L.; Plush, K.J.; Barton, M.; Kirkwood, R.N. Development and function of the intestinal microbiome and potential implications for pig production. Animals 2019, 9, 76. [Google Scholar]
- Rooks, M.G.; Garrett, W.S. Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 2016, 16, 341–352. [Google Scholar]
- Yao, Y.; Cai, X.; Ye, Y.; Wang, F.; Chen, F.; Zheng, C. The role of microbiota in infant health: From early life to adulthood. Front. Immunol. 2021, 12, 708472. [Google Scholar]
- Janzon, A.; Goodrich, J.K.; Koren, O.; Group, T.S.; Waters, J.L.; Ley, R.E. Interactions between the gut microbiome and mucosal immunoglobulins A, M, and G in the developing infant gut. Msystems 2019, 4, e00612–e00619. [Google Scholar]
- Chen, L.; Xu, Y.; Chen, X.; Fang, C.; Zhao, L.; Chen, F. The maturing development of gut microbiota in commercial piglets during the weaning transition. Front. Microbiol. 2017, 8, 1688. [Google Scholar]
- Arumugam, M.; Raes, J.; Pelletier, E.; Le Paslier, D.; Yamada, T.; Mende, D.R.; Fernandes, G.R.; Tap, J.; Bruls, T.; Batto, J.-M. Enterotypes of the human gut microbiome. Nature 2011, 473, 174–180. [Google Scholar]
- Sun, Y.; Zhang, S.; Nie, Q.; He, H.; Tan, H.; Geng, F.; Ji, H.; Hu, J.; Nie, S. Gut firmicutes: Relationship with dietary fiber and role in host homeostasis. Crit. Rev. Food Sci. Nutr. 2022, 1–16. [Google Scholar] [CrossRef]
- Azad, M.; Kalam, A.; Sarker, M.; Li, T.; Yin, J. Probiotic species in the modulation of gut microbiota: An overview. BioMed Res. Int. 2018, 2018, 9478630. [Google Scholar]
- Marcobal, A.; Sonnenburg, J. Human milk oligosaccharide consumption by intestinal microbiota. Clin. Microbiol. Inf. 2012, 18, 12–15. [Google Scholar]
- Li, M.; Bauer, L.L.; Chen, X.; Wang, M.; Kuhlenschmidt, T.B.; Kuhlenschmidt, M.S.; Fahey, G.C., Jr.; Donovan, S.M. Microbial composition and in vitro fermentation patterns of human milk oligosaccharides and prebiotics differ between formula-fed and sow-reared piglets. J. Nutr. 2012, 142, 681–689. [Google Scholar]
- Hu, J.; Nie, Y.; Chen, J.; Zhang, Y.; Wang, Z.; Fan, Q.; Yan, X. Gradual changes of gut microbiota in weaned miniature piglets. Front. Microbiol. 2016, 7, 1727. [Google Scholar]
- Kim, H.B.; Borewicz, K.; White, B.A.; Singer, R.S.; Sreevatsan, S.; Tu, Z.J.; Isaacson, R.E. Longitudinal investigation of the age-related bacterial diversity in the feces of commercial pigs. Vet. Microbiol. 2011, 153, 124–133. [Google Scholar]
- Wei, J.; Goldberg, M.; Burland, V.; Venkatesan, M.; Deng, W.; Fournier, G.; Mayhew, G.; Plunkett, G., III; Rose, D.; Darling, A. Complete genome sequence and comparative genomics of Shigella flexneri serotype 2a strain 2457T. Infect. Immun. 2003, 71, 2775–2786. [Google Scholar]
- Bettelheim, K.; Lennox-King, S. The acquisition of Escherichia coli by new-born babies. Infection 1976, 4, 174–179. [Google Scholar]
- Nowrouzian, F.; Hesselmar, B.; Saalman, R.; Strannegård, I.-L.; Åberg, N.; Wold, A.E.; Adlerberth, I. Escherichia coli in infants’ intestinal microflora: Colonization rate, strain turnover, and virulence gene carriage. Pediatr. Res. 2003, 54, 8–14. [Google Scholar]
- Bode, L. Human milk oligosaccharides: Every baby needs a sugar mama. Glycobiology 2012, 22, 1147–1162. [Google Scholar]
- LaTuga, M.S.; Stuebe, A.; Seed, P.C. A review of the source and function of microbiota in breast milk. Semin. Reprod. Med. 2014, 32, 68–73. [Google Scholar]
- Chen, W.; Mi, J.; Lv, N.; Gao, J.; Cheng, J.; Wu, R.; Ma, J.; Lan, T.; Liao, X. Lactation stage-dependency of the sow milk microbiota. Front. Microbiol. 2018, 9, 945. [Google Scholar]
- Wang, D.; Tang, G.; Zhao, L.; Wang, M.; Chen, L.; Zhao, C.; Liang, Z.; Chen, J.; Cao, Y.; Yao, J. Potential roles of the rectum keystone microbiota in modulating the microbial community and growth performance in goat model. J. Anim. Sci. Biotechnol. 2023, 14, 1–15. [Google Scholar]
- Stinson, L.F.; Payne, M.S.; Keelan, J.A. Planting the seed: Origins, composition, and postnatal health significance of the fetal gastrointestinal microbiota. Crit. Rev. Microbiol. 2017, 43, 352–369. [Google Scholar]
- Schubbert, R.; Hohlweg, U.; Renz, D.; Doerfler, W. On the fate of orally ingested foreign DNA in mice: Chromosomal association and placental transmission to the fetus. Mol. Gen. Genet. 1998, 259, 569–576. [Google Scholar]
- Liu, S.; Zhang, Z.; Ma, L. A Review Focusing on Microbial Vertical Transmission during Sow Pregnancy. Vet. Sci. 2023, 10, 123. [Google Scholar]
- Elolimy, A.; Alharthi, A.; Zeineldin, M.; Parys, C.; Helmbrecht, A.; Loor, J.J. Supply of methionine during late-pregnancy alters fecal microbiota and metabolome in neonatal dairy calves without changes in daily feed intake. Front. Microbiol. 2019, 10, 2159. [Google Scholar]
- Liu, H.; Zeng, X.; Zhang, G.; Hou, C.; Li, N.; Yu, H.; Shang, L.; Zhang, X.; Trevisi, P.; Yang, F. Maternal milk and fecal microbes guide the spatiotemporal development of mucosa-associated microbiota and barrier function in the porcine neonatal gut. BMC Biol. 2019, 17, 106. [Google Scholar]
- Pannaraj, P.S.; Li, F.; Cerini, C.; Bender, J.M.; Yang, S.; Rollie, A.; Adisetiyo, H.; Zabih, S.; Lincez, P.J.; Bittinger, K. Association between breast milk bacterial communities and establishment and development of the infant gut microbiome. JAMA Pediatr. 2017, 171, 647–654. [Google Scholar]
- Chavoya-Guardado, M.A.; Vasquez-Garibay, E.M.; Ruiz-Quezada, S.L.; Ramírez-Cordero, M.I.; Larrosa-Haro, A.; Castro-Albarran, J. Firmicutes, bacteroidetes and actinobacteria in human milk and maternal adiposity. Nutrients 2022, 14, 2887. [Google Scholar]
- Den Besten, G.; Van Eunen, K.; Groen, A.K.; Venema, K.; Reijngoud, D.-J.; Bakker, B.M. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 2013, 54, 2325–2340. [Google Scholar]
- Natividad, J.M.; Pinto-Sanchez, M.I.; Galipeau, H.J.; Jury, J.; Jordana, M.; Reinisch, W.; Collins, S.M.; Bercik, P.; Surette, M.G.; Allen-Vercoe, E. Ecobiotherapy rich in Firmicutes decreases susceptibility to colitis in a humanized gnotobiotic mouse model. Inflamm. Bowel Dis. 2015, 21, 1883–1893. [Google Scholar]
- Chen, D.; Wang, Y.-Y.; Li, S.-P.; Zhao, H.-M.; Jiang, F.-J.; Wu, Y.-X.; Tong, Y.; Pang, Q.-F. Maternal propionate supplementation ameliorates glucose and lipid metabolic disturbance in hypoxia-induced fetal growth restriction. Food Funct. 2022, 13, 10724–10736. [Google Scholar]
- Dogra, S.; Sakwinska, O.; Soh, S.-E.; Ngom-Bru, C.; Brück, W.M.; Berger, B.; Brüssow, H.; Karnani, N.; Lee, Y.S.; Yap, F. Rate of establishing the gut microbiota in infancy has consequences for future health. Gut Microbes 2015, 6, 321–325. [Google Scholar]
- Garcia-Mantrana, I.; Collado, M.C. Obesity and overweight: Impact on maternal and milk microbiome and their role for infant health and nutrition. Mol. Nutr. Food Res. 2016, 60, 1865–1875. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lim, J.-A.; Cha, J.; Choi, S.; Kim, J.-H.; Kim, D. Early Colonization of the Intestinal Microbiome of Neonatal Piglets Is Influenced by the Maternal Microbiome. Animals 2023, 13, 3378. https://doi.org/10.3390/ani13213378
Lim J-A, Cha J, Choi S, Kim J-H, Kim D. Early Colonization of the Intestinal Microbiome of Neonatal Piglets Is Influenced by the Maternal Microbiome. Animals. 2023; 13(21):3378. https://doi.org/10.3390/ani13213378
Chicago/Turabian StyleLim, Jin-A, Jihye Cha, Soyoung Choi, Jae-Hwan Kim, and Dahye Kim. 2023. "Early Colonization of the Intestinal Microbiome of Neonatal Piglets Is Influenced by the Maternal Microbiome" Animals 13, no. 21: 3378. https://doi.org/10.3390/ani13213378
APA StyleLim, J.-A., Cha, J., Choi, S., Kim, J.-H., & Kim, D. (2023). Early Colonization of the Intestinal Microbiome of Neonatal Piglets Is Influenced by the Maternal Microbiome. Animals, 13(21), 3378. https://doi.org/10.3390/ani13213378




