Effects of Different Generations and Sex on Physiological, Biochemical, and Growth Parameters of Crossbred Beef Cattle by Myostatin Gene-Edited Luxi Bulls and Simmental Cows
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
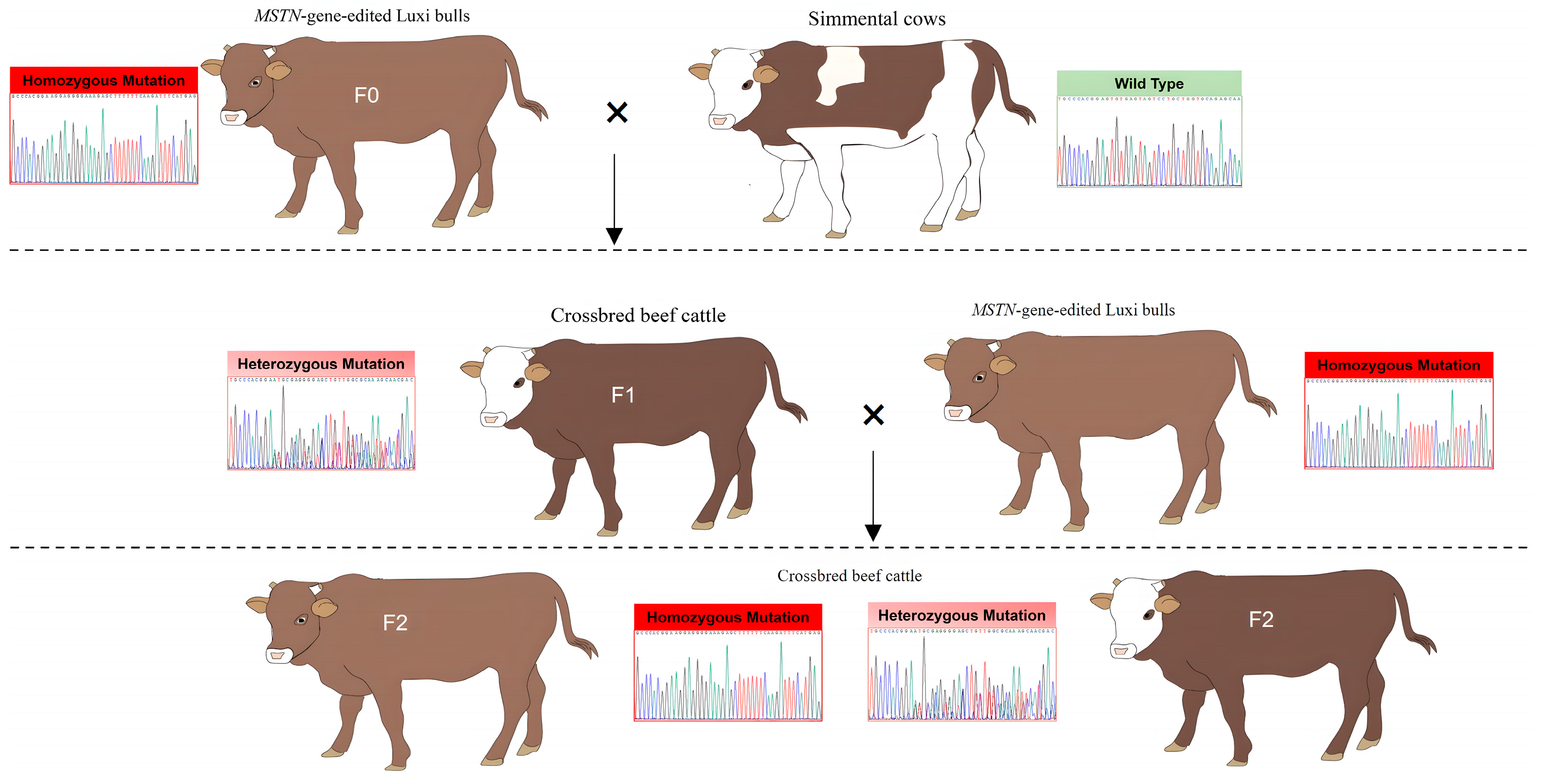
2.1. Animals
2.2. Body Weight and Body Size Measurement
2.3. Detection of Blood Physiological and Biochemical Parameters
2.4. Determination of Serum Insulin
2.5. Correlation Analysis
2.6. Statistical Analysis
3. Results
3.1. Physical Appearance and Growth Parameters
3.2. Hematological Parameters
3.3. Biochemical Parameters
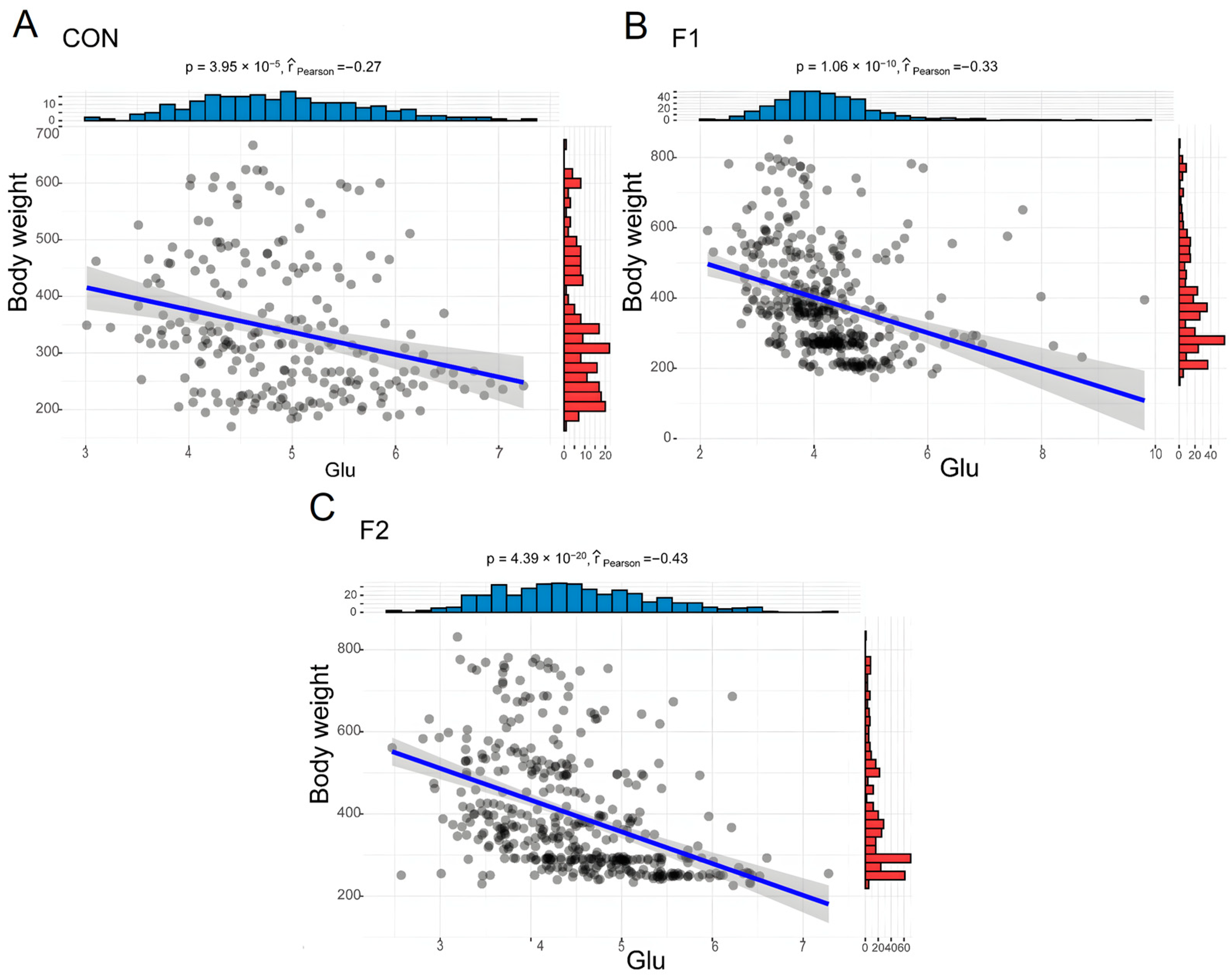
3.4. Relationship between BW and Hematological, and Biochemical Parameters
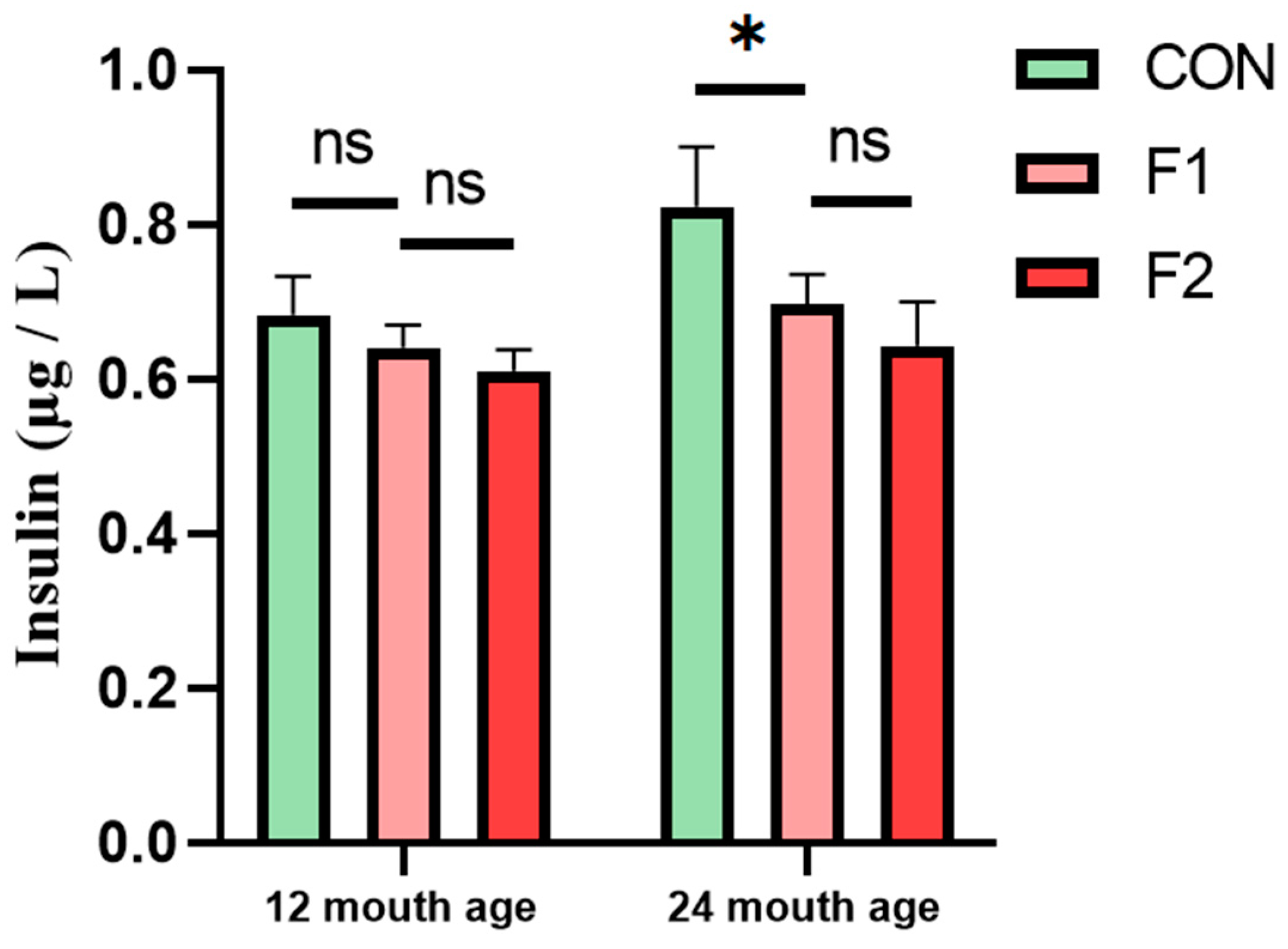
3.5. Determination of Serum Insulin
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fiems, L.O. Double Muscling in Cattle: Genes, Husbandry, Carcasses and Meat. Animals 2012, 2, 472–506. [Google Scholar] [CrossRef] [PubMed]
- McPherron, A.C.; Lee, S.J. Double muscling in cattle due to mutations in the myostatin gene. Proc. Natl. Acad. Sci. USA 1997, 94, 12457–12461. [Google Scholar] [CrossRef] [PubMed]
- Kambadur, R.; Sharma, M.; Smith, T.P.; Bass, J.J. Mutations in myostatin (GDF8) in double-muscled Belgian Blue and Piedmontese cattle. Genome Res. 1997, 7, 910–916. [Google Scholar] [CrossRef] [PubMed]
- Phocas, F. Genetic analysis of breeding traits in a Charolais cattle population segregating an inactive myostatin allele. J. Anim. Sci. 2009, 87, 1865–1871. [Google Scholar] [CrossRef]
- Esmailizadeh, A.K.; Bottema, C.D.; Sellick, G.S.; Verbyla, A.P.; Morris, C.A.; Cullen, N.G.; Pitchford, W.S. Effects of the myostatin F94L substitution on beef traits. J. Anim. Sci. 2008, 86, 1038–1046. [Google Scholar] [CrossRef]
- Bouyer, C.; Forestier, L.; Renand, G.; Oulmouden, A. Deep intronic mutation and pseudo exon activation as a novel muscular hypertrophy modifier in cattle. PLoS ONE 2014, 9, e97399. [Google Scholar] [CrossRef]
- Dierks, C.; Eder, J.; Glatzer, S.; Lehner, S.; Distl, O. A novel myostatin mutation in double-muscled German Gelbvieh. Anim. Genet. 2015, 46, 91–92. [Google Scholar] [CrossRef]
- Heaton, M.P.; Smith, T.P.L.; Bickhart, D.M.; Vander Ley, B.L.; Kuehn, L.A.; Oppenheimer, J.; Shafer, W.R.; Schuetze, F.T.; Stroud, B.; McClure, J.C.; et al. A Reference Genome Assembly of Simmental Cattle, Bos taurus taurus. J. Hered. 2021, 112, 184–191. [Google Scholar] [CrossRef]
- Zhu, B.; Guo, P.; Wang, Z.; Zhang, W.; Chen, Y.; Zhang, L.; Gao, H.; Wang, Z.; Gao, X.; Xu, L.; et al. Accuracies of genomic prediction for twenty economically important traits in Chinese Simmental beef cattle. Anim. Genet. 2019, 50, 634–643. [Google Scholar] [CrossRef]
- Gao, L.; Yang, M.; Wang, X.; Yang, L.; Bai, C.; Li, G. Mstn knockdown decreases the trans-differentiation from myocytes to adipocytes by reducing Jmjd3 expression via the SMAD2/SMAD3 complex. Biosci. Biotechnol. Biochem. 2019, 83, 2090–2096. [Google Scholar] [CrossRef]
- Gao, L.; Yang, M.; Wei, Z.; Gu, M.; Yang, L.; Bai, C.; Wu, Y.; Li, G. MSTN Mutant Promotes Myogenic Differentiation by Increasing Demethylase TET1 Expression via the SMAD2/SMAD3 Pathway. Int. J. Biol. Sci. 2020, 16, 1324–1334. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Yang, L.; Su, G.; Wei, Z.; Liu, X.; Song, L.; Hai, C.; Wu, D.; Hao, Z.; Wu, Y.; et al. Growth Traits and Sperm Proteomics Analyses of Myostatin Gene-Edited Chinese Yellow Cattle. Life 2022, 12, 627. [Google Scholar] [CrossRef] [PubMed]
- Mosher, D.S.; Quignon, P.; Bustamante, C.D.; Sutter, N.B.; Mellersh, C.S.; Parker, H.G.; Ostrander, E.A. A mutation in the myostatin gene increases muscle mass and enhances racing performance in heterozygote dogs. PLoS Genet. 2007, 3, e79. [Google Scholar] [CrossRef] [PubMed]
- Clop, A.; Marcq, F.; Takeda, H.; Pirottin, D.; Tordoir, X.; Bibé, B.; Bouix, J.; Caiment, F.; Elsen, J.M.; Eychenne, F.; et al. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat. Genet. 2006, 38, 813–818. [Google Scholar] [CrossRef]
- Li, X.L.; Wu, Z.L.; Liu, Z.Z.; Gong, Y.F.; Zhou, R.Y.; Zheng, G.R. SNP identification and analysis in part of intron 2 of goat MSTN gene and variation within and among species. J. Hered. 2006, 97, 285–289. [Google Scholar] [CrossRef]
- Schuelke, M.; Wagner, K.R.; Stolz, L.E.; Hübner, C.; Riebel, T.; Kömen, W.; Braun, T.; Tobin, J.F.; Lee, S.J. Myostatin mutation associated with gross muscle hypertrophy in a child. N. Engl. J. Med. 2004, 350, 2682–2688. [Google Scholar] [CrossRef]
- Han, H.; Ma, Y.; Wang, T.; Lian, L.; Tian, X.; Hu, R.; Deng, S.; Li, K.; Wang, F.; Li, N.; et al. One-step generation of myostatin gene knockout sheep via the CRISPR/Cas9 system. Front. Agric. Sci. Eng. 2014, 1, 2–5. [Google Scholar] [CrossRef]
- Luo, J.; Song, Z.; Yu, S.; Cui, D.; Wang, B.; Ding, F.; Li, S.; Dai, Y.; Li, N. Efficient generation of myostatin (MSTN) biallelic mutations in cattle using zinc finger nucleases. PLoS ONE 2014, 9, e95225. [Google Scholar] [CrossRef]
- Qian, L.; Tang, M.; Yang, J.; Wang, Q.; Cai, C.; Jiang, S.; Li, H.; Jiang, K.; Gao, P.; Ma, D.; et al. Targeted mutations in myostatin by zinc-finger nucleases result in double-muscled phenotype in Meishan pigs. Sci. Rep. 2015, 5, 14435. [Google Scholar] [CrossRef]
- Zou, Q.; Wang, X.; Liu, Y.; Ouyang, Z.; Long, H.; Wei, S.; Xin, J.; Zhao, B.; Lai, S.; Shen, J.; et al. Generation of gene-target dogs using CRISPR/Cas9 system. J. Mol. Cell Biol. 2015, 7, 580–583. [Google Scholar] [CrossRef]
- Lv, Q.; Yuan, L.; Deng, J.; Chen, M.; Wang, Y.; Zeng, J.; Li, Z.; Lai, L. Efficient Generation of Myostatin Gene Mutated Rabbit by CRISPR/Cas9. Sci. Rep. 2016, 6, 25029. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Cai, B.; Zhou, S.; Zhu, H.; Qu, L.; Wang, X.; Chen, Y. RNA-seq reveals transcriptome changes in goats following myostatin gene knockout. PLoS ONE 2017, 12, e0187966. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Zhang, T.; Jiang, L.; Zhou, M.; Wu, D.; Mei, J.; Cheng, Y. Use of CRISPR/Cas9 technology efficiently targetted goat myostatin through zygotes microinjection resulting in double-muscled phenotype in goats. Biosci. Rep. 2018, 38, BSR20180742. [Google Scholar] [CrossRef] [PubMed]
- Domingo, G.; Iglesias, A.; Monserrat, L.; Sanchez, L.; Cantalapiedra, J.; Lorenzo, J.M. Effect of crossbreeding with Limousine, Rubia Gallega and Belgium Blue on meat quality and fatty acid profile of Holstein calves. Anim. Sci. J. 2015, 86, 913–921. [Google Scholar] [CrossRef]
- Keady, S.M.; Waters, S.M.; Hamill, R.M.; Dunne, P.G.; Keane, M.G.; Richardson, R.I.; Kenny, D.A.; Moloney, A.P. Compensatory growth in crossbred Aberdeen Angus and Belgian Blue steers: Effects on the colour, shear force and sensory characteristics of longissimus muscle. Meat Sci. 2017, 125, 128–136. [Google Scholar] [CrossRef] [PubMed]
- Casas, E.; Bennett, G.L.; Smith, T.P.; Cundiff, L.V. Association of myostatin on early calf mortality, growth, and carcass composition traits in crossbred cattle. J. Anim. Sci. 2004, 82, 2913–2918. [Google Scholar] [CrossRef]
- Liu, X.; Pang, D.; Yuan, T.; Li, Z.; Li, Z.; Zhang, M.; Ren, W.; Ouyang, H.; Tang, X. N-3 polyunsaturated fatty acids attenuates triglyceride and inflammatory factors level in hfat-1 transgenic pigs. Lipids Health Dis. 2016, 15, 89. [Google Scholar] [CrossRef]
- Tang, L.; Yan, Z.; Wan, Y.; Han, W.; Zhang, Y. Myostatin DNA vaccine increases skeletal muscle mass and endurance in mice. Lipids Health Dis. 2007, 36, 342–348. [Google Scholar] [CrossRef]
- Li, Y.; Lian, D.; Deng, S.; Zhang, X.; Zhang, J.; Li, W.; Bai, H.; Wang, Z.; Wu, H.; Fu, J.; et al. Efficient production of pronuclear embryos in breeding and nonbreeding season for generating transgenic sheep overexpressing TLR4. J. Anim. Sci. Biotechnol. 2016, 7, 38. [Google Scholar] [CrossRef]
- Baron, M.; Maillet, J.; Huyvaert, M.; Dechaume, A.; Boutry, R.; Loiselle, H.; Durand, E.; Toussaint, B.; Vaillant, E.; Philippe, J.; et al. Loss-of-function mutations in MRAP2 are pathogenic in hyperphagic obesity with hyperglycemia and hypertension. Nat. Med. 2019, 25, 1733–1738. [Google Scholar] [CrossRef]
- Xin, X.B.; Yang, S.P.; Li, X.; Liu, X.F.; Zhang, L.L.; Ding, X.B.; Zhang, S.; Li, G.P.; Guo, H. Proteomics insights into the effects of MSTN on muscle glucose and lipid metabolism in genetically edited cattle. Gen. Comp. Endocrinol. 2020, 291, 113237. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Ye, J.; Cao, L.; Zhang, Y.; Xia, W.; Zhu, D. Myostatin regulates glucose metabolism via the AMP-activated protein kinase pathway in skeletal muscle cells. Int. J. Biochem. Cell Biol. 2010, 42, 2072–2081. [Google Scholar] [CrossRef] [PubMed]
- Park, S.W.; Goodpaster, B.H.; Strotmeyer, E.S.; Kuller, L.H.; Broudeau, R.; Kammerer, C.; de Rekeneire, N.; Harris, T.B.; Schwartz, A.V.; Tylavsky, F.A.; et al. Accelerated loss of skeletal muscle strength in older adults with type 2 diabetes: The health, aging, and body composition study. Diabetes Care 2007, 30, 1507–1512. [Google Scholar] [CrossRef] [PubMed]
- Morissette, M.R.; Stricker, J.C.; Rosenberg, M.A.; Buranasombati, C.; Levitan, E.B.; Mittleman, M.A.; Rosenzweig, A. Effects of myostatin deletion in aging mice. Aging Cell 2009, 8, 573–583. [Google Scholar] [CrossRef] [PubMed]
- Pabst, R. The pig as a model for immunology research. Cell Tissue Res. 2020, 380, 287–304. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Filippova, M.; Fagoaga, O.; Nehlsen-Cannarella, S.; Escher, A. Decreased insulitis and blood glucose levels after injection of GAD-transduced lymphocytes into NOD mice. Mol. Ther. 2002, 6, 701–709. [Google Scholar] [CrossRef]
- Riscal, R.; Skuli, N.; Simon, M.C. Even Cancer Cells Watch Their Cholesterol! Mol. Cell 2019, 76, 220–231. [Google Scholar] [CrossRef]
- Alvarado, M.M.; Kum, H.C.; Gonzalez Coronado, K.; Foster, M.J.; Ortega, P.; Lawley, M.A. Barriers to Remote Health Interventions for Type 2 Diabetes: A Systematic Review and Proposed Classification Scheme. J. Med. Internet Res. 2017, 19, e28. [Google Scholar] [CrossRef]
- Dupuis, J.; Langenberg, C.; Prokopenko, I.; Saxena, R.; Soranzo, N.; Jackson, A.U.; Wheeler, E.; Glazer, N.L.; Bouatia-Naji, N.; Gloyn, A.L.; et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat. Genet. 2010, 42, 105–116. [Google Scholar] [CrossRef]
- Cohen, D.H.; LeRoith, D. Obesity, type 2 diabetes, and cancer: The insulin and IGF connection. Endocr. Relat. Cancer 2012, 19, F27–F45. [Google Scholar] [CrossRef]
- Smith, T.M.; Austin, C.; Hinde, K.; Vogel, E.R.; Arora, M. Cyclical nursing patterns in wild orangutans. Sci. Adv. 2017, 3, e1601517. [Google Scholar] [CrossRef]
- Valdés, J.A.; Flores, S.; Fuentes, E.N.; Osorio-Fuentealba, C.; Jaimovich, E.; Molina, A. IGF-1 induces IP3 -dependent calcium signal involved in the regulation of myostatin gene expression mediated by NFAT during myoblast differentiation. J. Cell. Physiol. 2013, 228, 1452–1463. [Google Scholar] [CrossRef]
- Forbes, D.; Jackman, M.; Bishop, A.; Thomas, M.; Kambadur, R.; Sharma, M. Myostatin auto-regulates its expression by feedback loop through Smad7 dependent mechanism. J. Cell. Physiol. 2006, 206, 264–272. [Google Scholar] [CrossRef]



| Parameters | Sex | Age | Generations | p-Value | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M | F | 0 | 3 | 6 | 9 | 12 | 15 | 18 | 24 | WT | F1 | F2 | Sex | Age | Generation | Sex × Age | Sex × Generation | Age × Generation | Sex × Age × Generation | |
| BW (kg) | 305.7 ± 199.6 a | 259.7 ± 150.2 b | 41.1 ± 4 h | 117.2 ± 4.8 g | 182.5 ± 22 f | 237.5 ± 26.2 e | 276.5 ± 23.6 d | 352.4 ± 30.9 c | 439.6 ± 68.7 b | 605 ± 106.1 a | 252.9 ± 150.7 b | 285.8 ± 184.1 a | 293.3 ± 181.7 a | 0.000 | 0.000 | 0.461 | 0.000 | 0.876 | 0.000 | 0.162 |
| Shoulder height (cm) | 116.5 ± 26 a | 112.1 ± 21.8 b | 72.1 ± 5.3 h | 90.1 ± 2.2 g | 102.6 ± 4.9 f | 112.7 ± 5.5 e | 124 ± 5.4 d | 130.7 ± 6.7 c | 134.4 ± 6.8 b | 146.9 ± 7.1 a | 112.9 ± 23.8 a | 114.1 ± 23.8 a | 115 ± 24.2 a | 0.018 | 0.000 | 0.201 | 0.000 | 0.248 | 0.889 | 0.720 |
| Cross height (cm) | 119 ± 26.3 a | 114.7 ± 22.8 b | 74.1 ± 3.2 h | 89.5 ± 2.2 g | 105.5 ± 4.6 f | 116.1 ± 5.8 e | 128.1 ± 5.3 d | 132.2 ± 6.9 c | 138.6 ± 6.6 b | 149.7 ± 6.1 a | 114 ± 23.2 b | 117.2 ± 24.7 ab | 117.9 ± 25.2 a | 0.013 | 0.000 | 0.028 | 0.000 | 0.024 | 0.751 | 0.484 |
| Hip height (cm) | 118.3 ± 26.1 a | 114.7 ± 23.2 b | 72.5 ± 4 h | 89.9 ± 2.6 g | 105.8 ± 3.4 f | 115.7 ± 7.1 e | 129.5 ± 5.9 d | 132.4 ± 5.9 c | 136.8 ± 6.2 b | 148.7 ± 5.8 a | 113.9 ± 23.7 b | 116.8 ± 24.8 ab | 117.5 ± 25 a | 0.072 | 0.000 | 0.057 | 0.001 | 0.094 | 0.470 | 0.861 |
| Shoulder width (cm) | 35.7 ± 10.5 a | 32.3 ± 8 b | 20 ± 2.2 h | 24.9 ± 2.2 g | 28.2 ± 3.3 f | 33.8 ± 3.3 e | 38.2 ± 6.3 d | 38 ± 4.5 c | 40.3 ± 3.7 b | 48 ± 5.3 a | 32.9 ± 8.9 b | 33.3 ± 9.2 b | 35 ± 9.8 a | 0.108 | 0.000 | 0.639 | 0.000 | 0.580 | 0.918 | 0.870 |
| Cross width (cm) | 37.9 ± 11.7 a | 36 ± 8.8 b | 21.4 ± 2.9 h | 27 ± 2 | 30.4 ± 1.6 f | 36.7 ± 3.9 e | 39.3 ± 3.5 d | 42.2 ± 4.1 c | 45.6 ± 4.3 b | 52.6 ± 6.2 a | 35.7 ± 9.6 b | 36.8 ± 10.4 ab | 37.6 ± 10.6 a | 0.004 | 0.000 | 0.971 | 0.000 | 0.927 | 0.880 | 0.527 |
| Hip width (cm) | 37.6 ± 11.9 a | 35.4 ± 9.7 b | 19.9 ± 3.2 h | 25.6 ± 2.5 g | 29.8 ± 1.7 f | 36.1 ± 3.9 e | 40.2 ± 3.7 d | 41.4 ± 4 c | 46 ± 4.7 b | 52.5 ± 5.9 a | 35.5 ± 10.4 a | 36.3 ± 11 a | 37.1 ± 11 a | 0.000 | 0.000 | 0.749 | 0.000 | 0.857 | 0.177 | 0.720 |
| Chest circumference (cm) | 151.1 ± 44.9 a | 146.3 ± 39.6 b | 71.8 ± 3.3 h | 109.7 ± 3 g | 133.7 ± 3.9 f | 148 ± 6 e | 156.9 ± 6.3 d | 173.4 ± 9.6 c | 185.7 ± 16.1 b | 209.1 ± 20.8 a | 143.6 ± 39.6 b | 149.9 ± 43.1 ab | 150 ± 42.7 a | 0.039 | 0.000 | 0.529 | 0.000 | 0.450 | 0.159 | 0.894 |
| Abdominal circumference (cm) | 167.8 ± 53.8 a | 161.6 ± 45.6 b | 72.7 ± 3.2 h | 119.4 ± 4.2 g | 145.8 ± 5.1 f | 165.3 ± 9.7 e | 178.8 ± 11.5 d | 190.6 ± 20.5 c | 210.1 ± 16.5 b | 233.7 ± 14.3 a | 162.6 ± 48.4 a | 165.1 ± 49.3 a | 165.1 ± 50.8 a | 0.035 | 0.000 | 0.863 | 0.000 | 0.867 | 0.212 | 0.355 |
| Body oblique length (cm) | 124.6 ± 30.4 a | 120 ± 26.1 b | 74.2 ± 3.4 h | 91.1 ± 4.2 | 110.7 ± 4 f | 121.5 ± 7.4 e | 131.4 ± 4.2 d | 140.7 ± 8.9 c | 144.8 ± 9.7 b | 163.1 ± 10.1 a | 120.5 ± 26.8 a | 122.2 ± 28.1 a | 123 ± 29.2 a | 0.159 | 0.000 | 0.578 | 0.000 | 0.829 | 0.167 | 0.672 |
| Tube circumference (cm) | 17.4 ± 5.3 a | 15.8 ± 3.6 b | 10.1 ± 1.3 h | 12.8 ± 1.1 g | 14.2 ± 1.2 f | 15 ± 1.2 e | 16.6 ± 2.1 d | 19.3 ± 2.9 | 21.1 ± 2.8 b | 23 ± 2.4 a | 16.6 ± 4.6 a | 16.7 ± 4.7 a | 16.3 ± 4.4 a | 0.004 | 0.000 | 0.108 | 0.000 | 0.306 | 0.004 | 0.001 |
| Parameters | Sex | Age | Generation | p-Value | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M | F | 9 | 12 | 15 | 18 | 24 | WT | F1 | F2 | Sex | Age | Generation | Sex × Age | Sex × Generation | Age × Generation | Sex × Age × Generation | |
| WBC (×10 L−9) | 8.51 ± 2.65 | 8.48 ± 2.50 | 9.87 ± 2.66 a | 8.89 ± 2.52 b | 8.14 ± 2.54 c | 7.89 ± 2.41 c | 7.72 ± 2.23 c | 8.62 ± 2.57 a | 8.36 ± 2.64 a | 8.56 ± 2.58 a | 0.007 | 0.000 | 0.624 | 0.091 | 0.045 | 0.266 | 0.325 |
| LYM (%) | 4.36 ± 2.80 | 4.75 ± 3.25 | 6.71 ± 4.32 a | 4.75 ± 2.78 b | 4.21 ± 2.2 bc | 3.89 ± 1.44 c | 2.91 ± 1.64 d | 6.73 ± 3.24 a | 3.66 ± 1.71 b | 3.89 ± 3.01 b | 0.517 | 0.000 | 0.000 | 0.139 | 0.834 | 0.000 | 0.269 |
| RBC (×10 L−12) | 8.01 ± 1.69 | 8.10 ± 1.78 | 9.08 ± 1.81 a | 8.36 ± 1.3 b | 7.86 ± 1.39 b | 8.06 ± 1.77 b | 6.85 ± 1.47 c | 7.39 ± 1.36 b | 8.28 ± 1.73 a | 8.21 ± 1.81 a | 0.026 | 0.000 | 0.013 | 0.174 | 0.001 | 0.021 | 0.023 |
| HCT (%) | 3229.3 ± 514.6 | 3384.5 ± 511.5 | 3286.59 ± 562.49 ab | 3392.1 ± 393.59 a | 3357.56 ± 475.39 a | 3170.8 ± 552.82 b | 3203.15 ± 553.41 b | 3293.7 ± 384.96 b | 3113.57 ± 521.03 c | 3430.83 ± 541.38 a | 0.551 | 0.210 | 0.068 | 0.339 | 0.167 | 0.007 | 0.073 |
| PLT (×10 L−9) | 297.26 ± 140.83 | 291.76 ± 156.65 | 385.84 ± 138.81 a | 241.93 ± 96.79 c | 261.6 ± 161.47 bc | 286.56 ± 108.56 b | 301.03 ± 168.56 b | 283.91 ± 109.41 a | 299.52 ± 127.76 a | 298.65 ± 178.65 a | 0.499 | 0.013 | 0.539 | 0.383 | 0.517 | 0.250 | 0.809 |
| BAS (×10 L−9) | 0.09 ± 0.08 | 0.08 ± 0.06 | 0.09 ± 0.07 a | 0.11 ± 0.06 a | 0.08 ± 0.04 b | 0.08 ± 0.11 ab | 0.07 ± 0.06 b | 0.07 ± 0.04 b | 0.1 ± 0.1 a | 0.08 ± 0.06 b | 0.962 | 0.291 | 0.945 | 0.628 | 0.797 | 0.965 | 0.431 |
| HGB (g/dL) | 100.18 ± 14.04 | 105.07 ± 13.68 | 106.66 ± 15.81 a | 102.01 ± 14.11 b | 103.18 ± 13.06 ab | 97.65 ± 13.2 c | 99.7 ± 12.42 bc | 101.75 ± 10.55 b | 98.31 ± 13.49 c | 105.16 ± 15.7 a | 0.477 | 0.472 | 0.406 | 0.912 | 0.533 | 0.826 | 0.795 |
| Parameters | Sex | Age | Generations | p-Value | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M | F | 9 | 12 | 15 | 18 | 24 | WT | F1 | F2 | Sex | Age | Generation | Sex × Age | Sex × Generation | Age × Generation | Sex × Age × Generation | |
| Glu (mmol/L) | 4.23 ± 0.74 | 4.77 ± 1 | 5.19 ± 0.88 a | 4.75 ± 0.75 b | 4.39 ± 0.86 c | 4.15 ± 0.68 d | 3.99 ± 0.86 d | 4.94 ± 0.8 a | 4.22 ± 0.97 c | 4.47 ± 0.82 b | 0.000 | 0.000 | 0.003 | 0.267 | 0.470 | 0.385 | 0.132 |
| AST (U/L) | 62.71 ± 29.15 | 66.05 ± 28.12 | 65.69 ± 26.29 a | 63.31 ± 28.73 a | 64.55 ± 28.12 a | 64.65 ± 33.19 a | 63.45 ± 26.59 a | 63.04 ± 20.3 a | 63.64 ± 26.12 a | 65.76 ± 34.72 a | 0.661 | 0.551 | 0.075 | 0.651 | 0.110 | 0.112 | 0.153 |
| ALT (U/L) | 26.34 ± 7.26 | 24.28 ± 7.35 | 24.59 ± 7.6 b | 25.36 ± 7.23 ab | 24.66 ± 6.99 ab | 25.17 ± 7.75 ab | 26.9 ± 7.04 a | 24.7 ± 5.3 b | 26.63 ± 7.21 a | 24.53 ± 8.4 b | 0.663 | 0.783 | 0.432 | 0.998 | 0.368 | 0.374 | 0.509 |
| TP (g/L) | 67.26 ± 6.72 | 72.48 ± 7.87 | 67.75 ± 6.07 b | 71.29 ± 6.04 ab | 70.3 ± 7.39 ab | 68.02 ± 8.66 ab | 71.61 ± 9.18 a | 64.75 ± 7.23 b | 71.49 ± 6.66 a | 71.34 ± 7.66 a | 0.982 | 0.010 | 0.163 | 0.000 | 0.348 | 0.159 | 0.696 |
| ALB (g/L) | 32.34 ± 4.31 | 34.13 ± 4.3 | 33.37 ± 3.3 ab | 33.39 ± 3.65 ab | 33.25 ± 4.34 ab | 32.04 ± 5.15 b | 33.99 ± 5 a | 31.26 ± 3.76 a | 34.87 ± 3.84 a | 32.87 ± 4.66 a | 0.081 | 0.005 | 0.004 | 0.000 | 0.051 | 0.012 | 0.758 |
| CK (U/L) | 202.58 ± 143.25 | 218.07 ± 112.46 | 197.81 ± 125.04 a | 212.84 ± 127.42 a | 220.59 ± 124.5 a | 205.89 ± 136.12 a | 213.37 ± 132.66 a | 202.13 ± 59.42 a | 217.07 ± 189.2 a | 208.57 ± 84.93 a | 0.702 | 0.760 | 0.701 | 0.953 | 0.403 | 0.620 | 0.396 |
| HDL-C (mmol/L) | 3.27 ± 0.65 | 3.32 ± 1.22 | 2.91 ± 0.63 c | 3.13 ± 0.57 b | 3.49 ± 0.59 a | 3.33 ± 1.77 ab | 3.6 ± 0.51 a | 3 ± 0.54 b | 3.32 ± 0.72 ab | 3.45 ± 1.29 a | 0.002 | 0.094 | 0.073 | 0.015 | 0.071 | 0.096 | 0.188 |
| LDL-C (mmol/L) | 1.58 ± 0.44 | 1.6 ± 0.38 | 1.3 ± 0.4 d | 1.41 ± 0.39 c | 1.65 ± 0.33 b | 1.72 ± 0.39 b | 1.85 ± 0.29 a | 1.67 ± 0.28 a | 1.52 ± 0.51 c | 1.6 ± 0.37 b | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| AMY (U/L) | 28.21 ± 12.37 | 29.72 ± 12.86 | 19.68 ± 11.52 d | 22.76 ± 10.14 c | 31.31 ± 9.12 b | 34.54 ± 12.63 a | 36.43 ± 10.15 a | 26.07 ± 10.95 b | 30.61 ± 10.57 a | 29.18 ± 14.86 a | 0.446 | 0.000 | 0.814 | 0.111 | 0.643 | 0.664 | 0.409 |
| Ca (mmol/l) | 2.83 ± 0.35 | 2.96 ± 0.49 | 2.49 ± 0.22 d | 2.58 ± 0.25 c | 3.01 ± 0.4 b | 3.2 ± 0.31 a | 3.2 ± 0.31 a | 2.83 ± 0.39 b | 2.86 ± 0.4 b | 2.96 ± 0.47 a | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.001 | 0.000 |
| ALP (U/L) | 143.06 ± 62.69 | 150.09 ± 61.97 | 156.31 ± 59.79 a | 144.48 ± 63.18 a | 149.41 ± 63.46 a | 144.5 ± 61.61 a | 137.67 ± 62.57 b | 134.88 ± 57.63 b | 151.49 ± 68.34 a | 148.99 ± 58.56 a | 0.714 | 0.650 | 0.392 | 0.302 | 0.639 | 0.401 | 0.427 |
| LDH (U/L) | 1076.97 ± 164.15 | 1093.3 ± 213.98 | 1131.2 ± 190.47 a | 1060.43 ± 197.77 b | 1066.09 ± 143.67 b | 1102.58 ± 191.08 ab | 1064.19 ± 210.75 b | 1045.22 ± 118.53 b | 1158.09 ± 187.09 a | 1041.62 ± 207.55 b | 0.014 | 0.040 | 0.255 | 0.032 | 0.013 | 0.113 | 0.028 |
| CO2-L (mmol/L) | 20.27 ± 3.88 | 21.79 ± 4.06 | 17.24 ± 3.48 b | 20.59 ± 3.51 b | 22.01 ± 3.33 a | 22.24 ± 3.31 a | 22.96 ± 3.78 a | 21.36 ± 3.71 a | 21.21 ± 3.96 ab | 20.61 ± 4.27 b | 0.000 | 0.833 | 0.012 | 0.003 | 0.003 | 0.030 | 0.012 |
| CHE (U/L) | 135.83 ± 30.47 | 123.13 ± 31.39 | 143.93 ± 28.9 a | 134.59 ± 26.73 b | 138.69 ± 31.96 ab | 118.18 ± 29.51 c | 112.93 ± 28.59 c | 135.15 ± 30.79 a | 126.65 ± 33.45 b | 129.07 ± 29.74 b | 0.397 | 0.010 | 0.018 | 0.921 | 0.190 | 0.143 | 0.469 |
| CHOL (mmol/L) | 2.09 ± 0.74 | 1.85 ± 0.68 | 2.27 ± 0.71 a | 1.89 ± 0.66 b | 1.85 ± 0.64 b | 2.04 ± 0.87 b | 1.81 ± 0.6 c | 2.2 ± 0.58 a | 1.96 ± 0.85 b | 1.84 ± 0.62 b | 0.816 | 0.473 | 0.065 | 0.825 | 0.166 | 0.707 | 0.783 |
| LACT (mmol/L) | 3.38 ± 2.18 | 3.83 ± 2.79 | 4.37 ± 2.2 a | 3.91 ± 2.48 a | 3.38 ± 2.77 ab | 3.2 ± 2.12 b | 3.12 ± 2.67 b | 3 ± 1.75 b | 3.98 ± 3.29 a | 3.61 ± 1.93 a | 0.552 | 0.506 | 0.116 | 0.949 | 0.369 | 0.285 | 0.388 |
| TRIGL (mmol/L) | 0.38 ± 0.14 | 0.41 ± 0.14 | 0.35 ± 0.13 b | 0.42 ± 0.13 a | 0.39 ± 0.14 ab | 0.43 ± 0.15 a | 0.39 ± 0.14 ab | 0.43 ± 0.14 a | 0.37 ± 0.1 c | 0.4 ± 0.17 b | 0.707 | 0.570 | 0.278 | 0.670 | 0.601 | 0.735 | 0.858 |
| UREAL (mmol/L) | 5.6 ± 1.82 | 4.98 ± 1.87 | 3.69 ± 1.39 d | 5.16 ± 1.45 c | 5.65 ± 1.48 b | 5.6 ± 1.78 bc | 6.38 ± 2.02 a | 5.75 ± 1.38 a | 4.86 ± 1.74 c | 5.42 ± 2.14 c | 0.021 | 0.966 | 0.050 | 0.000 | 0.018 | 0.140 | 0.009 |
| LIPC (U/L) | 19.94 ± 8.92 | 23.53 ± 9.81 | 14.52 ± 9.1 c | 18.95 ± 9.61 b | 23.73 ± 9.64 a | 25.34 ± 6.58 a | 25.86 ± 7.02 a | 16.61 ± 6.9 c | 24.26 ± 7.18 a | 22.43 ± 11.46 b | 0.550 | 0.000 | 0.263 | 0.014 | 0.977 | 0.694 | 0.745 |
| CRE (μmol/L) | 99.17 ± 25.49 | 103.85 ± 27.72 | 89.94 ± 22.11 c | 100.69 ± 24.59 b | 105.76 ± 24.09 ab | 98.72 ± 26.89 b | 112.08 ± 29.91 a | 92.56 ± 13.68 c | 109.35 ± 35.96 a | 99.59 ± 19.73 b | 0.670 | 0.261 | 0.715 | 0.233 | 0.321 | 0.803 | 0.205 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hai, C.; Bai, C.; Yang, L.; Wei, Z.; Wang, H.; Ma, H.; Ma, H.; Zhao, Y.; Su, G.; Li, G. Effects of Different Generations and Sex on Physiological, Biochemical, and Growth Parameters of Crossbred Beef Cattle by Myostatin Gene-Edited Luxi Bulls and Simmental Cows. Animals 2023, 13, 3216. https://doi.org/10.3390/ani13203216
Hai C, Bai C, Yang L, Wei Z, Wang H, Ma H, Ma H, Zhao Y, Su G, Li G. Effects of Different Generations and Sex on Physiological, Biochemical, and Growth Parameters of Crossbred Beef Cattle by Myostatin Gene-Edited Luxi Bulls and Simmental Cows. Animals. 2023; 13(20):3216. https://doi.org/10.3390/ani13203216
Chicago/Turabian StyleHai, Chao, Chunling Bai, Lei Yang, Zhuying Wei, Hong Wang, Haoran Ma, Haibing Ma, Yuefang Zhao, Guanghua Su, and Guangpeng Li. 2023. "Effects of Different Generations and Sex on Physiological, Biochemical, and Growth Parameters of Crossbred Beef Cattle by Myostatin Gene-Edited Luxi Bulls and Simmental Cows" Animals 13, no. 20: 3216. https://doi.org/10.3390/ani13203216
APA StyleHai, C., Bai, C., Yang, L., Wei, Z., Wang, H., Ma, H., Ma, H., Zhao, Y., Su, G., & Li, G. (2023). Effects of Different Generations and Sex on Physiological, Biochemical, and Growth Parameters of Crossbred Beef Cattle by Myostatin Gene-Edited Luxi Bulls and Simmental Cows. Animals, 13(20), 3216. https://doi.org/10.3390/ani13203216





