Hybridization in Canids—A Case Study of Pampas Fox (Lycalopex gymnocercus) and Domestic Dog (Canis lupus familiaris) Hybrid
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Cell Culture, Chromosome Preparation, and Cytogenetics Analysis
2.3. Molecular Analysis
2.4. Photographs Analysis
3. Results
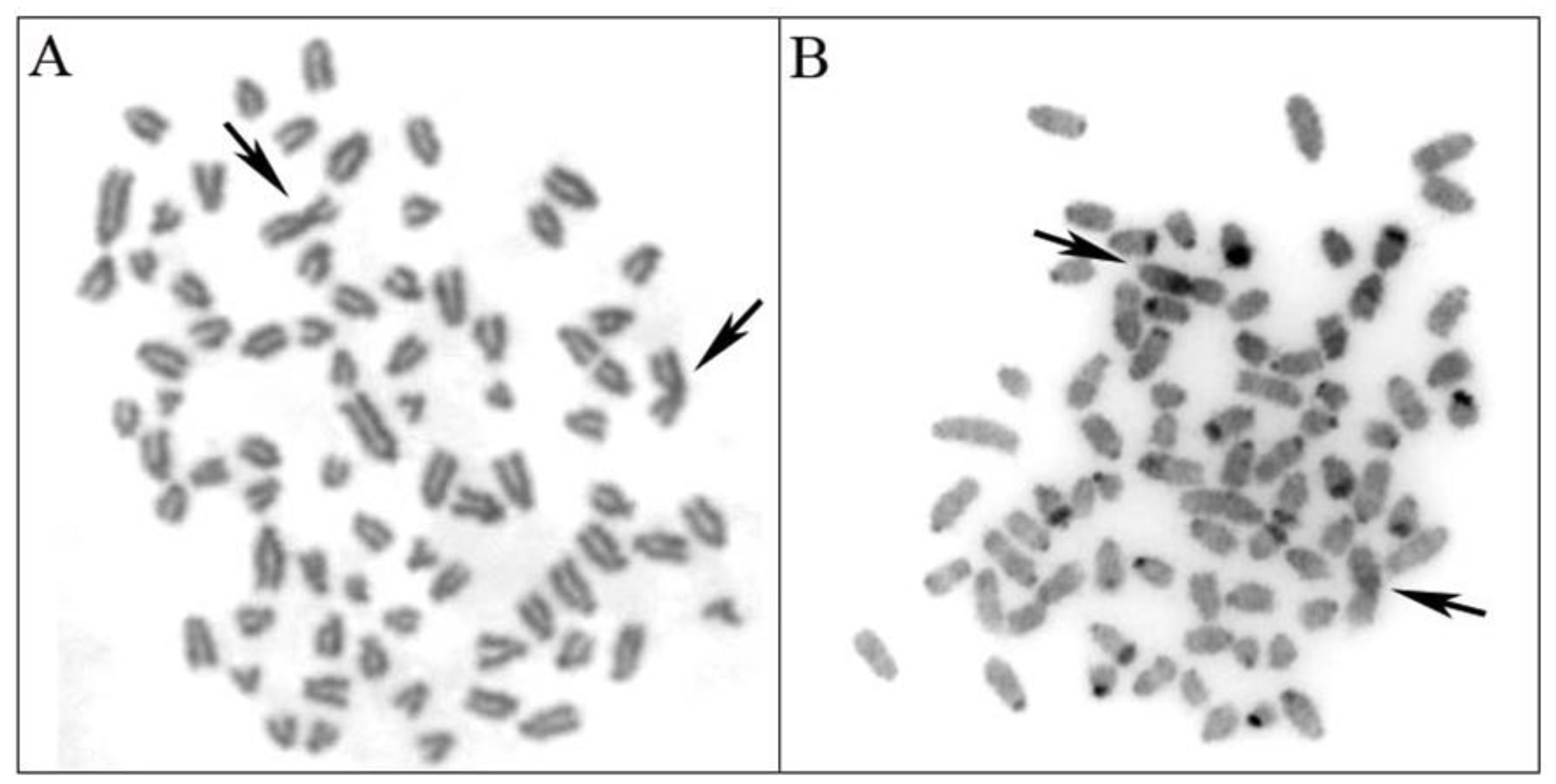
3.1. Chromosomal Analysis
3.2. MtDNA Analysis
3.3. Nuclear Analysis
3.4. Photographs Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Moroni, B.; Brambilla, A.; Rossi, L.; Meneguz, P.G.; Bassano, B.; Tizzani, P. Hybridization between Alpine Ibex and Domestic Goat in the Alps: A Sporadic and Localized Phenomenon? Animals 2022, 12, 751. [Google Scholar] [CrossRef]
- Hernández, F.; Brown, J.I.; Kaminski, M.; Harvey, M.G.; Lavretsky, P. Genomic Evidence for Rare Hybridization and Large Demographic Changes in the Evolutionary Histories of Four North American Dove Species. Animals 2021, 11, 2677. [Google Scholar] [CrossRef]
- Porto-Foresti, F.; Hashimoto, D.T.; Alves, A.L.; Almeida, R.B.C.; Senhorini, J.A.; Bortolozzi, J.; Foresti, F. Cytogenetic markers as diagnoses in the identification of the hybrid between Piauçu (Leporinus macrocephalus) and Piapara (Leporinus elongatus). Genet. Mol. Biol. 2008, 31, 195–202. [Google Scholar] [CrossRef][Green Version]
- Abbott, R.; Albach, D.; Ansell, S.; Arntzen, J.W.; Baird, S.J.E.; Bierne, N.; Boughman, J.; Brelsford, A.; Buerkle, C.A.; Buggs, R.; et al. Hybridization and speciation. J. Evol. Biol. 2013, 26, 229–246. [Google Scholar] [CrossRef]
- Edmands, S. Between a rock and a hard place: Evaluating the relative risks of inbreeding and outbreeding for conservation and management. Mol. Ecol. 2006, 16, 463–475. [Google Scholar] [CrossRef]
- Goldberg, T.L.; Grant, E.C.; Inendino, K.R.; Kassler, T.W.; Claussen, J.E.; Philipp, D.P. Increased Infectious Disease Susceptibility Resulting from Outbreeding Depression. Conserv. Biol. 2005, 19, 455–462. [Google Scholar] [CrossRef]
- Klemme, I.; Hendrikx, L.; Ashrafi, R.; Sundberg, L.-R.; Räihä, V.; Piironen, J.; Hyvärinen, P.; Karvonen, A. Opposing health effects of hybridization for conservation. Conserv. Sci. Pract. 2021, 3, e379. [Google Scholar] [CrossRef]
- Randi, E. Detecting hybridization between wild species and their domesticated relatives. Mol. Ecol. 2008, 17, 285–293. [Google Scholar] [CrossRef]
- Sartor, C.C.; Cushman, S.A.; Wan, H.Y.; Kretschmer, R.; Pereira, J.A.; Bou, N.; Cosse, M.; González, S.; Eizirik, E.; de Freitas, T.R.O.; et al. The role of the environment in the spatial dynamics of an extensive hybrid zone between two neotropical cats. J. Evol. Biol. 2021, 34, 591–722. [Google Scholar] [CrossRef]
- Grabenstein, K.C.; Otter, K.A.; Burg, T.M.; Taylor, S.A. Hybridization between closely related songbirds is related to human habitat disturbance. Glob. Chang. Biol. 2022, 29, 955–968. [Google Scholar] [CrossRef]
- Rhymer, J.M.; Simberloff, D. Extinction by Hybridization and Introgression. Annu. Rev. Ecol. Syst. 1996, 27, 83–109. [Google Scholar] [CrossRef]
- Wang, X.; Tedford, R.H.; Van Valkenburgh, B.; Wayne, R.K. Evolutionary history, molecular systematics, and evolutionary ecology of Canidae. In Biology and Conservation of Wild Canids; Oxford University Press: Oxford, UK, 2004. [Google Scholar]
- Wozencraft, W.C.; Wilson, D.E.; Reeder, D.M. Mammal species of the world. In A Taxonomic and Geographic Reference, 3rd ed.; Johns Hopkins University Press: Baltimore, MD, USA, 2005. [Google Scholar]
- Hübner, S.d.O.; Pappen, F.G.; Ruas, J.L.; Vargas, G.D.Á.; Fischer, G.; Vidor, T. Exposure of pampas fox (Pseudalopex gymnocercus) and crab-eating fox (Cerdocyon thous) from the Southern region of Brazil to Canine distemper virus (CDV), Canine parvovirus (CPV) and Canine coronavirus (CCoV). Braz. Arch. Biol. Technol. 2010, 53, 593–597. [Google Scholar] [CrossRef]
- Paula, R.C.; Dematteo, K. Chrysocyon brachyurus (Errata Version Published in 2016)—The IUCN Red List of Threatened Species 2015: E.T4819A88135664. Available online: https://doi.org/10.2305/IUCN.UK.2015-4.RLTS.T4819A82316878.en (accessed on 25 May 2022).
- Lucherini, M. Lycalopex gymnocercus—The IUCN Red List of Threatened Species 2016: E.T6928A85371194. Available online: https://www.iucnredlist.org/species/6928/85371194 (accessed on 25 May 2022).
- Courtenay, O.; Maffe, L. Crab-eating fox Cerdocyon thous (Linnaeus, 1766) Least Concern. In Canids: Foxes, Wolves, Jackals and Dogs—Status Survey and Conservation Action Plan; Sillero-Zubiri, C., Hoffmann, M., Macdonald, D.W., Eds.; IUCN/SSC Canid Specialist Group: Gland, Switzerland/Cambridge, UK, 2004; pp. 32–38. [Google Scholar]
- DeMatteo, K.E.; Loiselle, B.A. New data on the status and distribution of the bush dog (Speothos venaticus): Evaluating its quality of protection and directing research efforts. Biol. Conserv. 2008, 141, 2494–2505. [Google Scholar] [CrossRef]
- Songsasen, N.; Rodden, M.D. The role of the Species Survival Plan in Maned wolf Chrysocyon brachyurus conservation. Int. Zoo. Yearb. 2010, 44, 136–148. [Google Scholar] [CrossRef]
- Soler, L. Ecology and Conservation of the Maned Wolf: Multidisciplinary Perspectives; Consorte-McCrea, A.G., Santos, F.E., Eds.; CRC Press: Boca Raton, FL, USA, 2014; pp. 203–220. [Google Scholar]
- Edmands, S. Does parental divergence predict reproductive compatibility? Trends Ecol. Evol. 2002, 17, 520–527. [Google Scholar] [CrossRef]
- Seehausen, O. Conservation: Losing Biodiversity by Reverse Speciation. Curr. Biol. 2006, 16, R334–R337. [Google Scholar] [CrossRef]
- Bohling, J.H.; Waits, L.P. Assessing the prevalence of hybridization between sympatric Canis species surrounding the red wolf (Canis rufus) recovery area in North Carolina. Mol. Ecol. 2011, 20, 2142–2156. [Google Scholar] [CrossRef]
- Adams, J.R.; Kelly, B.T.; Waits, L.P. Using faecal DNA sampling and GIS to monitor hybridization between red wolves (Canis rufus) and coyotes (Canis latrans). Mol. Ecol. 2003, 12, 2175–2186. [Google Scholar] [CrossRef]
- Hinton, J.W.; Gittleman, J.L.; van Manen, F.T.; Chamberlain, M.J. Size-assortative choice and mate availability influences hybridization between red wolves (Canis rufus) and coyotes (Canis latrans). Ecol. Evol. 2018, 8, 3927–3940. [Google Scholar] [CrossRef]
- Roy, M.S.; Geffen, E.; Smith, D.; Ostrander, E.A.; Wayne, R.K. Patterns of differentiation and hybridization in North American wolflike canids, revealed by analysis of microsatellite loci. Mol. Biol. Evol. 1994, 11, 553–570. [Google Scholar] [CrossRef]
- Vila, C.; Wayne, R.K. Hybridization between Wolves and Dogs. Conserv. Biol. 1999, 13, 195–198. [Google Scholar] [CrossRef]
- Silva, F. Mamíferos Silvestres: Rio Grande do Sul, 3rd ed.; Via Sapiens: Porto Alegre, Brazil, 2014. [Google Scholar]
- DeMatteo, K.; Michalski, F.; Leite-Pitman, M.R.P. Speothos Venaticus—The IUCN Red List of Threatened Species 2011: E.T20468A9203243. Available online: https://doi.org/10.2305/IUCN.UK.2011-2.RLTS.T20468A9203243.en (accessed on 10 July 2023).
- De Mello Beisiegel, B.; Zuercher, G.L. Speothos venaticus. Mamm. Species 2005, 783, 1–6. [Google Scholar] [CrossRef]
- Rodden, M.; Rodrigues, F.; Bestelmeyer, S. Maned wolf (Chrysocyon brachyurus). In Canids: Foxes, Wolves, Jackals and Dogs—Status Survey and Conservation Action Plan; IUCN/SSC Canid Specialist Group: Gland, Switzerland; Cambridge, UK, 2004; pp. 38–43. [Google Scholar]
- Queirolo, D.; Moreira, J.R.; Soler, L.; Emmons, L.H.; Rodrigues, F.H.; Pautasso, A.A.; Cartes, J.L.; Salvatori, V. Historical and current range of the Near Threatened maned wolf Chrysocyon brachyurus in South America. Oryx 2011, 45, 296–303. [Google Scholar] [CrossRef]
- Lucherini, M.; Pessino, M.; Farias, A.A. Pampas fox Pseudalopex gymnocercus (Fischer, 1814). In Canids: Foxes, Wolves, Jackals and Dogs—Status Survey and Conservation Action Plan; IUCN/SSC Canid Specialist Group: Gland, Switzerland; Cambridge, UK, 2004; pp. 63–68. [Google Scholar]
- Zurano, J.P.; Ojeda, D.S.; Bidau, C.J.; Molina, W.F.; Ledesma, M.A.; Martinez, P.A. A comparison of heterochromatic regions in three species of neotropical canids. Zool. Anz. A J. Comp. Zool. 2015, 254, 1–7. [Google Scholar] [CrossRef]
- Fujinaga, T.; Yamashita, M.; Yoshida, M.C.; Mizuno, S.; Tajima, M.; Okamoto, Y.; Otomo, K. The banding patterns of normal canine chromosomes. Jpn. J. Vet. Sci. 1989, 51, 294–299. [Google Scholar] [CrossRef] [PubMed]
- Verma, R.; Babu, A. Human Chromosomes: Principles & Techniques, 2nd ed.; McGraw-Hill: New York, NY, USA, 1996. [Google Scholar]
- Sumner, A.T. A simple technique for demonstrating centromeric heterochromatin. Exp. Cell Res. 1972, 75, 304–306. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Lab Press: New York, NY, USA, 1989. [Google Scholar]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar] [PubMed]
- Amrine-Madsen, H.; Koepfli, K.-P.; Wayne, R.K.; Springer, M.S. A new phylogenetic marker, apolipoprotein B, provides compelling evidence for eutherian relationships. Mol. Phylogenet. Evol. 2003, 28, 225–240. [Google Scholar] [CrossRef]
- Murphy, W.J.; Eizirik, E.; Johnson, W.E.; Zhang, Y.P.; Ryder, O.A.; O’Brien, S.J. Molecular phylogenetics and the origins of placental mammals. Nature 2001, 409, 614–618. [Google Scholar] [CrossRef]
- Lyons, L.A.; Laughlin, T.F.; Copeland, N.G.; Jenkins, N.A.; Womack, J.E.; O’Brien, S.J. Comparative anchor tagged sequences (CATS) for integrative mapping of mammalian genomes. Nat. Genet. 1997, 15, 47–56. [Google Scholar] [CrossRef]
- Venta, P.J.; Brouillette, J.A.; Yuzbasiyan-Gurkan, V.; Brewer, G.J. Gene-specific universal mammalian sequence-tagged sites: Application to the canine genome. Biochem. Genet. 1996, 34, 321–341. [Google Scholar] [CrossRef] [PubMed]
- Eizirik, E.; Murphy, W.J.; Koepfli, K.-P.; Johnson, W.E.; Dragoo, J.W.; Wayne, R.K.; O’Brien, S.J. Pattern and timing of diversification of the mammalian order Carnivora inferred from multiple nuclear gene sequences. Mol. Phylogenet. Evol. 2010, 56, 49–63. [Google Scholar] [CrossRef] [PubMed]
- Eizirik, E. Molecular Dating and Biogeography of the Early Placental Mammal Radiation. J. Hered. 2001, 92, 212–219. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.; Doyle, J. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Bandelt, H.J.; Forster, P.; Rohl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef]
- Leigh, J.W.; Bryant, D. Popart: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Bardeleben, C.; Moore, R.L.; Wayne, R.K. A molecular phylogeny of the Canidae based on six nuclear loci. Mol. Phyl. Evol. 2005, 37, 815–831. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Suleski, M.; Hedges, S.B. TimeTree: A Resource for Timelines, Timetrees, and Divergence Times. Mol. Biol. Evol. 2017, 34, 1812–1819. [Google Scholar] [CrossRef] [PubMed]
- Kubiak, B.B.; Kretschmer, R.; Leipnitz, L.T.; Maestri, R.; Almeida, T.S.; Borges, L.R.; Galiano, D.; Pereira, J.C.; Oliveira, E.H.C.; Ferguson-Smith, M.; et al. Hybridization between subterranean tuco-tucos (Rodentia, Ctenomyidae) with contrasting phylogenetic positions. Sci. Rep. 2020, 10, 1502. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, E.H.C.; Gomes, A.J.B.; Costa, A.F.; Emin-Lima, R.; Bonvicino, C.R.; Viana, M.C.; Reis, L.M.A.; Vidal, M.D.; Cavalcanti, M.V.G.; Attademo, F.L.N.; et al. Karyotypical Confirmation of Natural Hybridization between Two Manatee Species, Trichechus manatus and Trichechus inunguis. Life 2022, 12, 616. [Google Scholar] [CrossRef] [PubMed]
- Nash, W.G.; Menninger, J.C.; Wienberg, J.; Padilla-Nash, H.M.; O’Brien, S.J. The pattern of phylogenomic evolution of the Canidae. Cytogenet. Genome Res. 2001, 95, 210–224. [Google Scholar] [CrossRef] [PubMed]
- Graphodatsky, A.S.; Perelman, P.L.; Sokolovskaya, N.V.; Beklemisheva, V.R.; Serdukova, N.A.; Dobigny, G.; O’Brien, S.J.; Ferguson-Smith, M.A.; Yang, F. Phylogenomics of the dog and fox family (Canidae, Carnivora) revealed by chromosome painting. Chromosome Res. 2008, 16, 129–143. [Google Scholar] [CrossRef]
- Perelman, P.L.; Beklemisheva, V.R.; Yudkin, D.V.; Petrina, T.N.; Rozhnov, V.V.; Nie, W.; Graphodatsky, A.S. Comparative Chromosome Painting in Carnivora and Pholidota. Cytogenet. Genome Res. 2012, 137, 174–193. [Google Scholar] [CrossRef]
- Manolache, M.; Ross, W.M.; Schmid, M. Banding analysis of the somatic chromosomes of the domestic dog (Canis familiaris). Can. J. Genet. Cytol. 1976, 18, 513–518. [Google Scholar] [CrossRef]
- Lucherini, M.; Vidal, E.M.L. Lycalopex gymnocercus (Carnivora: Canidae). Mamm. Species 2008, 820, 1. [Google Scholar] [CrossRef]
- Ribeiro, M.C.; Metzger, J.P.; Martensen, A.C.; Ponzoni, F.J.; Hirota, M.M. The Brazilian Atlantic Forest: How much is left, and how is the remaining forest distributed? Implications for conservation. Biol. Conserv. 2009, 142, 1141–1153. [Google Scholar] [CrossRef]
- Atlântica, S.M. Atlas dos Remanescentes Florestais da Mata Atlântica, Período de 2000 a 2005; SOS Mata Atlântica and INPE: São Paulo, Brazil, 2011. [Google Scholar]
- Allendorf, F.W.; Leary, R.F.; Spruell, P.; Wenburg, J.K. The problems with hybrids: Setting conservation guidelines. Trends Ecol. Evol. 2001, 16, 613–622. [Google Scholar] [CrossRef]
- Godinho, R.; Llaneza, L.; Blanco, J.C.; Lopes, S.; Álvares, F.; García, E.J.; Palacios, V.; Cortés, Y.; Talegón, J.; Ferrand, N. Genetic evidence for multiple events of hybridization between wolves and domestic dogs in the Iberian Peninsula. Mol. Ecol. 2011, 20, 5154–5166. [Google Scholar] [CrossRef]
- Monteiro, G.; Fleck, J.; Kluge, M.; Rech, N.K.; Soliman, M.C.; Staggemeier, R.; Rodrigues, M.T.; Barros, M.P.; Heinzelmann, L.S.; Spilki, F.R. Adenoviruses of canine and human origins in stool samples from free-living pampas foxes (Lycalopex gymnocercus) and crab-eating foxes (Cerdocyon thous) in São Francisco de Paula, Rio dos Sinos basin. Brazilian J. Biol. 2015, 75, 11–16. [Google Scholar] [CrossRef] [PubMed]


| Hybridization Hypothesis | Expected Number of Chromosomes | Cytogenetics Reference |
|---|---|---|
| C. lupus familiaris (2n = 78) X L. gymnocercus (2n = 74) | 76 | [34,35] |
| C. lupus familiaris (2n = 78) X C. thous (2n = 74) | 76 | [34,35] |
| C. lupus familiaris (2n = 78) X C. brachyurus (2n = 76) | 77 | [34,35] |
| C. brachyurus (2n = 76) X L. gymnocercus (2n = 74) | 75 | [34] |
| C. brachyurus (2n = 76) X C. thous (2n = 74) | 75 | [34] |
| C. thous (2n = 74) X L. gymnocercus (2n = 74) | 74 | [34] |
| Polymorphic Sites | ||||||
|---|---|---|---|---|---|---|
| 161 | ||||||
| APOB | Pampas fox 1 | * | ||||
| Pampas fox 2 | G | |||||
| Canid | R | |||||
| Domestic dog 1 | A | |||||
| Domestic dog 2 | A | |||||
| 110 | 192 | 267 | 287 | |||
| CHRNA1 | Pampas fox 1 | T | C | G | C | |
| Pampas fox 2 | T | C | G | C | ||
| Canid | T | C | G | Y | ||
| Domestic dog 1 | K | Y | G | C | ||
| Domestic dog 2 | K | T | A | C | ||
| 88 | 231 * | 308 * | ||||
| FES | Pampas fox 1 | A | G | S | ||
| Pampas fox 2 | A | G | S | |||
| Canid | M | K | G | |||
| Domestic dog 1 | C | T | G | |||
| Domestic dog 2 | C | T | G | |||
| 99 | 446 | 569 | 638 | 671 | ||
| GHR | Pampas fox 1 | T | T | W | Y | C |
| Pampas fox 2 | T | T | W | Y | C | |
| Canid | Y | W | T | T | C | |
| Domestic dog 1 | C | A | W | C | Y | |
| Domestic dog 2 | C | A | T | C | C | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Szynwelski, B.E.; Kretschmer, R.; Matzenbacher, C.A.; Ferrari, F.; Alievi, M.M.; de Freitas, T.R.O. Hybridization in Canids—A Case Study of Pampas Fox (Lycalopex gymnocercus) and Domestic Dog (Canis lupus familiaris) Hybrid. Animals 2023, 13, 2505. https://doi.org/10.3390/ani13152505
Szynwelski BE, Kretschmer R, Matzenbacher CA, Ferrari F, Alievi MM, de Freitas TRO. Hybridization in Canids—A Case Study of Pampas Fox (Lycalopex gymnocercus) and Domestic Dog (Canis lupus familiaris) Hybrid. Animals. 2023; 13(15):2505. https://doi.org/10.3390/ani13152505
Chicago/Turabian StyleSzynwelski, Bruna Elenara, Rafael Kretschmer, Cristina Araujo Matzenbacher, Flávia Ferrari, Marcelo Meller Alievi, and Thales Renato Ochotorena de Freitas. 2023. "Hybridization in Canids—A Case Study of Pampas Fox (Lycalopex gymnocercus) and Domestic Dog (Canis lupus familiaris) Hybrid" Animals 13, no. 15: 2505. https://doi.org/10.3390/ani13152505
APA StyleSzynwelski, B. E., Kretschmer, R., Matzenbacher, C. A., Ferrari, F., Alievi, M. M., & de Freitas, T. R. O. (2023). Hybridization in Canids—A Case Study of Pampas Fox (Lycalopex gymnocercus) and Domestic Dog (Canis lupus familiaris) Hybrid. Animals, 13(15), 2505. https://doi.org/10.3390/ani13152505








